Abstract
The mechanisms involved in the ability of Bacillus cereus to multiply at low temperatures were investigated. It was assumed that many genes involved in cold acclimation would be upregulated at low temperatures. Recombinase-based in vivo expression technology (IVET) was adapted to the detection of the transient activation of B. cereus promoters during growth at 10°C. Four independent screenings of a promoter library from type strain ATCC 14579 were performed, and 17 clones were isolated. They corresponded to 17 promoter regions that displayed reproducibly elevated expression at 10°C relative to expression at 30°C. This analysis revealed several genes that may be important for B. cereus to grow successfully under the restrictive conditions of cold habitats. Among them, a locus corresponding to open reading frames BC5402 to BC5398, harboring a lipase-encoding gene and a putative transcriptional regulator, was identified three times. While a mutation in the putative regulator-encoding gene did not cause any particular phenotype, a mutant deficient in the lipase-encoding gene showed reduced growth abilities at low temperatures compared with the parental strain. The mutant did not change its fatty acid profiles in the same way as the wild type when grown at 12°C instead of 37°C. This study demonstrates the feasibility of a promoter trap strategy for identifying cold-induced genes. It outlines a first picture of the different processes involved in B. cereus cold acclimation.
The food-borne disease agent Bacillus cereus is an endospore-forming bacterium belonging to the B. cereus group (B. cereus sensu lato). B. cereus sensu lato has recently been divided into seven major phylogenetic groups (I to VII) with clear-cut differences in growth temperature ranges, suggesting that the genetic structure corresponds to “thermotypes” and showing the emergence of multiple psychrotrophic groups within B. cereus sensu lato (26). Temperature adaptation has thus presumably played a major role in B. cereus evolution. B. cereus is also a human pathogen, causing local and systemic infections. Most outbreaks of food-borne poisoning have been caused by mesophilic strains (26) that can grow at temperatures as low as 10°C. This characteristic enables initially relatively low levels of B. cereus in foods to increase greatly under commonly reported suboptimal refrigeration conditions (20). Understanding the ability of B. cereus to grow at low temperatures will help to control multiplication in refrigerated food and prevent outbreaks of food-borne poisoning.
At low temperatures, bacteria undergo various modifications in cellular physiology, with effects such as decreased membrane fluidity and inefficient folding of proteins and secondary structures of RNA and DNA (43). Bacterial responses can be divided into low-temperature responses (or acclimation, also called low-temperature adaptation) and cold shock responses (43). Both types of responses include a vast array of structural and physiological adjustments to cope with the reduction in biochemical reaction rates induced by low temperatures. Previous studies of cold responses in bacteria focused mainly on cold shock responses after a temperature drop (43, 51) rather than on long-term adaptive responses. Cold shock responses of many bacteria occur as a change in the fatty acid (FA) profile of the bacterial cell membrane to maintain optimal fluidity or in the expression of small RNA-binding cold shock proteins (Csps) that mediate transcription elongation and message stability, as investigated for instance in Listeria monocytogenes (12). Studies of B. subtilis, reviewed by Weber and Marahiel (51), account for most of the work on cold adaptation in the Bacillus genus. Because of the psychrotrophy of many strains, B. cereus sensu lato is a good model for research on the molecular mechanisms of low-temperature adaptation in Bacillus species and still offers wide scope for investigation. Previous work is scant and piecemeal, dealing for instance with the analysis of cold shock proteins (23) or changes in membrane FA profiles at a suboptimal temperature (21 or 15°C) (29, 33). In fact, the establishment of B. cereus in the food environment under refrigeration probably requires the coordinated activity of many genes whose identities and modes of action are still largely unknown.
Assuming that many, if not most, of the genes involved in cold acclimation are induced or upregulated at low temperatures, we hypothesized that ecologically significant genes allowing B. cereus to survive at or adapt to low temperatures could be identified using a promoter trap strategy to capture promoters specifically activated during growth at low temperatures. We therefore adapted a recombinase-based in vivo expression technology (IVET) to study the genes activated in B. cereus ATCC 14579 during growth at low temperatures. The advantages of the IVET approach over other genetic methods (screening of a mutant library, for instance) reside in its sensitivity, since it enables the detection of genes that are only slightly or transiently induced, and its allowance of gene selection independent of whether the loss of the gene sequence would be lethal. Thus, IVET helps to recover both essential and nonessential genes that contribute to the ecological success of B. cereus under the conditions tested. Another advantage is that this approach allows the detection of promoters expressed, even transiently, at any growth stage. Therefore, it is well adapted to investigate acclimation, which may involve phenomena occurring from lag phase to the end of growth.
