Abstract
To evaluate the potential for organic nitrogen addition to stimulate the in situ growth of ammonia oxidizers during a field scale bioremediation trial, samples collected from the Eastern Snake River Plain Aquifer in Idaho before, during, and after the addition of molasses and urea were subjected to PCR analysis of ammonia monooxygenase subunit A (amoA) genes. Ammonia-oxidizing bacteria (AOB) and archaea (AOA) were present in all of the samples tested, with AOA amoA genes outnumbering AOB amoA genes in all of the samples. Following urea addition, nitrate levels rose and bacterial amoA copy numbers increased dramatically, suggesting that urea hydrolysis stimulated nitrification. Bacterial amoA diversity was limited to two Nitrosomonas phylotypes, whereas archaeal amoA analyses revealed 20 distinct operational taxonomic units, including several that were markedly different from all previously reported sequences. Results from this study demonstrate the likelihood of stimulating ammonia-oxidizing communities during field scale manipulation of groundwater conditions to promote urea hydrolysis.
Subsurface calcite precipitation driven by microbial urea hydrolysis has been proposed as a means of remediating trace metal or radionuclide contaminants (e.g., strontium-90) that can be coprecipitated and retained in the solid phase (11, 12, 42). Urea hydrolysis generates carbonate alkalinity and raises pH, both of which promote calcite precipitation. However, another product of urea hydrolysis is ammonium, as shown in the following equation:
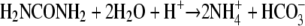 |
In low-nutrient groundwater, the ammonium resulting from urea hydrolysis can have a number of fates, including uptake by nitrogen-limited microorganisms or transformation to nitrite by ammonia-oxidizing microorganisms. Microbial oxidation of ammonia is a net acid-yielding process. The resultant acidity from this reaction could inhibit calcite precipitation or promote destabilization of preexisting calcite, potentially liberating contaminants from the solid phase. In addition, the further transformation of nitrite by nitrite-oxidizing bacteria leads to the formation of nitrate, a regulated contaminant of drinking water.
The first step of bacterial ammonia oxidation, the conversion of ammonia to hydroxylamine, is catalyzed by the membrane-bound enzyme ammonia monooxygenase. The gene coding for the catalytic α subunit of this enzyme, amoA, has proven to be an effective molecular marker for ammonia-oxidizing bacteria (AOB) (20, 34). All of the currently known chemoautotrophic AOB are associated with the Nitrosomonas and Nitrosospira genera within the Betaproteobacteria or the genus Nitrosococcus within the Gammaproteobacteria (15, 32). Although ammonia oxidation was long believed to be carried out exclusively by members of the domain Bacteria, considerable evidence now suggests that recently discovered ammonia-oxidizing archaea (AOA) (18) are key players in this critical step of the microbial nitrogen cycle (8).
The archaeal amoA gene has been found in a wide range of environments (9; reviewed in references 8 and 31), and its expression has been documented in enrichment cultures (35) and soil microcosms (40), as well as in marine and terrestrial environments (21, 23). Reported quantitative PCR (qPCR) analyses of amoA in marine and terrestrial environments suggest that AOA typically outnumber AOB by orders of magnitude (23, 26, 44), and AOA abundance has also recently been shown to be highly correlated with water column 15NH4+ oxidation rates (1). However, some recent studies have reported that AOB are more abundant under certain conditions (6, 27, 35, 43, 45).
In an effort to better understand the fate of ammonium generated from urea hydrolysis, we monitored the abundance and diversity of bacterial and archaeal amoA genes during a field experiment designed to test stimulation of urea hydrolysis in groundwater. Dilute molasses and urea were sequentially introduced into a well in the Eastern Snake River Plain Aquifer (ESRPA) in Idaho (13). Previous laboratory experiments indicated that molasses, an inexpensive and commonly used bioremediation amendment (14), was effective in increasing overall microbial populations, as well as total ureolytic activity (13, 39). The ESRPA is a deep basalt aquifer and is considered oligotrophic (4, 22, 29); however, previous work has demonstrated the presence of ureolytic microbes in this environment (11, 13). Erwin et al. also reported evidence of AOB during the analysis of methane monooxygenase clone libraries from ESRPA samples (7), but in general, the structure and function of ammonia-oxidizing microbial communities (and especially AOA) in deep aquifers like the ESRPA have been relatively unexplored.
MATERIALS AND METHODS
Site, treatment, and sample collection description.
