Abstract
Anaerobic ammonium oxidation (anammox) is a promising new process to treat high-strength nitrogenous wastewater. Due to the low growth rate of anaerobic ammonium-oxidizing bacteria, efficient biomass retention is essential for reactor operation. Therefore, we studied the settling ability and community composition of the anaerobic ammonium-oxidizing granules, which were cultivated in an upflow anaerobic sludge blanket (UASB) reactor seeded with aerobic granules. With this seed, the start-up period was less than 160 days at a NH4+-N removal efficiency of 94% and a loading rate of 0.064 kg N per kg volatile suspended solids per day. The formed granules were bright red and had a high settling velocity (41 to 79 m h−1). Cells and extracellular polymeric substances were evenly distributed over the anaerobic ammonium-oxidizing granules. The high percentage of anaerobic ammonium-oxidizing bacteria in the granules could be visualized by fluorescent in situ hybridization and electron microscopy. The copy numbers of 16S rRNA genes of anaerobic ammonium-oxidizing bacteria in the granules were determined to be 4.6 × 108 copies ml−1. The results of this study could be used for a better design, shorter start-up time, and more stable operation of anammox systems for the treatment of nitrogen-rich wastewaters.
The anaerobic ammonia oxidation (anammox) process is a recently discovered biological nitrogen removal technology in which ammonia is oxidized to nitrogen gas with nitrite as the electron acceptor (5, 29, 32). In contrast to heterotrophic denitrification (6, 26), the anammox process does not require external electron donors (e.g., methanol) due to their chemolithoautotrophic lifestyle. Furthermore, if this process is combined with a partial nitrification step, only half of the ammonium needs to be nitrified to nitrite, which together with the remaining ammonium can subsequently be converted into nitrogen through the anammox process. This reduces the oxygen demand of the system and leads to further reduction in operational costs (27).
The anaerobic ammonium-oxidizing bacteria (anammox bacteria) have a low growth rate (18), with a doubling time at best estimated as 7 to 11 days (18, 28). The yield of the anammox bacteria has been determined to be 0.066 mol C biomass mol−1 ammonium consumed, and the maximum ammonium consumption rate is ∼45 nmol mg−1 protein min−1 (18). Given the low growth rate and low yield, very efficient biomass retention is essential to retain the anammox bacteria within the reactor systems during cultivation (19). The enrichment of anammox bacteria from a mixed inoculum requires the optimization of conditions favorable for the anammox bacteria and generally takes 200 to 300 days (5, 6, 27). Thus, conditions that would reduce the start-up time of anammox reactors would positively effect the implementation of the process. Several sources of inocula, such as activated sludge (4), nitrifying activated sludge (27), and anaerobic sludge (6), have been used for the start-up of anammox reactors with start-up times of as long as 1,000 days (27).
Aerobic granules have been reported to have high microbial diversity (31) and compact structure with very good settling properties resulting in an efficient means of biomass retention. These properties, including interspecies competition and mass transfer, result in the stratification of microbial species with anoxic pockets in the interior of the granules that may be suitable to harbor anammox bacteria. Therefore, the main objective of this study was to investigate the feasibility of start-up of the anammox process by seeding the reactor with aerobic granular sludge by using an upflow anaerobic sludge blanket (UASB) reactor. After the successful start-up and the formation of anammox granules, the structure and physicochemical properties of the anammox granules and the reactor performance were characterized. Microbial community analysis revealed that the dominant anammox species was related to a species of anammox bacteria present in anammox biofilms.
MATERIALS AND METHODS
Reactor operation.
A plexiglass UASB reactor (2.1 liters; 10 cm in inner diameter) and a gas-solids separator portion (2.6 liters) was used in this study. The seeding aerobic granular sludge was taken from a lab-scale sequencing batch reactor treating soybean-processing wastewater (21). The volatile suspended solid (VSS) concentration of the seeding aerobic granules was 11.12 ± 0.15 g liter−1. Sludge (1.0 liter) was seeded to the UASB reactor, resulting in an initial VSS level of 7.85 g liter−1 in the reactor.
