Abstract
The fermentative metabolism of Escherichia coli was reengineered to efficiently convert glycerol to succinate under anaerobic conditions without the use of foreign genes. Formate and ethanol were the dominant fermentation products from glycerol in wild-type Escherichia coli ATCC 8739, followed by succinate and acetate. Inactivation of pyruvate formate-lyase (pflB) in the wild-type strain eliminated the production of formate and ethanol and reduced the production of acetate. However, this deletion slowed growth and decreased cell yields due to either insufficient energy production or insufficient levels of electron acceptors. Reversing the direction of the gluconeogenic phosphoenolpyruvate carboxykinase reaction offered an approach to solve both problems, conserving energy as an additional ATP and increasing the pool of electron acceptors (fumarate and malate). Recruiting this enzyme through a promoter mutation (pck*) to increase expression also increased the rate of growth, cell yield, and succinate production. Presumably, the high NADH/NAD+ ratio served to establish the direction of carbon flow. Additional mutations were also beneficial. Glycerol dehydrogenase and the phosphotransferase-dependent dihydroxyacetone kinase are regarded as the primary route for glycerol metabolism under anaerobic conditions. However, this is not true for succinate production by engineered strains. Deletion of the ptsI gene or any other gene essential for the phosphotranferase system was found to increase succinate yield. Deletion of pflB in this background provided a further increase in the succinate yield. Together, these three core mutations (pck*, ptsI, and pflB) effectively redirected carbon flow from glycerol to succinate at 80% of the maximum theoretical yield during anaerobic fermentation in mineral salts medium.
Renewable bioenergy offers the potential to solve many environmental problems associated with petroleum-based fuels and chemicals. Biodiesel is produced by reacting vegetable oil or animal fat with alcohol (methanol or ethanol) and used as a transportation fuel in many countries (33). Glycerol is formed as an abundant waste product with limited commercial uses. As the worldwide production of biodiesel continues to increase, the development of effective uses for glycerol may prove essential for the economics and competitiveness of the biodiesel industry. The value of glycerol waste from biodiesel is similar to that of sugars currently used to produce fuel ethanol. Bioconversion of glycerol to higher-value products that replace petroleum, such as polymers, surfactants, solvents, and chemical intermediates, represents an opportunity to decrease waste and improve the economics of the biodiesel industry (5).
Many previous investigations have focused on the fermentative production of 1,3-propanediol (1,3-PD) from glycerol (2, 26, 35). Microorganisms including Klebsiella (14), Citrobacter (6), Enterobacter (1), Lactobacillus (29), and Clostridium (10, 28) have the native ability to ferment glycerol into this product. Dupont and Genencor have commercialized a 1,3-PD-based polyester, a condensation product of 1,3-PD and terephthalic acid using glucose as the feedstock. Potential demand for this polymer is estimated to be 1 billion to 2 billion pounds per year over the next 10 years (26). Other investigations of glycerol fermentation have described the production of hydrogen and ethanol (15), polyhydroxyalkanoates (PHAs) (20, 27), glyceric acid (13), and small amounts of succinate (21).
Succinic acid is currently used as a specialty chemical in the agricultural, food, and pharmaceutical industries (24, 34). It has also been identified by the U.S. Department of Energy as one of the top 12 building block chemicals (31) because it can be converted into a wide variety of products, including green solvents, pharmaceutical products, and biodegradable plastics (24, 34). Succinate is primarily produced from petroleum-derived maleic anhydride. Recent increases in the petroleum price have generated considerable interest in the fermentative production of succinate from sugars using either natural succinate-producing rumen bacteria or metabolically engineered Escherichia coli strains (24, 36, 38). Succinate can also be produced from glycerol by rumen bacteria, such as Anaerobiospirillum succiniciproducens (21). However, these strains require complex nutrients that increase costs of production, purification, and waste treatment.