The field experiment was conducted at well UP-1 in Idaho Falls, ID. The ESRPA lies approximately 50 m below the land surface (mbls) at this location. The site characteristics were described in detail elsewhere (13). Amendments were injected into and water was collected from a 2.1-m vertical interval located approximately 82 mbls; the interval, in a zone of consolidated basalt, was isolated within the open borehole by inflatable packers. During the first phase of the field experiment (pretreatment; 13 days), groundwater samples for microbiological and chemical analyses were collected on five separate occasions (days 0, 6, 8, 10, and 13) prior to the addition of any nutrient amendments, in order to characterize the background conditions. On day 13, following background sample collection, the first injection solution (650 liters) consisting of groundwater amended with 7.5 mg liter−1 food-grade molasses (Grandma's Molasses, Riverton, NJ) was introduced into UP-1. On day 16, after sample collection, another 1,460 liters of the molasses solution (7.5 mg liter−1) was introduced; the same procedure was repeated on day 20. On day 22, after the last of three molasses treatment phase samples had been collected, 580 liters of groundwater containing 50 mM urea (Falls Fertilizer, Idaho Falls, ID) and 100 mg liter−1 bromide (as KBr) as a conservative tracer were added. Post-urea phase samples were collected on days 30 and 35. Microbial cells (from 20 liters of groundwater) were collected on filters simultaneously in triplicate from each sampling date and immediately frozen by using methods similar to those described elsewhere (28).
Water chemistry characterization.
Methods for the comprehensive water chemistry characterization were described in detail previously (13), but protocols for some of the parameters are briefly summarized here. Dissolved oxygen (DO) and pH were measured in the field using a multiprobe (DataSonde 4/minisonde; Hydrolab, Austin, TX), common anions (e.g., Cl−, NO3−, NO2−, SO42−, PO43−, F−, and Br−) and ammonium were measured by ion chromatography (Dionex, Sunnyvale, CA), and metals (Ca2+, Mg2+, Na+, K+, Sr2+, and Ni2+) were measured by inductively coupled plasma mass spectrometry (ICP-MS model 7500a; Agilent, Palo Alto, CA). Water samples and the molasses were characterized for organic carbon and total nitrogen content using a total organic carbon/total nitrogen instrument (Shimadzu Scientific Instruments, Columbia, MD). All of the analyses were conducted in accordance with the manufacturer recommendations. Difference in median values in the data (averages of replicate measurements) across the three phases was tested using the nonparametric Kruskal-Wallis test, and pairwise comparisons between the pretreatment and molasses treatment phases were made using Wilcoxon rank-sum tests. A P value of <0.05 was considered the threshold of significance in the Kruskal-Wallis test, and a Bonferroni adjustment was used for the pairwise tests, resulting in a threshold of 0.05/3 = 0.017.
DNA extractions, PCR amplification, and cloning.
DNA from groundwater samples was extracted as described previously (13). Briefly, filters (containing 20 liters of groundwater cell biomass) were fragmented and thawed, and cells were lysed by treatment at 50°C with a proteinase K, EDTA, and sodium dodecyl sulfate solution. Proteins were removed by phenol-chloroform extraction, and the DNA was precipitated with sodium acetate and isopropanol, followed by suspension in 500 μl 10 mM Tris (pH 8.0) and storage at −20°C. DNA concentrations were determined by NanoDrop spectrophotometric analysis. The diversity of bacterial amoA genes was evaluated by analysis of PCR products derived from one biological replicate sample taken from each of the sample collection days. As described elsewhere (10), a 672-bp fragment of bacterial amoA was PCR amplified with primers A189DR (5′ GGNGACTGGGAYTTCTGG) and amoA-2R′DR (5′ CCCCTSBGSRAAVCCTTYTTC) for each of the sample collection days, and the amplified products were pooled into three groups for constructing three distinct clone libraries (one library representing each of the study periods: pretreatment, molasses treatment, and post-urea). An additional bacterial amoA library limited to the day 35 samples (post-urea phase) was constructed with shorter gene fragments (491 bp) generated by using commonly used primers AmoA1F* (38) and AmoA2R (34); this library was constructed to verify that the low AOB amoA diversity observed in the first library was not due to primer bias. The diversity of the archaeal amoA genes was assessed using PCR products derived from the biological triplicate samples taken on days 10, 20, and 35, time points representing the three study periods (pretreatment, molasses treatment, and post-urea). Archaeal amoA clone libraries were constructed from a 635-bp segment of amoA genes generated by using primers Arch-amoAF and Arch-amoAR (9). The PCR conditions for the 491-bp AOB amoA fragment and the AOA amoA libraries were identical to those described previously by Francis et al. (9), except that the PCR was carried out for 37 cycles. Clone libraries were generated using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). Sequences were aligned, and neighbor-joining trees were constructed from Juke-Cantor-corrected distance matrices using ARB (24). ARB-generated, Jukes-Cantor-corrected distance matrices were used for rarefaction analysis with DOTUR (36) employing a 5% DNA sequence identity cutoff to define operational taxonomic units (OTUs). Abundance-based Sørensen-type similarities among clone libraries were also calculated using DOTUR.
qPCR analysis of archaeal and bacterial amoA genes.