Synthetic wastewater consisting of (NH4)2SO4, 25 to 200 mg N liter−1; NaNO2, 25 to 220 mg N liter−1; CaCl2·2H2O, 0.18 g liter−1; MgSO4·7H2O, 0.12 g liter−1; KH2PO4, 0.027 g liter−1; KHCO3, 0.5 g liter−1; and 1 ml liter−1 trace element solution I and II, in which trace element solution I consisted of EDTA, 5.0 g liter−1; FeSO4, 5.0 g liter−1, and trace element solution II consisted of EDTA, 5.0 g liter−1; ZnSO4·7H2O, 0.43 g liter−1; CoCl2·6H2O, 0.24 g liter−1; MnCl2·4H2O, 0.99 g liter−1; CuSO4·5H2O, 0.25 g liter−1; Na2MoO4·2H2O, 0.22 g liter−1; NiCl2·6H2O, 0.19 g liter−1; Na2SeO4·10H2O, 0.21 g liter−1; H3BO4, 0.014 g liter−1, was used as the feed for the enrichment culture described in the present study. The influent pH value was adjusted to 7.0 by adding 3 M NaOH or 3 M HCl.
After inoculation, the reactor was sparged with nitrogen gas for 30 min at a flow rate of 0.1 liter min−1. In the start-up, the UASB reactor was initially fed with the synthetic wastewater with a relatively low nitrogen loading rate (NLR) of 0.042 g liter−1 per day at a hydraulic retention time (HRT) of 30 h for 1 month. Later, the HRT was shortened to 24 h for another 1 month of operation. Thereafter, the HRT was kept constant at 24 h, but the NLR was elevated by increasing the influent ammonium and nitrite concentrations, as shown in Fig. 1. The ammonium and nitrite concentrations in the influent were increased up to 0.21 g liter−1 of NH4+-N and 0.24 g liter−1 of NO2−-N, respectively. The applied NLR was increased from 0.042 to 0.5 g liter−1 per day. The reactor temperature was maintained at 30°C by using a ribbon heater and a temperature controller throughout the experiments.
FIG. 1.
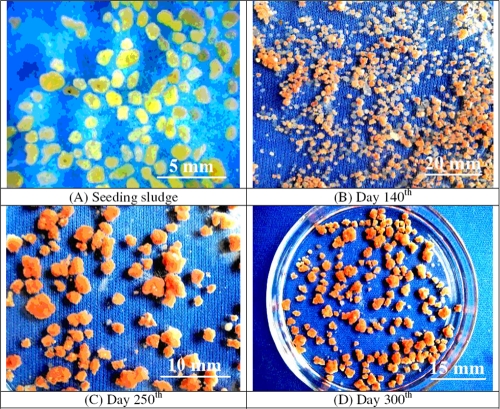
Images of sludge samples in the granulation process. (A) Seeding aerobic granular sludge. (B) Small granules after 140 days of operation. (C) Granules on day 250. (D) Granules on day 300.
DNA extraction.
Biomass was harvested from the reactor (2 ml) and centrifuged. The supernatant was discarded, and the pellet was resuspended in 0.75 ml sodium phosphate buffer (120 mM; pH 8). Glass beads (∼0.3 g; 0.25-mm diameter) were added, and cells were disrupted by bead beating for 30 s. Then, the protocol described by Kowalchuk et al. (10) was followed. Isolated DNA was then suspended in 50 μl ultrapure water and kept at 4°C for 24 h until further analysis. DNA quality was assessed by agarose (1%) gel electrophoresis, and concentrations were measured with the NanoDrop ND-1000 (Isogen Life Science, The Netherlands).
16S rRNA sequences and phylogenetic analysis.
A primer combination of Pla46F (Escherichia coli positions 46 to 63) forward and universal reverse (630R) (E. coli positions 1529 to 1545) primers was used for the preferential amplification of 16S rRNA genes of the members of the Planctomycetes (14). PCR fragments were cloned with the pGEM-T Easy cloning kit (Promega, Madison, WI) and XL1-Blue E. coli competent cells. Plasmid DNA was isolated and purified with the gene JETTM plasmid miniprep kit (Fermentas, Canada). The near-complete sequences of the 16S rRNA gene fragments were obtained by using M13 forward and reverse primers targeting vector sequences adjacent to the multiple cloning site. Phylogenetic analyses were performed with maximum likelihood, neighbor joining, and maximum parsimony methods with 50% sequence conservation filters for Planctomycetes. Sequencing and retrieval of the cloned 16S rRNA genes and phylogenetic analyses were performed with MEGA 4 as described by Kartal et al. (8).