E. coli has been previously engineered for the commercial production of 1,3-PD from sugars by Dupont and Genecor (26). It is an excellent organism for biotechnology applications but was long thought incapable of anaerobic growth on glycerol (23). Recent studies demonstrated that E. coli can ferment glycerol anaerobically (8, 11, 25, 33), and a new model was proposed for glycerol fermentation (11). In this model, glycerol is metabolized through the glycerol dehydrogenase (encoded by gldA) and dihydroxyacetone kinase (encoded by dhaKLM) pathway with the production of ethanol and acetate as primary fermentation products (11). Small amounts of succinate and 1,2-propanediol were also produced. Native genes encoding glycerol dehydrogenase and dihydroxyacetone kinase were expressed from a plasmid to increase the rates of glycerol metabolism and ethanol production (32). Succinate production has also been increased by expressing Clostridium freundii dihydroxyacetone kinase (encoded by dhaKL) (11). However, neither of these enhanced pathways would appear suitable for efficient succinate production due to the absence of net ATP production and the requirement for phosphoenolpyruvate as a phosphoryl donor for dihydroxyacetone, limiting the carboxylation of this intermediate (Fig. 1).
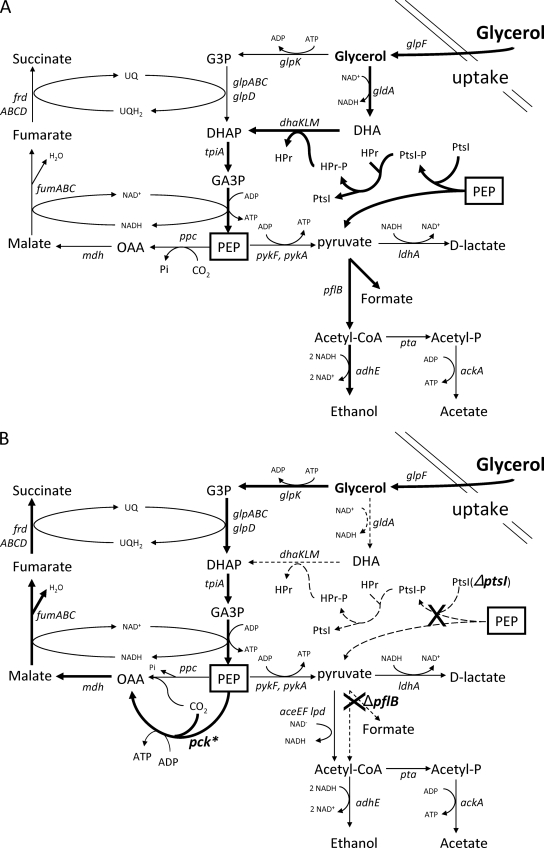
FIG. 1.
Glycerol uptake and fermentation by E. coli. (A) Native E. coli pathways. Bold black arrows represent dominant fermentation reactions prior to engineering; thin black arrows represent minor fermentation reactions. GlpK and GlpD are thought to function primarily during aerobic metabolism. Pathways are based on current reviews in EcoSal (3, 4, 22), data available in Ecocyc (19), and primary literature (11, 12, 18, 25, 30). (B) Engineered pathway for the fermentative metabolism of glycerol to succinate. Bold black arrows represent the engineered reactions for glycerol fermentation to succinate as the dominant product; thin black arrows represent minor fermentation reactions in the engineered strain. Dashed arrows represent reactions that are not functional due to deletions in ptsI and pflB. Deleted genes are marked with a black X. In native E. coli strains, phosphoenolpyruvate carboxykinase functions during gluconeogenesis to produce phosphoenolpyruvate. Mutational activation of the pck gene (denoted pck*) allows this enzyme to function in the reverse direction and to serve as the dominant carboxylation step, conserving energy as ATP. With this engineered pathway, competing needs for PEP have been eliminated and net ATP production has been increased. PEP is boxed to indicate a common pool. Abbreviations: DHA, dihydroxyacetone; DHAP, dihydroxyacetone 3-phosphate; PEP, phosphoenolpyruvate; G3P, glycerol 3-phosphate; GA3P, glyceraldehydes 3-phosphate.