Bacterial and archaeal amoA genes were quantified in groundwater DNA extracts by qPCR with primer sets A189DR/amoA-2R′DR and Arch-amoAF/Arch-amoAR and a Platinum qPCR SuperMix-UDG kit (1×; Invitrogen) on a Rotor-Gene 3000 (source, 470 nm; detector, 510 nm; Corbett Research, Mortlake, Australia) and using methods modified from Okano et al. (30). For AOB amoA, each reaction mixture contained DNA extract (2 μl pretreatment or 0.2 μl molasses treatment/post-urea sample, corresponding to ∼6 to 25 ng genomic DNA per reaction), 0.5× SYBR green I (Molecular Probes, Eugene, OR), 4 mM MgCl2, 400 ng/μl bovine serum albumin, 300 nM A189DR, and 900 nM amoA2R′DR. The cycling conditions were 2 min at 50°C, 5 min at 95°C and 40 cycles of 95°C for 45 s, 61°C for 60 s, and 72°C for 45 s. The AOA amoA amplicon was generated under similar conditions, with the following modifications: the concentration of Arch-amoAF and Arch-amoAR was 300 nM each, and the annealing temperature was 58°C. Melt curve analysis was utilized to determine an optimal temperature for measuring amoA gene product amplification in qPCR; therefore, in addition to measuring fluorescence at 72°C, the qPCR cycling program was adjusted for a second 15-s hold during which fluorescence data were acquired at 86°C and 81°C for the AOB and AOA, respectively. For determining gene copy numbers in the qPCR assays, a plasmid (33) containing a fragment of the amoA gene from Nitrosomonas europaea (accession no. AF058691) served as the template for AOB, whereas a crenarchaeal amoA clone/plasmid (accession no. EU651199) was used for the AOA. The standard curves for the AOB and AOA assays were linear from approximately 101 to 107 and 102 to 107 gene copies, respectively, assay efficiencies ranged from 0.85 to 0.88 (AOB) and 0.84 to 0.93 (AOA), and correlation coefficients (R2) were greater than 0.99. As with the water chemistry data, the variance of the qPCR data from the pretreatment, molasses treatment, and post-urea phases was examined using Kruskal-Wallis and Wilcoxon rank-sum tests.
Nucleotide sequence accession numbers.
The amoA sequences obtained in this study were deposited in GenBank under accession numbers FJ546426 to FJ546456 (for AOB) and FJ543209 to FJ543376 (for AOA).
RESULTS
Water chemistry.
An extensive suite of chemical parameters was monitored over the course of the field experiment in order to discern the effect of the nutrient additions on groundwater biogeochemistry (13); only a subset is discussed here. Table 1 presents data for pH, DO, ammonium, and nitrate. The ammonium, indicative of urea hydrolysis, was only detected in the samples collected after urea addition. The Kruskal-Wallis tests and subsequent pairwise comparisons using Wilcoxon rank-sum tests indicated that the pHs of the post-urea and molasses treatment phases differed from the pretreatment phase pH; however, due to limited sample size, none of the pairwise tests were significant. The observed pH change was very slight, likely reflecting the buffering capacity of the rock matrix. Surprisingly, DO concentrations did not exhibit a statistically significant decrease following the addition of molasses, suggesting that the small amount of organic carbon added (the nominal mass of molasses added was 7.5 mg molasses per liter of injection water; triplicate measurements of the molasses-amended water indicated a final dissolved organic carbon content of 7.02 ± 0.12 mg C liter−1) did not induce sufficient increased oxygen consumption to overcome the effect of the recharge of the treatment zone by oxygenated water. However, following addition of the urea solution, measured DO values were lower (three- to sixfold) than in the pretreatment and molasses treatment phases, although anaerobic conditions were not attained. For nitrate, there were insufficient data to attain statistical significance, although the available data suggest that nitrate concentrations did increase over the course of the field experiment, particularly after urea addition.
TABLE 1.
Selected field water parametersb
| Parameter | Pretreatment, daya: |
Molasses treatment, daya: |
Post-urea, daya: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 8 | 10 | 13 | 16 | 20 | 22 | 30 | 35 | |
| pH | 7.01 | 7.02 | 6.91 | 6.96 | 6.98 | 7.06 | 7.12 | 7.10 | 7.12 | 7.11 |
| DO concn (mg/liter) | 7.3 | 6.9 | 6.3 | 6.4 | 6.5 | 6.9 | 7.6 | 7.8 | 1.3 | 1.8 |
| NH4 concn (mg/liter) | BDc | BD | BD | BD | BD | BD | BD | BD | 1.03 | 0.73 |
| NO3 concn (mg/liter) | NDd | 6.62 | 6.45 | 6.66 | 6.74 | 6.56 | 7.23 | 7.54 | 8.81 | 9.85 |
| Cell density (104/ml) | 1.54 | 1.46 | 1.67 | 1.29 | 1.58 | 8.10 | 19.0 | 21.8 | 6.46 | 6.60 |
| DNA density (μg/ml) | 6.3 | 3.0 | 3.8 | 2.9 | 4.3 | 39.3 | 99.6 | 128.2 | 126.8 | 96.4 |
Days elapsed from start of field experiment.