Quantitative PCR.
A primer combination of forward Amx694F (Escherichia coli positions 694 to 713; GGGGAGAGTGGAACTTCGG) and reverse Amx960R (E. coli positions 960 to 979; GCTCGCACAAGCGGTGGAGC) primers was used for quantifying anammox bacteria.
Real-time gradient PCR was performed with an iCycler iQ5 thermocycler and real-time detection system (Bio-Rad, Berkeley, CA). For the standard curves, PCR fragments were cloned with the pGEM-T Easy cloning kit (Promega, Madison, WI) and XL1-Blue E. coli competent cells. The plasmids were isolated and purified with the Gene JETTM Plasmid Miniprep kit (Fermentas, Canada). Each PCR mixture (25 μl) was composed of 12.5 μl of 1× SYBR green PCR master mix (Finnzymes, Finland), 1 μl of each forward and reverse primer (20 pmol μl−1), and either 1 μl of template DNA or 100 to 109 copies per well of the standard vector plasmid of the clone “Candidatus Kuenenia stuttgartiensis” (AF375995) grown as single cellular suspensions (28). PCR amplification and detection were performed in MicroAmp optical 96-well reaction plates. The PCR temperature program was initiated with 3 min at 96°C, followed by 40 cycles of 1 min at 96°C, 1 min at a different annealing temperature, and 1 min at 72°C. For 16S rRNA primers, the annealing temperature was increased from 55 to 65°C using the iQ Custom SYBR green supermix (Bio-Rad, Berkeley, CA). A melting curve analysis for SYBR green assay was done after amplification to distinguish the targeted PCR product from the nontargeted PCR product (24).
The primers used for quantitative PCR were designed based on the near-full-length 16S rRNA gene sequences of anammox bacteria from the NCBI database and nine different reactor cultures. The specificity of the primers was checked using the PROBE_MATCH tool of the ARB program. The primers were tested on environmental samples and enrichment cultures available in our laboratory. The standard curves for anammox 16S rRNA gene copies were constructed from a series of 10-fold dilutions (from 3.6 × 101 copies to 3.6 × 107 copies) of plasmid DNA carrying the 16S rRNA gene of “Candidatus Kuenenia stuttgartiensis” for the primer sets of AMX658F-AMX924R. The range test was 2.6 × 10−8 to 2.6 × 10−2 ng of DNA per well for each primer set. Amplification efficiency was calculated from the equation, ɛc = 101/s − 1. The real-time PCR assay with these primers was very consistent, as shown by the strong inverse linear relationship between the threshold cycle numbers and the copy numbers of 16S rRNA gene (R2 = 0.990). The amplification efficiencies were between 95% and 105%, with slopes of −3.30.
Fluorescence in situ hybridization (FISH).
Biomass (1 ml) was harvested from the enrichment culture and hybridized with fluorescent probe Amx368; the probe was purchased as Cy3-labeled derivatives from Thermo Electron Corporation (Ulm, Germany). The hybridization conditions and approaches used were as described by Kartal et al. (8).
Other analyses.
The morphology of the anammox granules was analyzed by using an image analysis system (Image-Pro Express 4.0; Media Cybernetics) equipped with an Olympus CX41 microscope and a digital camera (Olympus C5050 Zoom). Their microstructure and predominant bacterial morphologies were observed using scanning electron microscopy (SEM; Quantn 200; FEI Ltd., China). The structure, particularly the distribution of microbial cells and extracellular polymeric substances (EPS) within granules, was examined using a confocal laser scanning microscope (CLSM) (LSM 5 Pa; Zeiss Inc., Jena, Germany). The hydrophobicity of the granules was determined by measuring the contact angle following the axisymmetric drop shape method described by Sheng et al. (16). The extraction and determination of EPS in granules were conducted according to Sheng and Yu (15). Their settling velocity was measured by recording the time taken for an individual granule to fall from a certain height in a measuring cylinder. Measurement of total suspended solids (TSS), VSS, NH4+-N, NO2−-N, and NO3−-N was performed according to the standard methods (1).
Nucleotide sequence accession number.
The 16S rRNA gene was deposited into GenBank as accession number GQ175382.
RESULTS AND DISCUSSION
Formation of the anammox granules.