Previous studies in our laboratory (16, 17, 36, 38) have engineered E. coli ATCC 8739 for the efficient production of succinate from glucose by recruiting genes from alternative pathways (36, 38). In this paper, we report the use of a similar approach to engineer strains for succinate production from glycerol in mineral salts medium.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used in this study are listed in Table 1. During strain construction, cultures were grown aerobically at 30, 37, or 39°C in Luria broth (10 g liter−1 Difco tryptone, 5 g liter−1 Difco yeast extract, and 5 g liter−1 NaCl) containing 2% (wt/vol) glucose or 5% (wt/vol) arabinose. Ampicillin (50 mg liter−1), kanamycin (50 mg liter−1), or chloramphenicol (40 mg liter−1) was added as needed during construction.
TABLE 1.
Sources and characteristics of E. coli strains used in this study
| Strain | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| ATCC 8739 | Wild type | Lab collection |
| XZ17 | ΔpflB | This study |
| XZ462b | pck* ΔglpA | This study |
| XZ464 | pck* ΔgldA | This study |
| XZ465b | pck* ΔdhaKL | This study |
| XZ466b | pck* ΔdhaM | This study |
| XZ467b | pck* ΔptsA | This study |
| XZ468b | pck* ΔptsH | This study |
| XZ632 | pck* | This study |
| XZ647 | pck* ΔptsI | This study |
| XZ721 | pck* ΔptsI ΔpflB | This study |
Genetic methods.
Chromosomal genes were deleted either seamlessly without leaving segments of foreign DNA as described previously (17, 37) or through one-step gene deletion methods (7). Red recombinase technology (Gene Bridges GmbH, Dresden, Germany) was used to facilitate chromosomal integration. Plasmids and primers used during construction are listed in Supplement S1 in the supplemental material.
Glycerol fermentation.
Strains were grown without antibiotics at 37°C (150 rpm) in NBS (New Brunswick Scientific) mineral salts medium supplemented with 5% (wt/vol) glycerol and 100 mM potassium bicarbonate as previously described (38). Inocula were prepared by transferring fresh colonies into a 250-ml flask (100 ml NBS mineral salts medium, 2% glycerol). After 16 h (37°C, 120 rpm), this culture was diluted into a pH-controlled fermentation vessel containing 300 ml NBS medium (5% glycerol, 100 mM potassium bicarbonate) to provide an inoculum of 0.033 g cell dry weight (CDW) liter−1. Fermentations were automatically maintained at pH 7.0 by adding base containing additional CO2 (2.4 M potassium carbonate in 1.2 M potassium hydroxide).
Analysis.
Cell mass was estimated by measuring the optical density at 550 nm (OD550). Organic acids and the glycerol concentration were measured by high-performance liquid chromatography (37).
RESULTS
Inactivating competing pathway.
Wild-type E. coli ATCC 8739 grew very slowly during glycerol fermentation. After 6 days, 153 mM glycerol was metabolized with a cell yield of 0.55 g liter−1 CDW (Table 2). Formate and ethanol were the major products, with smaller amounts of succinate (38 mM). Although no lactate was detected, a small amount of acetate was produced. To minimize production of ethanol and formate, the pflB gene was deleted (Fig. 1), with unexpected results. Although the production of formate and ethanol was eliminated in the resulting mutant (strain XZ17), glycerol metabolism and cell yield were lowered (45% and 69%, respectively). Succinate production and succinate yield (5 mM succinate after 6 days; 0.11 mol succinate per mol glycerol) were reduced by the pflB deletion. Lactate was produced as the dominant fermentation product (Table 2). Since lactate does not provide redox balance with glycerol, small amounts of dissolved oxygen and air leakage are presumed to have also contributed to NADH oxidation.
TABLE 2.