Reported in more detail elsewhere (13).
BD, below detection limit.
ND, not determined.
Quantification of ammonia-oxidizing microorganisms.
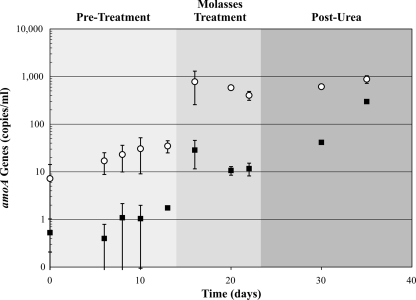
To corroborate the apparent stimulation of ammonia oxidation in the aquifer following urea addition, bacterial and archaeal amoA genes were enumerated via qPCR over the course of the experiment. Genes associated with both domains were detectable by qPCR at all points during the study, and the AOB and AOA amoA gene copies were each quantified on a per-milliliter basis (Fig. 1).
FIG. 1.
Abundance of betaproteobacterial and archaeal amoA genes at multiple time points over the course of this study. Samples were collected in the pretreatment (days 0, 6, 8, 10, and 13), molasses treatment (days 16 and 20), and post-urea (days 30 and 35) phases. qPCR analysis of the AOA (open circle) and AOB (solid square) amoA genes within DNA extracted per milliliter of filtered groundwater samples was carried out. Error bars (1 standard deviation of triplicate biological samples) are shown for amoA values; error bars fall within the symbols for some coordinates (e.g., in the post-urea phase).
Analysis of the water samples prior to molasses treatment suggested that the total microbial cell numbers were consistent (∼104 cells/ml) with those measured previously in the low-nutrient ESRPA environment (4, 29); following molasses addition, the total cell numbers in the water phase increased by over an order of magnitude (Table 1). During the pretreatment phase, the AOB and AOA amoA genes were determined to be <10 and <100 copies each per ml of groundwater, respectively, together constituting less than 1% of the total microbial cell numbers. However, AOA and AOB amoA gene copy numbers both increased by over an order of magnitude following molasses addition (Fig. 1), mirroring the observed increase in total cell numbers.
In the samples collected after urea addition, the total cell numbers were lower than during the molasses phase but were still approximately fourfold above the pretreatment levels. AOB amoA gene copy numbers increased further after urea addition. In contrast, AOA amoA gene copy numbers in the post-urea samples were statistically indistinguishable from their levels in the molasses treatment phase, although they remained higher than in the pretreatment phase. Overall, the data suggest that ammonia oxidizers came to comprise a greater percentage of the microbial population.
Bacterial and archaeal amoA-based community structure.
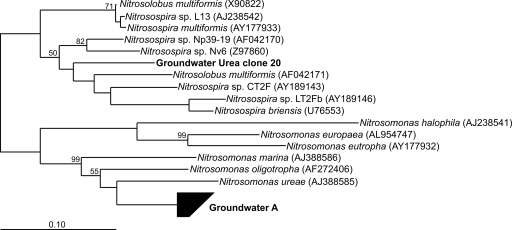
Bacterial and archaeal amoA clone libraries were constructed from samples from each of the monitoring periods (pretreatment, molasses treatment, and post-urea; see Materials and Methods). Only two bacterial amoA sequence types were obtained from the 175 clones screened in this study (Fig. 2). Initial AOB clone libraries constructed using amoA primers developed for this experiment revealed the presence of a single phylotype/OTU (5% nucleotide sequence identity cutoff) in samples from all of the incubation periods. Phylogenetic analysis indicated this genotype to be most closely related to Nitrosomonas ureae-like sequences previously obtained from groundwater (17). To further investigate the strikingly low level of AOB diversity in our samples, an additional clone library was constructed using a different primer set (34, 38). This library, consisting of sequences obtained from urea-treated water sampled on day 35, was dominated by the same Nitrosomonas lineage found in the original set of libraries (Fig. 2). However, a single Nitrosospira sequence (groundwater urea clone 20), similar to that of an environmental clone obtained from an unpublished investigation of soil (accession no. EU620222), was also obtained in this library.
FIG. 2.
Neighbor-joining Jukes-Cantor-corrected phylogenetic tree (based on a 468-bp alignment) of betaproteobacterial amoA sequences from this study, compared to sequences from cultivated members available in GenBank.