A 2.1-liter UASB reactor was seeded with 7.9 g VSS liter−1 aerobic granular sludge. The aerobic granules were regular spheres and yellow, with an average diameter of approximately 1.2 mm (Fig. 1A). One week after seeding of the UASB reactor, the granules began to shrink, and then they partially disintegrated to diameters of 0.3 to 0.5 mm, probably due to the anaerobic conditions. In the subsequent 40 days after seeding, a part (∼50%) of the suspended biomass was washed out, and new granulation occurred. About 100 days after seeding, the outer surface of the particles turned red, suggesting that the anammox microorganisms became dominant in the reactor. In this phase, the cells on the outer surface began to grow, and particles became bigger. Brownish-red anammox granules with diameters of 0.7 to 0.8 mm were visible at the bottom of the reactor after 160 days of operation (Fig. 1B). These granules developed rapidly, and large red granules with diameters of over 2.5 mm were formed after 250 days of operation (Fig. 1C). After day 280, the granule growth slowed down, followed by a decrease in the granule formation. As a result, stable red granules formed a stratified layer in the reactor (Fig. 1D).
Performance of the anammox reactor.
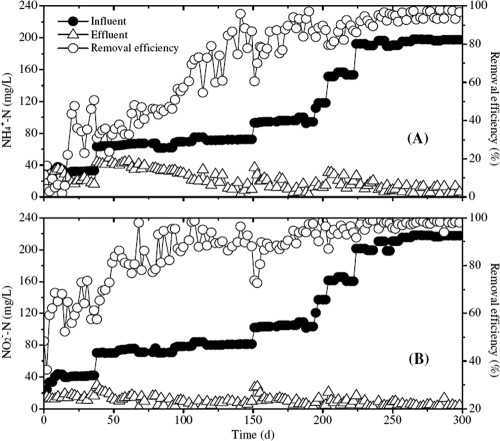
The anammox reactor was continuously operated for more than 300 days (Fig. 2). After day 20, the NH4+-N removal efficiency gradually increased (Fig. 2A), reaching 90% after 200 days of operation. The total nitrogen removal efficiency was 94% after 250 days of operation.
FIG. 2.
Reactor performance. (A) Concentration profiles of NH4+-N. (B) Concentration profiles of NO2−-N.
As depicted in Fig. 1, bright-red anammox granules were apparent within 140 days. The start-up time in this work was similar to that of those seeded with activated sludge (23) and anaerobic digestion sludge (150 days) (3) and was significantly faster than the reactors inoculated with nitrifying (250 days) (13) or denitrifying (392 days) (25) sludge.
Based on the observation and studies of the microstructure of the granules, the granule formation after seeding with aerobic granules could proceed as follows: (i) disintegration of the seeded aerobic granules into small particles due to the anaerobic conditions; (ii) growth of the anammox bacteria; (iii) attachment and growth of biofilm on the small particles; (iv) growth of the biofilm; and (v) development of new granules from the detached biofilm. In this process, it is clear that the first step of providing a suitably large attachment surface may be important for enhanced granulation.
To confirm the stoichiometry of the anammox activity in our reactors, nitrogen balances were performed. The mean ratio of NO2−-N consumed to NH4+-N consumed was about 1.21, and the mean ratio of NO3−-N produced to NH4+-N consumed was about 0.22. These values are in good agreement with those reported by Strous et al. (18) for the anammox process, in which the ratio of NO2−-N consumed to NH4+-N consumed and the ratio of NO3−-N produced to NH4+-N consumed were 1.3 and 0.26, respectively. In addition, the pH of the effluent (7.5 to 8.0) in the anammox reactor was always higher than that of the influent (7.0).
Characteristics of the anammox granules.
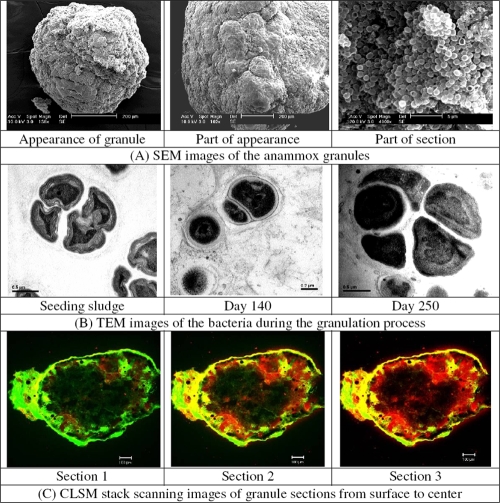
The mature anammox granules had diameters between 1.0 and 4.5 mm, with the majority (approximately 67%) having a diameter of 2.0 to 3.5 mm (Fig. 3). Their porous inner structure could facilitate the diffusion of substrates as well as the release of N2. The scanning electron micrographs showed typical coccoid-shaped cells as the dominant microorganisms on the granule surface embedded in an extracellular polysaccharide (EPS) matrix (Fig. 4A), very similar to those reported by others (7, 18, 26). Figure 4B shows the typical crescent-shaped anammox cells obtained via transmission electron micrography of chemically fixed anammox cells (30).