Succinate production from glycerol by engineered E. coli strains in NBS mineral salts medium
| Strain | Genetic modificationa | Time (days) | Cell mass (g liter−1) | Glycerol used (mM) | Fermentation product (mM)b |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Suc | Suc yield | For | EtOH | Lac | Ace | |||||
| ATCC 8739 | Wild type | 6 | 0.55 | 153 | 38 ± 6 | 0.25 ± 0.03 | 110 | 81 | 16 | |
| XZ17 | ΔpflB | 6 | 0.3 | 48 | 5 ± 1 | 0.11 ± 0.02 | 20 | 5 | ||
| XZ632 | pck* | 6 | 0.7 | 149 | 64 ± 13 | 0.44 ± 0.05 | 83 | 46 | 20 | |
| XZ647 | pck* ΔptsI | 6 | 0.4 | 89 | 63 ± 4 | 0.71 ± 0 | 25 | 5 | 13 | |
| XZ721 | pck* ΔpflB ΔptsI | 6 | 0.5 | 128 | 102 ± 25 | 0.80 ± 0.01 | 6 | 12 | ||
| XZ632 | pck* | 6 | 0.7 | 149 | 64 ± 13 | 0.44 ± 0.05 | 83 | 46 | 20 | |
| XZ467 | pck* ΔptsA | 6 | 0.79 | 168 | 72 ± 16 | 0.43 ± 0.03 | 85 | 56 | 16 | |
| XZ462 | pck* ΔglpA | 6 | 0.5 | 77 | 28 ± 4 | 0.36 ± 0.05 | 50 | 22 | 17 | |
| XZ464 | pck* ΔgldA | 6 | 0.47 | 150 | 112 ± 29 | 0.75 ± 0.03 | 37 | 11 | 15 | |
| XZ465 | pck* ΔdhaKL | 6 | 0.43 | 125 | 95 ± 25 | 0.77 ± 0.02 | 35 | 8 | 16 | |
| XZ466 | pck* ΔdhaM | 6 | 0.5 | 152 | 108 ± 33 | 0.71 ± 0.05 | 30 | 9 | 14 | |
| XZ468 | pck* ΔptsH | 6 | 0.5 | 125 | 85 ± 22 | 0.68 ± 0.07 | 32 | 14 | 10 | |
pck* denotes a mutated form of pck (G to A at −64 relative to the ATG start). This mutation increases expression by 8-fold (36, 38).
Fermentations were carried out in NBS medium with 5% glycerol and 100 mM potassium bicarbonate (37°C, pH 7.0, 150 rpm). Succinate yield was calculated as moles succinate produced per mole glycerol metabolized. Data represent an average of 4 experiments (±SD). Abbreviations: Suc, succinate; For, formate; EtOH, ethanol; Lac, lactate; Ace, acetate.
Recruiting phosphoenolpyruvate carboxykinase.
No net ATP would be produced during the anaerobic metabolism of glycerol to succinate using the native phosphoenolpyruvate (PEP) carboxylase for carboxylation (Fig. 1).
In contrast, the high energy of PEP can be conserved by replacing this enzyme with phosphoenolpyruvate carboxykinase (36, 38) to produce 1 mole of ATP per mole of succinate (Fig. 1B). A chromosomal promoter mutation was used to upregulate pck expression by approximately 8-fold (36, 38). In the resulting mutant strain, XZ632, cell yield and glycerol metabolism were increased by 27% compared to those of the wild-type parent (ATCC 8739) (Table 2). Production of formate and ethanol were also lower, with a corresponding increase in the yield of succinate (from 0.25 to 0.44 mol per mol glycerol; P < 0.05), a 75% increase in carbon partitioning into product.
Inactivating the glycerol dehydrogenase-dihydroxyacetone kinase (encoded by gldA and dhaKLM) pathway.
Glycerol enters the cell by facilitated diffusion using the GlpF permease. Two pathways have been proposed for the initial metabolism of glycerol in E. coli (9, 11). During aerobic (oxidative) metabolism (Fig. 1), glycerol is phosphorylated using ATP as the phosphoryl donor (GlpK) before being oxidized to phosphorylated dihydroxyacetone by GlpD. Quinones serve as electron acceptors. In the absence of oxygen, glycerol is oxidized to dihydroxyacetone by GldA using NAD+ as the electron acceptor (11). Dihydroxyacetone is subsequently phosphorylated by an intracellular phosphorelay system (PtsI, Hpr, and DhaKLM) using phosphoenolpyruvate as the phosphoryl donor (Fig. 1). Glycerol may also be metabolized through GlpK and GlpABC as a minor pathway during anaerobic conditions (Fig. 1).