In sharp contrast to bacterial amoA, a far greater level of diversity was evident in archaeal amoA clone libraries. Rarefaction analyses comparing the richness within clone libraries during the discrete phases revealed a total of 20 unique OTUs across all of the libraries. In comparing the different treatment phases of the study by rarefaction analysis, the pretreatment community was slightly more diverse than the others, with 13 OTUs, compared to 10 OTUs each for the molasses treatment and post-urea phases (data not shown). Small differences were observed in the percentages of shared OTUs among the three treatment phases. In particular, abundance-based Sørensen-type similarities (based on a 5% OTU cutoff) among the three archaeal amoA clone libraries were as follows: pretreatment and molasses, 0.9793; molasses and urea, 0.9379; pretreatment and urea, 0.9019. Thus, despite fairly comparable richness, subtle compositional shifts were also detectable over time.
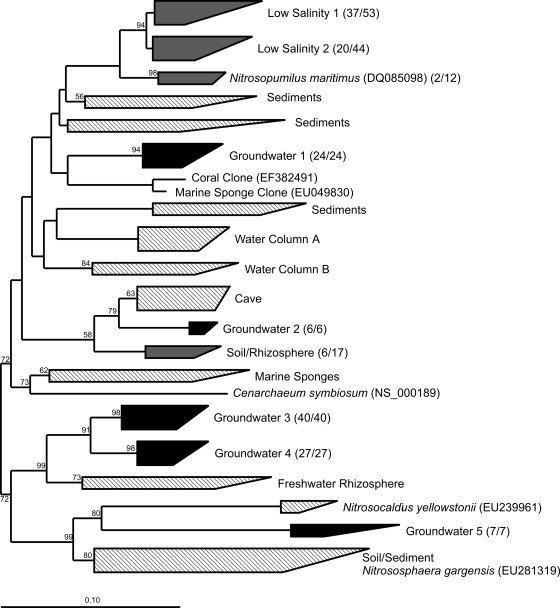
Overall, phylogenetic analysis of 425 archaeal amoA sequences (including 169 sequences obtained from this study) suggested that the AOA community native to the ESRPA groundwater environment was unique (Fig. 3); only a portion of the sequences obtained were notably (>85%) similar to other environmental sequences in GenBank. Nine of the 19 phylogenetic clusters shown in Fig. 3 contained sequences from this study, and interestingly, 5 of these clusters (representing 62% of our 169 total sequences) were made up exclusively of amoA sequences from the ESRPA. Low-salinity clusters 1 and 2, dominated by ESRPA sequences, were most closely related to those obtained from sediments in oligohaline regions of San Francisco Bay (27), freshwater environments (16), and groundwater (41). The other major clusters containing our sequences, designated groundwater clusters 3 and 4, contained 67 sequences from this study. These sequences showed little similarity to others in the GenBank database, and both groups were only distantly related to archaeal amoA genes from freshwater sediments.
FIG. 3.
Neighbor-joining Juke-Cantor-corrected phylogenetic tree for a 585-bp fragment of 425 crenarchaeal amoA genes from this study (169 from ESRPA groundwater) and GenBank. Fractions in parentheses indicate the number of ESRPA groundwater sequences out of the total number of sequences in each cluster. Clusters colored black are composed only of sequences from this study, gray clusters include other environmental sequences, as well as sequences from this study, and line-patterned clusters do not contain sequences from this study.
Interestingly, 21 archaeal amoA sequences in groundwater clusters 3 and 4 had a 3-bp deletion at position 96 of the nucleotide sequence, corresponding to a missing alanine in the deduced amino acid sequence. Sequences containing this deletion were obtained only from molasses treatment samples. Further, the seven amoA sequences in highly divergent groundwater cluster 5 (also from the molasses treatment sample) contained a 3-bp insertion at position 90. Predictive translation of these sequences produced a glycine insertion.
DISCUSSION
Groundwater chemistry.
The stimulation of urea-hydrolyzing microorganisms in order to promote calcite precipitation for contaminant immobilization results in the production of ammonium, an important component of the nitrogen cycle. By enzymatically oxidizing ammonia, AOB and AOA have the potential to cause changes in water chemistry (specifically, a decreased pH) that could be detrimental to remediation performance.
Although in this study the pH increased only very slightly following nutrient addition, DO values decreased three- to sixfold following urea addition. A possible reason for the lower DO concentrations is enhanced microbial metabolic activity due to alleviation of nitrogen limitation; the measured ratio of total organic carbon to total nitrogen in the molasses was 239:1, far greater than the C/N ratio of 8.6:1 recently reported for soil microbial biomass (3). Another potential cause of the observed decrease in DO was a change in formation permeability, due to mineral precipitation and/or biomass formation, that inhibited recharge of the treated zone by fresh oxygenated water; a dramatic decrease in well productivity consistent with reduced permeability was observed after urea addition (13). The increased ammonium concentrations observed following urea addition were likely due to hydrolysis of urea; indeed, a separate study conducted during this experiment demonstrated a marked increase in the abundance of bacterial urease genes (13). In an oligotrophic environment, such a spike in ammonium was likely sufficient to stimulate ammonia-oxidizing microbial communities. This is corroborated by the observed increase in nitrate concentrations following urea addition. The inability to detect an increase in nitrite concentrations during a period of increased ammonia oxidation is likely attributable to tightly coupled rates of ammonia and nitrite oxidation.