FIG. 3.
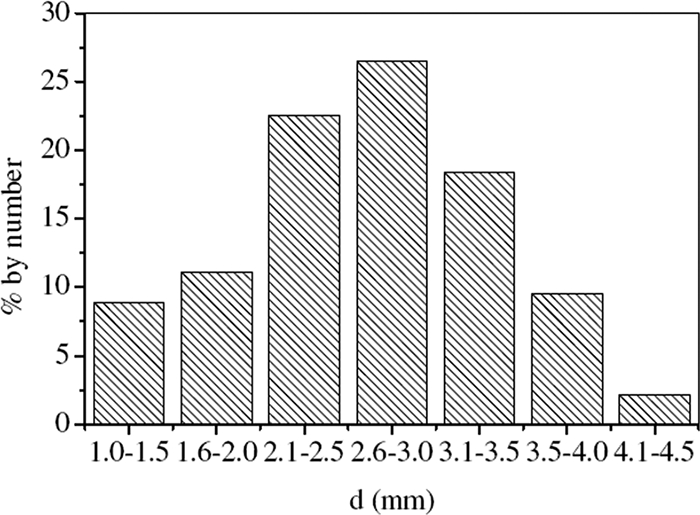
Size distribution of the anammox granules by number on day 250.
FIG. 4.
(A) SEM images of the anammox granular sludge. (B) Transmission electron microscopy (TEM) images of the anammox bacteria in sludge in the granulation process. (C) CLSM images of the 50-μm cryosections through the anammox granules from surface to center. Cells were stained with SYTO9 (green), and polysaccharides were stained with concanavalin A (red).
The mature anammox granules settled very well at velocities of 41 to 79 m h−1, which were similar to the settling velocities of the methanogenic granules (e.g., 52.9 m h−1) and at least two times greater than those of the flocculated anammox sludge (4). The significant increase in settling velocities indicated that the anammox granules had a highly dense and compact structure. This property would allow a greater biomass accumulation in the reactor and more-effective sludge-effluent separation in the treatment system.
The EPS of each gram of anammox granule (in VSS) contained 83.2 ± 7.9 mg carbohydrates and 42.7 ± 6.5 mg proteins, with a ratio of proteins/carbohydrates of 0.51. The CLSM analysis (Fig. 4C) showed mixed patterns of cells and EPS distributions in the anammox granules. The EPS were distributed throughout the granules, while the bacteria were mainly situated in the outer layer of the granule. The total EPS content in methanogenic granules was substrate dependent, ranging from 10 to 91 mg EPS g−1 VSS (11). This was substantially lower than that for the anammox granules (total EPS content was about 125 mg EPS g−1 VSS) found in this study. The ratio of proteins/carbohydrates was 1.2 to 4.0 for the EPS of methanogenic granules (11), much greater than that of the anammox ones. This significant difference suggested that proteins might be the key EPS constituents for the methanogenic granules, but that carbohydrates, rather than proteins, might play a more important role in the formation of the anammox granules. The large amount of glycosylation proteins encoded by the “Candidatus Kuenenia” genome is supportive of this suggestion (9, 20).
After granulation, the contact angle became 49 ± 3o, which was higher than that of the seeding sludge of 44 ± 2o, indicating a high hydrophobicity. Usually, a higher hydrophobicity of the cell surfaces would result in a stronger cell-to-cell interaction and is considered beneficial for the formation of more dense and stable structures (22). From a thermodynamic point of view, an increase in the hydrophobicity of cell surface causes a decrease in the excess Gibbs energy of the surface (2), which is in favor of the formation of granules.
Phylogenetic analysis and quantification of anammox.