An IVET library constructed in the B. cereus type strain ATCC 14579 has already permitted the identification of genes specifically expressed during virulence in insect larvae (22). We used this library in the present study and herein describe the application of IVET to detect promoters specifically expressed, even transiently, during the growth of B. cereus at 10°C with the aim of gaining a better understanding of the mechanisms involved in its adaptive response to low temperatures.
MATERIALS AND METHODS
Strains and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. B. cereus cells were grown aerobically in Luria broth (LB) or brain heart infusion (BHI) with vigorous shaking (200 rpm) at 30 or 10°C. Escherichia coli cells were routinely grown in LB medium with shaking at 37°C. When required, erythromycin (Em) at 10 μg ml−1, kanamycin (Km) at 100 μg ml−1, and spectinomycin (Sp) at 275 μg ml−1 for B. cereus and ampicillin at 100 μg ml−1 for E. coli were used for bacterial selection.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| B. cereus ATCC 14579 | WT | Laboratory collection |
| B. cereus R2SK | Chromosomal reporter strain harboring R2SK cassette | 22 |
| B. cereus ΔBC5402 | ATCC 14579 BC5402::Kmr | This work |
| B. cereus ΔBC5401 | ATCC 14579 BC5401::Kmr | This work |
| E. coli TG1 | Δ(lac-proAB) supE thi hsd-5 (F′ traD36 proA+proB+lacIqZΔM15) | Laboratory collection |
| E. coli ET12567 | F−dam-13::Tn9 dcm-6 hsdM hsdR recF143 zjj-202::Tn10 galK2 galT22 ara-14 pacY1 xyl-5 leuB6 thi-1 | Laboratory collection |
| Plasmids | ||
| pHT304-I | Apr Emr shuttle vector used for B. cereus genomic library | 22 |
| pRN5101 | Apr Emr shuttle vector | 49 |
| pRN5101ΔBC5401 | Recombinant pRN5101 plasmid harboring BC5401::Kmr | This work |
| pRN5101ΔBC5402 | Recombinant pRN5101 plasmid harboring BC5402::Kmr | This work |
| pHT304 | Apr Emr cloning vector | 4 |
| pDG783 | Apr Kmr cloning vector | 25 |
| pHT304-Km | pHT304 carrying the aphA3 Kmr gene with its own promoter cloned between the XbaI and PstI sites of pHT304 | This work |
Ap, ampicillin.
E. coli ET12567, with mutations in dam and dcm, was used to generate unmethylated plasmid DNA for the electrotransformation of B. cereus. B. cereus and E. coli strains were transformed by electroporation as described previously (19, 36).
Growth experiments were performed with an automated turbidometer, the Bioscreen C microbiology reader (Labsystems, Uxbridge, United Kingdom), in 100-well sterile microplates. Volumes of 0.1 ml of overnight cultures at 30°C were inoculated into 10 ml of fresh LB and incubated at 30°C with shaking until an optical density at 600 nm (OD600) of 0.8 was obtained. These cultures were used to inoculate 1 ml of fresh LB to reach a concentration of 6 × 106 to 1 × 107 CFU per ml (depending on the experiments). Three replicate wells of the microplate were filled with these dilutions of inoculated medium to a final volume of 300 μl per well. A negative control consisting of uninoculated LB was prepared. The cultures were incubated with vigorous constant shaking, and the OD600 was measured at 15-min intervals at 30 and 45°C or 1-h intervals at 12°C over an incubation period of 48 h or 10 days, respectively. The temperature of 12°C was used here instead of 10°C to avoid the flocculation of the cells, which sometimes occurs under these culture conditions. At least three independent experiments for each growth condition were performed.