Ammonia oxidizer abundance.
The results of this study suggest that both ammonia-oxidizing bacterial and archaeal populations were stimulated by the addition of molasses to groundwater and that ammonia oxidizers constituted a more significant fraction of the total microbial community after urea addition than before the amendment. The second observation is consistent with the hypothesis that the ammonia liberated into the environment via ureolysis provided sufficient energy (i.e., an electron donor) to sustain a larger population of ammonia-oxidizing microorganisms. The increased ammonia-oxidizing population was, in turn, consistent with enhanced overall nitrification, as evidenced by the apparent increase in nitrate concentrations. On the other hand, the observed increase in AOB and AOA amoA gene copy numbers after molasses addition alone was unexpected, as increased carbon availability would have been expected to result in increased nitrogen demand by the overall microbial community and consequently less nitrogen availability for ammonia oxidizers. While molasses-stimulated growth of AOA and AOB would also be consistent with mixotrophy (i.e., organic carbon acquisition by chemoautrophs), further studies are clearly needed to determine the underlying reason(s) for this unanticipated finding.
Prior to urea addition, the AOA gene copy numbers were approximately 15- to 55-fold greater than the AOB gene copy numbers in every sample tested (Fig. 1). These data are consistent with many recent studies which have shown that the AOA amoA gene copy numbers typically far outnumber those of the AOB in soil, oceans, and freshwater by 1 to 4 orders of magnitude, often with the AOB hardly above the detection limit (16, 23, 26, 44). However, it has been reported that in some localized environments, AOB may play a more significant role. Santoro et al. showed that the AOB were 30 times more abundant than the AOA in the more saline, oxygenated portions of a subterranean estuary (coastal aquifer) but up to 10 times less abundant than AOA in the lower-oxygen freshwater portions; interestingly, the AOA numbers remained fairly constant across the transect (35). Likewise, in the San Francisco Bay estuary, Mosier and Francis reported that the ammonia oxidizers exhibit a distinct spatial structure; AOB are more abundant than AOA throughout most of the estuary, especially where salinity is high (22 to 31 practical salinity units [PSU]) and C/N ratios are low, while AOA predominate at sites with low salinity (0.2 to 9 PSU) and high C/N ratios (27). Thus, the numerical dominance of AOA over AOB in the low-salinity groundwater of the ESRPA is consistent with these two estuarine studies.
In the post-urea phase of this study, while AOA amoA abundance did not appear to show a clear response to urea addition, the AOB responded positively (with amoA copy numbers increasing 25-fold between days 22 and 35). The underlying reasons for the shift toward higher AOB gene copy numbers are not entirely clear. However, the greater apparent response of the AOB population numbers than the AOA population numbers may be consistent with recent physiological evidence that, in contrast to known AOB, “Candidatus Nitrosopumilus”-like AOA are specifically adapted to life under extreme nutrient limitation, sustaining high specific oxidation rates at nanomolar-level ammonium concentrations (25). It is also possible that, like many other described AOB (19), the ESRPA groundwater AOB have the capacity to directly utilize urea via ureases; interestingly, there is no evidence for urease genes in the completed genome of “Candidatus Nitrosopumilus maritimus” SCM1.
Community composition.
The AOB and AOA populations within the ESRPA also differed significantly in terms of overall amoA gene diversity, with AOA exhibiting far greater diversity than AOB. The dearth of AOB diversity was similar to that observed previously in sandy sediments spanning the freshwater-seawater interface of a coastal aquifer, where only two amoA phylotypes were identified from 205 clones sequenced at a 5% OTU cutoff (35), whereas AOA amoA diversity was high across the transect.
Archaeal amoA sequences from this study were related to those from a variety of different environments, including freshwater and estuarine sediments (9, 16, 27), corals and marine sponges (2), and caves (37), but the majority fell into unique ESRPA groundwater-specific clusters. In addition, a number of the archaeal amoA sequences from the molasses treatment samples had a 3-bp deletion or insertion at positions 96 and 90, respectively, of the nucleotide sequence. Other reported amoA sequences containing the 3-bp insertion, including that of the thermophilic AOA isolate “Candidatus Nitrosocaldus yellowstonii” and other hot spring amoA clones (5), showed little overall similarity to our ESRPA sequences. Whether these amino acid deletions and insertions impact the function of archaeal ammonia monooxygenase is unknown, but the fact that these particular sequences were obtained only from a sample collected after exposure to molasses is intriguing.