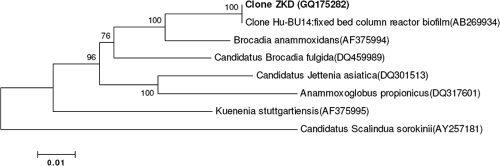
After the anammox activity was observed in the anammox granules, we applied a Planctomycetes-specific full-cycle rRNA approach to identify the 16S rRNA gene sequence of the dominant bacteria. Primers Pla46 and 630R were used to amplify near-complete 16S rRNA genes from total genomic DNA extracted from the granules at day 250. Ten clones were randomly selected for sequencing. All of the analyzed 16S rRNA genes had the same sequence, and subsequent phylogenetic analysis (Fig. 5) showed that the phylotype present in the reactor had 99% sequence similarity to an anammox clone obtained from a Japanese wastewater plant (AB269934) and was 95% similar to “Candidatus Brocadia anammoxidans” (24).
FIG. 5.
Phylogenetic tree of the clones obtained from the reactor enrichment culture, based on neighbor-joining analysis of almost the full length of 16S rRNA gene sequences. PCR amplification of 16S rRNA genes was conducted with the primer set of Pla46 and 630R. Bootstrap values (>50%) are indicated at branch points. The scale bar represents 10% estimated sequence divergence.
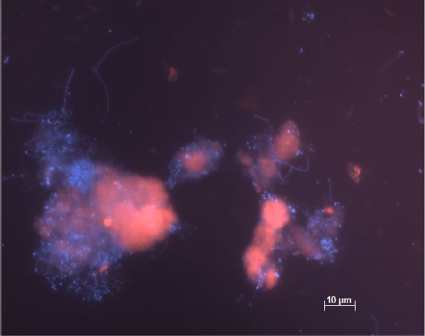
Based on the 16S rRNA gene sequences obtained from the anammox granules, a real-time PCR primer set (AMX658F-AMX924R) was designed to quantify the anammox bacteria in the sludge (Table 1). The copy numbers of 16S rRNA gene of anammox bacteria in the reactor enrichment were quantified as 4.6 × 108 copies ml−1 (Table 2). This was corroborated by FISH analysis using the general anammox probe S-*-AMX-0368-a-A-18 (Fig. 6).
TABLE 1.
List of primers used in this study
| Target | Primer name | Sequence (5′-3′) | Target site (position) | Reference |
|---|---|---|---|---|
| Planctomycetes 16S rRNA gene | Pla46 | GGATTAGGCATGCAAGTC | 43-63 | 12 |
| Planctomycetes 16S rRNA gene | 630R | CAKAAAGGAGGTGATCC | 1529-1545 | 17 |
| Anammox 16S rRNA gene | Amx694F | GGGGAGAGTGGAACTTCGG | 694-713 | This study |
| Anammox 16S rRNA gene | Amx960R | GCTCGCACAAGCGGTGGAGC | 960-979 | This study |
TABLE 2.
16S rRNA gene copies of the granular sludge
| Source | No. of 16S rRNA gene copies of anammox granules |
|
|---|---|---|
| ml−1 | g−1 VSS | |
| Seeding sludge | 1.9 × 105 | 2.4 × 107 |
| Day 140 sludge | 2.3 × 107 | 3.8 × 109 |
| Day 250 sludge | 3.5 × 108 | 5.4 × 1010 |
| Day 300 sludge | 4.6 × 108 | 7.1 × 1010 |
FIG. 6.
FISH micrographs with Cy3-labeled amx368 (targeting all anammox bacteria), counterstained with DAPI (4′,6-diamidino-2-phenylindole), for the enriched anammox granules—total active bacteria (blue) and anammox bacteria (purple).
Conclusions.
In this study, the anammox granules were successfully cultivated in a UASB reactor by seeding with aerobic granular sludge. The average total nitrogen removal efficiency exceeded 94% after the formation of the granules. The mature granules with porous inner structure, which were dominated by coccoid microorganisms, had a diameter ranging from 1.0 to 4.5 mm with red color and had good settling ability and a high settling velocity of 41.4 to 79.4 m/h. Cells and EPS were well mixed and distributed in the granules. The anammox phylotype in the reactor was 99% similar to a clone retrieved from a Japanese wastewater treatment plant (AB269934.1) and was 95% similar to “Candidatus Brocadia anammoxidans.” PCR and FISH analysis shows that 4.6 × 108 cells ml−1 were present. The results reported in this work may contribute to better design, shorter start-up time, and more stable operation of anammox reactors for the treatment of nitrogen-rich wastewaters.