DNA manipulation.
Plasmid DNA was extracted from B. cereus and E. coli by a standard alkaline lysis procedure using the Wizard SV miniprep purification system (Promega, Charbonnières, France), with additional incubation with lysozyme for the lysis of B. cereus cells as described previously (9). Chromosomal DNA was extracted from B. cereus cells harvested in mid-log phase as described previously (7). Restriction enzymes and T4 DNA ligase were used as recommended by the manufacturer (Promega). Oligonucleotide primers (Table 2) were synthesized by Eurogentec (Seraing, Belgium). PCR was performed in a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer, Coutaboeuf, France) using Expand high-fidelity DNA polymerase (Roche Applied Science, Meylan, France). Amplified DNA fragments were purified using a PCR purification kit (Roche) and separated on 0.7% agarose gels after digestion. Digested DNA fragments were extracted from agarose gels with a centrifugal filter device (Montage DNA gel extraction kit; Millipore, Molsheim, France). All constructions were confirmed by DNA sequencing by GenomeExpress, Grenoble, France.
TABLE 2.
Primers used in this study
| Primer name | 5′-3′ sequencea | Restriction site |
|---|---|---|
| 5Up-01_Bam | CGGGATCCATAGCGCTATTATCGAGGGG | BamHI |
| 3Up-01_Pst | AAAACTGCAGCGCCCAACATAGCCAATTCC | PstI |
| 5Dn-01_Xba | GCTCTAGAACAAGGAGGGGTAAAATGACG | XbaI |
| 3Dn-01_Eag | CGCGGCCGGCCTTCTACACTCACTTCCC | EagI |
| 5Up-02_Bam | CGGGATCCGCGAACTATCGAATTGAAGGG | BamHI |
| 3Up-02_Pst | AAACTGCAGGCTGTCTCTTTTGATATCCCC | PstI |
| 5Dn-02_Xba | GCTCTAGAGCGGGGATGGTTACAGAAGC | XbaI |
| 3Dn-02_Eag | CGCGGCCGCTCTGTGTCAGGACGTACG | EagI |
| Km5in | TCTGTCTAGACATTTGAGGTGATAGG | XbaI |
| Km3in | GCTACTGCAGATCGATACAAATTCCTCGTAGGCG | PstI |
| Km5out | CGGTATAATCTTACCTATCACC | |
| Km3out | TACTCTGATGTTTTATATCTTTTCTAA | |
| IVET-I1 | CCCTGAACAGTGTTCTCGG | |
| IVET-I2 | GGCGATTAAGTTGGGTAAC | |
| RT-5402-F | TTATTGCCACACGTAAATGA | |
| RT-5402-R | CCCACGTGAAAATACTTTGT | |
| RT-5402-01-F | ACAAGGAGTTCGTATTCCAG | |
| RT-5402-01-R | GCCAATTCCTTCTTTATCAC | |
| RT-5401-F | GATGTAGTGGCAAAAGAGAA | |
| RT-5401-R | CAGGACGATACGTCTCTAAA | |
| RT-5401-00-F | AGCGCTAAACGATAAAAATG | |
| RT-5401-00-R | TCTGGGTTTTCAACGATACT | |
| RT-5400-F | CGAATGTTAGTCGGATTGAT | |
| RT-5400-R | TAATCCCTGCTGGATCTAAA | |
| RT-5400-5399-F | GAAGGCGATCGATTACTATG | |
| RT-5400-5399-R | CGGTTCTGAGAATCAACAAT | |
| RT-5399-98-F | AGGTTTATCGATGGGATTTT | |
| RT-5399-98-R | CAGAAAAGCTTAAACCATGC | |
| RT-5398-F | TTGTCATTTTAGGGCAAGAT | |
| RT-5398-R | CCCCCACAATATGAAAATAA | |
| LC-BC5402-F | TTATTGCCACACGTAAATGA | |
| LC-BC5402-R | CCCACGTGAAAATACTTTGT | |
| LC-BC5401-F | CATATGGCTTTGTTTCAGG | |
| LC-BC5401-R | TTTGCACCACTAACTGCTAA | |
| LC-16S-F | GGTAGTCCACGCCGTAAACG | |
| LC-16S-R | GACAACCATGCACCACCTG |
Restriction enzyme sites are underlined.