Based on this model, a mutation in ptsI (strain XZ632) would be expected to impede the fermentative production of succinate. However, the opposite was found (Table 2; Fig. 1). Deletion of ptsI significantly increased the succinate yield (from 0.44 to 0.71 mol succinate per mol glycerol; P < 0.05). The shift in fermentation products resulting from a ptsI deletion confirms that glycerol dehydrogenase and dihydroxyacetone kinase (encoded by gldA and dhaKLM) represent the primary pathway in native strains, as proposed elsewhere (11). However, this native anaerobic pathway does not appear desirable for succinate production and limits the usage of phosphoenolpyruvate for oxaloacetic acid production (Fig. 1).
These two glycerol metabolism pathways were further investigated by deleting single genes in XZ632 containing the pck* up-mutation (Table 2). No growth was observed in mineral salts medium after deletion of glpK, suggesting that the gldA-dhaKLM pathway alone could not support anaerobic growth with succinate production. A glpA deletion was accompanied by lower succinate production and cell yield (Table 2). With this deletion (XZ462), glycerol could be metabolized by the gldA-dhaKLM pathway, GlpD, or both. In this pck* background, deletion of any single gene concerned with the gldA-dhaKLM pathway, including ptsH (encoding Hpr of the phosphorelay system) and ptsI, resulted in a similar increase in succinate yield. Deletion of ptsA, a cryptic gene in the ptsI-glpA-ptsA operon, had little effect on glycerol metabolism and may not be functional.
Core mutations (pck*, ΔptsI, and ΔpflB).
The pflB gene was deleted in strain XZ647 (pck* ΔptsI) to eliminate formate and ethanol accumulation. In this strain, yield was further increased from 0.71 to 0.80 mol succinate per mol glycerol with small amounts of lactate and acetate as side products (Table 2). These three core mutations were sufficient for the fermentation to redirect the fermentative metabolism of glycerol to succinate at 80% of the maximum theoretical yield in mineral salts medium. Growth and metabolism were slow, however, requiring 6 days to produce 102 mM succinate.
DISCUSSION
Previous studies have demonstrated that glycerol can be effectively metabolized to succinate by the rumen bacterium Anaerobiospirillum succiniproducens using complex medium containing yeast extract and peptone (21). Succinate yields based on glycerol alone exceeded the maximum theoretical yield, with product titers of 200 mM succinate (5 days). Gonzalez and colleagues (8, 11, 25, 32) recently discovered that E. coli can ferment glycerol as a sole carbon source under anaerobic conditions, eliminating the need for complex nutrients. Ethanol and formate were the most abundant products, together with lower levels of succinate and acetate. The succinate yield was increased to 0.4 mol per mol glycerol by expressing the Clostridium freundii dihydroxyacetone kinase (dhaKL) and providing elevated levels of CO2 (11).
The anaerobic pathway for glycerol dissimilation described by Gonzalez and colleagues (11, 25, 32) works well for ethanol production but may not be optimal for succinate production using the fermentative phosphoenolpyruvate carboxylase (Fig. 1). No net ATP would be produced. Oxaloacetate production for redox balance would be limited by the competing requirement for phosphoenolpyruvate as the phosphoryl donor of dihydroxyacetone phosphate. We have shown that succinate production can be substantially increased by conserving energy during the carboxylation of phosphoenolpyruvate using the gluconeogenic phosphoenolpyruvate carboxykinase (36, 38), disrupting the primary pathway for anaerobic glycerol metabolism (deletion of any step), and recruiting the minor (native) anaerobic pathways for glycerol metabolism to dihydroxyacetone phosphate (Fig. 1B). With this assembled pathway for succinate, the ATP yield was increased from 0 to 1 per glycerol and the conflict with phosphoenolpyruvate usage was eliminated. With the further deletion of pflB, XZ721 containing three mutations (pck*, ΔptsI, and ΔpflB) achieved a succinate yield of 0.8 mol per mol glycerol. This represents 80% of the maximum theoretical yield for glycerol. Related strains have been shown to produce 1.4 mol succinate per mol glucose (38), also 80% of the maximum theoretical yield (1.7 mol succinate/mol glucose).
Growth and metabolism with glycerol remained slow, however, hindering most biotechnology applications for succinate. Slow anaerobic growth with glycerol has been previously attributed to a redox imbalance resulting from the use of intermediates for biosynthesis (11). Additional potential causes include the limited availability of energy for gluconeogesis (required for cell envelope biosynthesis).