Acknowledgments
We thank the Natural Science Foundation of China (50625825 and 50738006) for the partial support of this study. M.J. is supported by ERC advanced grant 232937.
Erwin van de Biezen is acknowledged for technical assistance.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.APHA. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, New York, NY.
- 2.Bos, R., H. C. van der Mei, and H. J. Busscher. 1999. Physico-chemistry of initial microbial adhesive interactions—its mechanisms and methods for study. FEMS Microbiol. Rev. 23:179-230. [DOI] [PubMed] [Google Scholar]
- 3.Chamchoi, N., and S. Nitisoravut. 2007. Anammox enrichment from different conventional sludges. Chemosphere 66:2225-2232. [DOI] [PubMed] [Google Scholar]
- 4.Dapena-Mora, A., S. W. H. Van Hulle, J. L. Campos, R. Mendez, P. A. Vanrolleghem, and M. Jetten. 2004. Enrichment of anammox biomass from municipal activated sludge: experimental and modelling results. J. Chem. Technol. Biotechnol. 79:1421-1428. [Google Scholar]
- 5.Jetten, M. S. M., S. J. Horn, and M. C. M. van Loosdrecht. 1997. Towards a more sustainable municipal wastewater treatment system. Water Sci. Technol. 35:171-180. [Google Scholar]
- 6.Jetten, M. S. M., M. C. Schmid, K. van de Pas-Schoonen, J. S. Sinninghe Damsté, and M. Strous. 2005. Anammox organisms: enrichment, cultivation and environmental analysis. Methods Enzymol. 397:34-57. [DOI] [PubMed] [Google Scholar]
- 7.Jetten, M. S. M., M. Wagner, J. Fuerst, M. van Loosdrecht, G. Kuenen, and M. Strous. 2001. Microbiology and application of the anaerobic ammonium oxidation (‘anammox') process. Curr. Opin. Biotechnol. 12:283-288. [DOI] [PubMed] [Google Scholar]
- 8.Kartal, B., J. Rattray, L. A. van Niftrik, J. van de Vossenberg, M. C. Schmid, and R. I. Webb. 2007. Candidatus ‘Anammoxoglobus propionicus’ a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 30:39-49. [DOI] [PubMed] [Google Scholar]
- 9.Kartal, B., L. van Niftrik, J. Rattray, J. van de Vossenberg, M. Schmid, J. S. Sinninghe Damsté, M. S. M. Jetten, and M. Strous. 2008. Candidatus ‘Brocadia fulgida’: an autofluorescent anaerobic ammonium oxidizer. FEMS Microbiol. Ecol. 63:46-55. [DOI] [PubMed] [Google Scholar]
- 10.Kowalchuk, G. A., F. J. de Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.). 2004. Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 11.Morgan, J. W., C. F. Forster, and L. M. A. Evison. 1990. Comparative-study of the nature of biopolymers extracted from anaerobic and activated sludges. Water Res. 24:743-750. [Google Scholar]
- 12.Neef, A., R. Amann, H. Schlesner, and K. H. Schleifer. 1998. Monitoring a widespread bacterial group in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 13.Pynaert, K., B. F. Smets, D. Beheydt, and W. Verstraete. 2004. Start-up of autotrophic nitrogen removal reactors via sequential biocatalyst addition. Environ. Sci. Technol. 38:1228-1235. [DOI] [PubMed] [Google Scholar]
- 14.Schmid, M., K. Walsh, R. Webb, W. I. C. Rijpstra, K. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Damste, J. Harris, P. Shaw, M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae,” sp nov., Candidatus “Scalindua wagneri,” sp nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 15.Sheng, G. P., and H. Q. Yu. 2006. Characterization of extracellular polymeric substances of aerobic and anaerobic sludge using 3-dimensional fluorescence spectroscopy. Water Res. 40:1233-1239. [DOI] [PubMed] [Google Scholar]
- 16.Sheng, G. P., H. Q. Yu, and Z. Yu. 2005. Extraction of the EPS from a photosynthetic bacterium Rhodopseudomonas acidophila. Appl. Microbiol. Biotechnol. 67:125-130. [DOI] [PubMed] [Google Scholar]