Conclusions.
Taken together, the results of our study demonstrate the utility of examining functional gene abundance and diversity, together with geochemical data, for better understanding the ecological impacts of biogeochemical manipulations of the subsurface for contaminant remediation. Our data showed that before and after nutrient addition, the AOA were more abundant than the AOB in the groundwater and that the AOA population exhibited far greater amoA gene diversity than did the AOB population. We observed that both the AOB and AOA populations were stimulated by the addition of molasses to groundwater and that the proportion of ammonia oxidizers in the total microbial community increased after urea amendment (and ureolysis); the AOB, in particular, appeared to respond more positively to urea than did the AOA. However, no adverse impacts on groundwater quality were observed, suggesting that, in this system at least, these dynamic changes in the ammonia-oxidizing microbial communities should not hinder the success of a remediation scheme based on ureolytically driven calcite precipitation. Future work will include the application of amoA gene assays to other environmental systems where this remediation approach is tested, along with nitrification activity measurements, in order to gauge the extent to which these findings are representative.
Acknowledgments
We express appreciation to Molly Leecaster at the University of Utah for assistance with statistical analyses. We thank INL students and technical staff Stephanie Freeman, Tina Tyler-Gresham, Lynn Wendt, Mark Delwiche, Alison Rope, Joanna Taylor, Sindy Byington, and Michele Bernal for assistance with sample collections, DNA extractions, standards, and assay development. N. europaea cells were generously provided by Daniel J. Arp from Oregon State University.
This work was supported by the U.S. Department of Energy, Office of Science under DOE Idaho Operations Office contract DE-AC07-05ID14517.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Beman, J. M., B. N. Popp, and C. A. Francis. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine crenarchaeota in the Gulf of California. ISME J. 2:429-441. [DOI] [PubMed] [Google Scholar]
- 2.Beman, J. M., K. J. Roberts, L. Wegley, F. Rohwer, and C. A. Francis. 2007. Distribution and diversity of archaeal ammonia monooxygenase genes associated with corals. Appl. Environ. Microbiol. 73:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleveland, C. C., and D. Liptzin. 2007. C:N:P stoichiometry in soil: is there a “Redfield ratio” for the microbial biomass? Biogeochemistry 85:235-252. [Google Scholar]
- 4.Colwell, F. S., and R. M. Lehman. 1997. Carbon source utilization profiles for microbial communities from hydrologically distinct zones in a basalt aquifer. Microb. Ecol. 33:240-251. [DOI] [PubMed] [Google Scholar]
- 5.de la Torre, J. R., C. B. Walker, A. E. Ingalls, M. Könneke, and D. A. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 6.Di, H. J., K. C. Cameron, J. P. Shen, C. S. Winefield, M. O'Callaghan, S. Bowatte, and J. Z. He. 2009. Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nat. Geosci. 2:621-624. [Google Scholar]
- 7.Erwin, D. P., I. K. Erickson, M. E. Delwiche, F. S. Colwell, J. L. Strap, and R. L. Crawford. 2005. Diversity of oxygenase genes from methane- and ammonia-oxidizing bacteria in the Eastern Snake River Plain aquifer. Appl. Environ. Microbiol. 71:2016-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 9.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman, S., D. Reed, and Y. Fujita. 2006. Testing the specificity of primers to environmental ammonia monooxygenase (amoA) genes in groundwater treated with urea to promote calcite precipitation. U.S. DOE J. Undergrad. Res. 6:114-118. [Google Scholar]
- 11.Fujita, Y., F. G. Ferris, D. L. Lawson, and F. S. Colwell. 2000. Calcium carbonate precipitation by ureolytic subsurface bacteria. Geomicrobiol. J. 17:305-318. [Google Scholar]
- 12.Fujita, Y., G. D. Redden, J. C. Ingram, M. M. Cortez, F. G. Ferris, and R. W. Smith. 2004. Strontium incorporation into calcite generated by bacterial ureolysis. Geochim. Cosmochim. Acta 68:3261-3270. [Google Scholar]
- 13.Fujita, Y., J. L. Taylor, T. L. T. Gresham, M. E. Delwiche, F. S. Colwell, T. L. McLing, L. M. Petzke, and R. W. Smith. 2008. Stimulation of microbial urea hydrolysis in groundwater to enhance calcite precipitation. Environ. Sci. Technol. 42:3025-3032. [DOI] [PubMed] [Google Scholar]
- 14.Hazen, T. C. 1997. Bioremediation, p. 247-266. In P. S. Amy and D. L. Haldeman (ed.), The microbiology of the terrestrial deep subsurface. CRC Press LLC, Boca Raton, FL.