Supplementary Material
Acknowledgments
We gratefully acknowledge research support by grants from the U.S. Department of Energy (DE-FG36-08GO88142).
This work was facilitated by the EcoCyc database (19).
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barbirato, F., C. Camarasaclaret, J. P. Grivet, and A. Bories. 1995. Glycerol fermentation by a new 1,3-propanediol-producing microorganism Enterobacter agglomerans. Appl. Microbiol. Biotechnol. 43:786-793. [Google Scholar]
- 2.Biebl, H., K. Menzel, A. P. Zeng, and W. D. Deckwer. 1999. Microbial production of 1,3-propanediol. Appl. Microbiol. Biotechnol. 52:289-297. [DOI] [PubMed] [Google Scholar]
- 3.Bock, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 4.Booth, I. R. 2005. Chapter 3.4.3, Glycerol and methylglyoxal metabolism. In A. Bock, R. Curtiss III, J. B. Kaper, F. C. Neidhardt, T. Nystrom, K. E. Rudd, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org/.
- 5.Claude, S. 1999. Research of new outlets for glycerol—recent developments in France. Fett-Lipid 101:101-104. [Google Scholar]
- 6.Daniel, R., and G. Gottschalk. 1992. Growth temperature-dependent activity of glycerol dehydratase in Escherichia coli expressing the Citrobacter freundii dha regulon. FEMS Microbiol. Lett. 100:281-285. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dharmadi, Y., A. Murarka, and R. Gonzalez. 2006. Anaerobic fermentation of glycerol by Escherichia coli: a new platform for metabolic engineering. Biotechnol. Bioeng. 94:821-829. [DOI] [PubMed] [Google Scholar]
- 9.Durnin, G., J. Clomburg, Z. Yeates, P. J. Alvarez, K. Zygourakis, P. Campbell, and R. Gonzalez. 2009. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol. Bioeng. 103:148-161. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg, C. W. 1987. Production of 1,3-propanediol from glycerol by Clostridium acetobutylicum and other Clostridium species. Appl. Environ. Microbiol. 53:639-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, R., A. Murarka, Y. Dharmadi, and S. S. Yazdani. 2008. A new model for the anaerobic fermentation of glycerol in enteric bacteria: trunk and auxiliary pathways in Escherichia coli. Metab. Eng. 10:234-245. [DOI] [PubMed] [Google Scholar]
- 12.Gutknecht, R., R. Beutler, L. F. Garcia-Alles, U. Baumann, and B. Erni. 2001. The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J. 20:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habe, H., T. Fukuoka, D. Kitamoto, and K. Sakaki. 2009. Biotransformation of glycerol to d-glyceric acid by Acetobacter tropicalis. Appl. Microbiol. Biotechnol. 81:1033-1039. [DOI] [PubMed] [Google Scholar]
- 14.Homann, T., C. Tag, H. Biebl, W. D. Deckwer, and B. Schink. 1990. Fermentation of glycerol to 1,3-propanediol by Klebsiella and Citrobacter strains. Appl. Microbiol. Biotechnol. 33:121-126. [Google Scholar]
- 15.Ito, T., Y. Nakashimada, K. Senba, T. Matsui, and N. Nishio. 2005. Hydrogen and ethanol production from glycerol-containing wastes discharged after biodiesel manufacturing process. J. Biosci. Bioeng. 100:260-265. [DOI] [PubMed] [Google Scholar]
- 16.Jantama, K., M. J. Haupt, S. A. Svoronos, X. Zhang, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng. 99:1140-1153. [DOI] [PubMed] [Google Scholar]
- 17.Jantama, K., X. Zhang, J. C. Moore, K. T. Shanmugam, S. A. Svoronos, and L. O. Ingram. 2008. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol. Bioeng. 101:881-893. [DOI] [PubMed] [Google Scholar]
- 18.Jin, R. Z., J. C. Tang, and E. C. Lin. 1983. Experimental evolution of a novel pathway for glycerol dissimilation in Escherichia coli. J. Mol. Evol. 19:429-436. [DOI] [PubMed] [Google Scholar]