- 17.Stephen, J. R., G. A. Kowalchuk, M. A. V. Bruns, A. E. McCaig, C. J. Philips, T. M. Embley, and J. I. Prosser. 1998. Analysis of beta-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strous, M., J. A. Fuerst, E. H. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 19.Strous, M., J. G. Kuenen, J. A. Fuerst, M. Wagner, and M. S. M. Jetten. 2002. The anammox case—a new experimental manifesto for microbiological eco-physiology. Antonie van Leeuwenhoek 81:693-702. [DOI] [PubMed] [Google Scholar]
- 20.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. M. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. M. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440:790-794. [DOI] [PubMed] [Google Scholar]
- 21.Su, K. Z., and H. Q. Yu. 2005. Formation and characterization of aerobic granules in a sequencing batch reactor treating soybean-processing wastewater. Environ. Sci. Technol. 39:2818-2827. [DOI] [PubMed] [Google Scholar]
- 22.Thaveesri, J., D. Daffonchio, B. Liessens, P. Vandemeren, and W. G. Verstraete. 1995. Granulation and sludge bed stability in upflow anaerobic sludge bed reactors in relation to surface thermodynamics. Appl. Environ. Microbiol. 61:3681-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Third, K. A., J. Paxman, M. Schmid, M. Strous, M. S. M. Jetten, and R. Cord-Ruwisch. 2005. Enrichment of anammox from activated sludge and its application in the CANON process. Microb. Ecol. 49:236-244. [DOI] [PubMed] [Google Scholar]
- 24.Tsushima, I., T. Kindaichi, and S. Ohabe. 2007. Quantification of anaerobic ammonium oxidising bacteria in enrichment cultures by real-time PCR. Water Res. 41:785-794. [DOI] [PubMed] [Google Scholar]
- 25.Tsushima, I., Y. Ogasawara, T. Kindaichi, H. Satoh, and S. Okabe. 2007. Development of high-rate anaerobic ammonium-oxidizing (anammox) biofilm reactors. Water Res. 41:1623-1634. [DOI] [PubMed] [Google Scholar]
- 26.van de Graaf, A. A., P. de Bruijn, L. A. Robertson, M. S. M. Jetten, and J. G. Kuenen. 1996. Autotrophic growth of anaerobic ammonium-oxidizing micro-organisms in a fluidized bed reactor. Microbiology 142:2187-2196. [Google Scholar]
- 27.van der Star, W. R. L., W. R. Abma, D. Blommers, J. W. Mulder, T. Tokutomi, M. Strous, C. Picioreanu, and M. C. M. van Loosdrecht. 2007. Startup of reactors for anoxic ammonium oxidation. Experiences from the first full-scale anammox reactor in Rotterdam. Water Res. 41:4149-4163. [DOI] [PubMed] [Google Scholar]
- 28.van der Star, W. R. L., A. I. Miclea, U. G. J. M. van Dongen, G. Muyzer, C. Picioreanu, and M. C. M. van Loosdrecht. 2008. The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotechnol. Bioeng. 101:286-294. [DOI] [PubMed] [Google Scholar]
- 29.van Dongen, U. G. J. M., M. S. M. Jetten, and M. C. M. van Loosdrecht. 2001. The SHARON((R))-Anammox((R)) process for treatment of ammonium rich wastewater. Water Sci. Technol. 44:153-160. [PubMed] [Google Scholar]
- 30.van Niftrik, L., W. J. C. Geerts, E. G. van Donselaar, B. M. Humbel, A. Yakushevska, A. J. Verkleij, M. S. M. Jetten, and M. Strous. 2008. Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J. Struct. Biol. 161:401-410. [DOI] [PubMed] [Google Scholar]
- 31.Xavier, J. B., M. K. de Kreuk, C. Picioreanu, and M. C. M. van Loosdrecht. 2007. Multi-scale individual-based model of microbial and bioconversion dynamics in aerobic granular sludge. Environ. Sci. Technol. 41:6410-6417. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, Y., X. H. Ruan, H. J. M. Op den Camp, T. J. M. Smits, M. S. M. Jetten, and M. C. Schmid. 2007. Diversity and abundance of aerobic and anaerobic ammonium-oxidizing bacteria in freshwater sediments of the Xinyi River (China). Environ. Microbiol. 9:2375-2382. [DOI] [PubMed] [Google Scholar]