- 15.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal-RNA gene-sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, M., A. M. Saunders, and A. Schramm. 2008. Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl. Environ. Microbiol. 74:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova, I. A., J. R. Stephen, Y. J. Chang, J. Bruggemann, P. E. Long, J. P. McKinley, G. A. Kowalchuk, D. C. White, and S. J. Macnaughton. 2000. A survey of 16S rRNA and amoA genes related to autotrophic ammonia-oxidizing bacteria of the beta-subdivision of the class proteobacteria in contaminated groundwater. Can. J. Microbiol. 46:1012-1020. [DOI] [PubMed] [Google Scholar]
- 18.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 19.Koper, T. E., A. F. El-Sheikh, J. M. Norton, and M. G. Klotz. 2004. Urease-encoding genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 70:2342-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 21.Lam, P., M. M. Jensen, G. Lavik, D. F. McGinnis, B. Muller, C. J. Schubert, R. Amann, B. Thamdrup, and M. M. M. Kuypers. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman, R. M., S. P. O'Connell, A. Banta, J. K. Fredrickson, A. L. Reysenbach, T. L. Kieft, and F. S. Colwell. 2004. Microbiological comparison of core and groundwater samples collected from a fractured basalt aquifer with that of dialysis chambers incubated in situ. Geomicrobiol. J. 21:169-182. [Google Scholar]
- 23.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens-Habbena, W., P. M. Berube, H. Urakawa, J. R. de la Torre, and D. A. Stahl. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976-979. [DOI] [PubMed] [Google Scholar]
- 26.Mincer, T. J., M. J. Church, L. T. Taylor, C. Preston, D. M. Kar, and E. F. DeLong. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162-1175. [DOI] [PubMed] [Google Scholar]
- 27.Mosier, A. C., and C. A. Francis. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002-3016. [DOI] [PubMed] [Google Scholar]
- 28.Newby, D. T., D. W. Reed, L. M. Petzke, A. L. Igoe, M. E. Delwiche, F. F. Roberto, J. P. McKinley, M. J. Whiticar, and F. S. Colwell. 2004. Diversity of methanotroph communities in a basalt aquifer. FEMS Microbiol. Ecol. 48:333-344. [DOI] [PubMed] [Google Scholar]
- 29.O'Connell, S. P., R. M. Lehman, O. Snoeyenbos-West, V. D. Winston, D. E. Cummings, M. E. Watwood, and F. S. Colwell. 2003. Detection of euryarchaeota and crenarchaeota in an oxic basalt aquifer. FEMS Microbiol. Ecol. 44:165-173. [DOI] [PubMed] [Google Scholar]
- 30.Okano, Y., K. R. Hristova, C. M. Leutenegger, L. E. Jackson, R. F. Denison, B. Gebreyesus, D. Lebauer, and K. M. Scow. 2004. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosser, J. I., and G. W. Nicol. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931-2941. [DOI] [PubMed] [Google Scholar]
- 32.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rope, A. M., D. W. Reed, and Y. Fujita. Development of an improved standard for quantification of the amoA gene in environmental samples. U.S. DOE J. Undergrad. Res., in press.
- 34.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoro, A. E., C. A. Francis, N. R. de Sieyes, and A. B. Boehm. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068-1079. [DOI] [PubMed] [Google Scholar]
- 36.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spear, J. R., H. A. Barton, C. E. Robertson, C. A. Francis, and N. R. Pace. 2007. Microbial community biofabrics in a geothermal mine adit. Appl. Environ. Microbiol. 73:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephen, J. R., Y. J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, J. L. 2003. Stimulation of microbial urea hydrolysis in groundwater for remediation of metal contaminants. M.S. thesis. Idaho State University, Pocatello, ID.
- 40.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H. P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 41.van der Wielen, P. W. J. J., S. Voost, and D. van der Kooij. 2009. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 75:4687-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren, L. A., P. A. Maurice, N. Parmar, and F. G. Ferris. 2001. Microbially mediated calcium carbonate precipitation: implications for interpreting calcite precipitation and for solid-phase capture of inorganic contaminants. Geomicrobiol. J. 18:93-115. [Google Scholar]
- 43.Wells, G. F., H. D. Park, C. H. Yeung, B. Eggleston, C. A. Francis, and C. S. Criddle. 2009. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ. Microbiol. 11:2310-2328. [DOI] [PubMed] [Google Scholar]
- 44.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. S. Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, L. M., M. Wang, J. I. Prosser, Y. M. Zheng, and J. Z. He. 2009. Altitude ammonia-oxidizing bacteria and archaea in soils of Mount Everest. FEMS Microbiol. Ecol. 70:52-61. [DOI] [PubMed] [Google Scholar]