- 19.Keseler, I. M., J. Collado-Vides, S. Gama-Castro, J. Ingraham, S. Paley, I. T. Paulsen, M. Peralta-Gil, and P. D. Karp. 2005. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 33:D334-D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koller, M., R. Bona, G. Braunegg, C. Hermann, P. Horvat, M. Kroutil, J. Martinz, J. Neto, L. Pereira, and P. Varila. 2005. Production of polyhydroxyalkanoates from agricultural waste and surplus materials. Biomacromolecules 6:561-565. [DOI] [PubMed] [Google Scholar]
- 21.Lee, P. C., W. G. Lee, S. Y. Lee, and H. N. Chang. 2001. Succinic acid production with reduced by-product formation in the fermentation of Anaerobiospirillum succiniciproducens using glycerol as a carbon source. Biotechnol. Bioeng. 72:41-48. [PubMed] [Google Scholar]
- 22.Lin, E. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 23.Lin, E. C. 1976. Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30:535-578. [DOI] [PubMed] [Google Scholar]
- 24.McKinlay, J. B., C. Vieille, and J. G. Zeikus. 2007. Prospects for a bio-based succinate industry. Appl. Microbiol. Biotechnol. 76:727-740. [DOI] [PubMed] [Google Scholar]
- 25.Murarka, A., Y. Dharmadi, S. S. Yazdani, and R. Gonzalez. 2008. Fermentative utilization of glycerol by Escherichia coli and its implications for the production of fuels and chemicals. Appl. Environ. Microbiol. 74:1124-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura, C. E., and G. M. Whited. 2003. Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14:454-459. [DOI] [PubMed] [Google Scholar]
- 27.Nikel, P. I., M. J. Pettinari, M. A. Galvagno, and B. S. Mendez. 2008. Poly(3-hydroxybutyrate) synthesis from glycerol by a recombinant Escherichia coli arcA mutant in fed-batch microaerobic cultures. Appl. Microbiol. Biotechnol. 77:1337-1343. [DOI] [PubMed] [Google Scholar]
- 28.Raynaud, C., P. Sarcabal, I. Meynial-Salles, C. Croux, and P. Soucaille. 2003. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. U. S. A. 100:5010-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schutz, H., and F. Radler. 1984. Anaerobic reduction of glycerol to propanediol-1.3 by Lactobacillus brevis and Lactobacillus buchneri. Syst. Appl. Microbiol. 5:169-178. [Google Scholar]
- 30.Tang, J. C., R. G. Forage, and E. C. C. Lin. 1982. Immunochemical properties of NAD+-linked glycerol dehydrogenases from Escherichia coli and Klebsiella pneumoniae. J. Bacteriol. 152:1169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werpy, T., and G. Petersen (ed.). 2004. Top value added chemicals from biomass. U.S. Department of Energy, Washington, DC. http://www1.eere.energy.gov/biomass/pdfs/35523.pdf.
- 32.Yazdani, S., and R. Gonzalez. 2008. Engineering Escherichia coli for the efficient conversion of glycerol to ethanol and co-products. Metab. Eng. 10:340-351. [DOI] [PubMed] [Google Scholar]
- 33.Yazdani, S. S., and R. Gonzalez. 2007. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 18:213-219. [DOI] [PubMed] [Google Scholar]
- 34.Zeikus, J. G., M. K. Jain, and P. Elankovan. 1999. Biotechnology of succinic acid production and markets for derived industrial products. Appl. Microbiol. Biotechnol. 51:545-552. [Google Scholar]
- 35.Zeng, A. P., and H. Biebl. 2002. Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. Adv. Biochem. Eng. Biotechnol. 74:239-259. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, X., K. Jantama, J. C. Moore, L. R. Jarboe, K. T. Shanmugam, and L. O. Ingram. 2009. Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. doi: 10.10173/pnas.0905396106. [DOI] [PMC free article] [PubMed]
- 37.Zhang, X., K. Jantama, J. C. Moore, K. T. Shanmugam, and L. O. Ingram. 2007. Production of L-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77:355-366. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, X., K. Jantama, K. T. Shanmugam, and L. O. Ingram. 16 October 2009. Re-engineering Escherichia coli for succinate production in mineral salts medium. Appl. Environ. Microbiol. doi: 10.1128/AEM.01758-09. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.