Screening of the IVET library.
We have adapted recombinase-based IVET to study the genes activated in B. cereus ATCC 14579 during its growth at low temperatures. In this IVET system, constructed as described previously (22), B. cereus Sau3A chromosomal fragments were fused to a promoterless resolvase tnpI gene from Tn4430 harbored by the promoter trap-IVET vector pHT304-I. The reporter strain, named B. cereus R2SK, carries a Kmr-res-Spr-res-pKm cassette (the R2SK construct) integrated into its chromosome at the tetB locus. This cassette carries two selectable resistance genes: a nonfunctional aphA3 kanamycin resistance (Kmr) gene and a spectinomycin resistance (Spr) gene flanked by two internal resolvase recognition sequences (res). The coding sequence of the aphA3 gene is thus separated from its promoter (pKm) by the res-Spr-res DNA fragment, at which TnpI can catalyze recombination (39). After the introduction of the promoter trap vector pHT304-I constructs carrying the genomic library of chromosomal fragments from B. cereus fused to tnpI into B. cereus R2SK, the activation of a promoter cloned upstream of the tnpI gene would result in TnpI production and excision of the spectinomycin resistance marker from the chromosome. This event also results in the restoration of a functional aphA3 gene and marks the bacterium by endowing it with an inheritable Sp-sensitive and Km-resistant phenotype. The IVET library was screened as described below. Most of the promoters expressed at the reference incubation temperature of 30°C were removed using the following procedure: 1-ml aliquots of the frozen library stock (22) were grown in 50 ml of BHI supplemented with Sp in 250-ml flasks at 30°C until an OD600 of 1.0 was obtained. One percent of the culture was used for inoculation of fresh BHI-Sp medium to increase the number of generations during the exponential growth phase. This step was repeated four times to maximize the elimination of the majority of cells for which a resolution event occurred at 30°C (i.e., for which the phenotype switched from Km sensitive and Sp resistant to Km resistant and Sp sensitive). Even after these removal steps, in a total population of 7 × 107 CFU/ml, 10 to 50 CFU/ml for which a resolution event had occurred (resolution-positive CFU) were usually found, corresponding to the background level for this technique. The resulting depleted IVET library was frozen at −80°C as 1-ml aliquots in glycerol to be used as frozen inocula.
Four independent screenings at 10°C were performed as follows: 1 ml of frozen inoculum was used to inoculate 50 ml of BHI in a 250-ml flask, and the flask was incubated at 10°C with shaking. As a control, an identically inoculated flask was grown at 30°C. This step allowed the estimation of the number of residual promoters activated under usual culture conditions that escaped the prescreening depletion step. During these control experiments, the background level of resolution-positive CFU at 30°C remained at 10 to 50 CFU/ml. Cultures were then diluted and spread either onto LB agar-Em to determine the total harvested bacterial population or onto LB agar-Km to isolate Km-resistant clones. Depending on the experiments, between 5 × 102 and 1 × 104 CFU/ml for which a resolution event had occurred were isolated from a total population ranging from 5 × 107 to 9 × 107 CFU/ml. To confirm that the Km-resistant clones arose from the resolution of the R2SK chromosomal cassette, selected Km-resistant clones were checked for their spectinomycin sensitivity. At this stage, less than 10% of the clones were discarded.
To remove potential replicates of the same clone, the following procedure was used: amplification of the DNA fragments cloned into pHT304-I from selected Km-resistant, Sp-sensitive colonies was performed by PCR using the IVET-I1 and IVET-I2 primers (Table 2). After electrophoresis, the clones displaying exactly the same migration profile on both 0.7% and 1% agarose gels were considered to be exactly the same size and thus to be potential copies of the same clone. Only one copy was kept, and the others were discarded. In addition, the clones for which no PCR amplification was obtained were removed. Altogether, about 25% of the clones were selected for further analysis during this step. The selected PCR products were then purified and sequenced.
After DNA sequencing, two of the remaining clones were removed: one contained multiple DNA fragments originating from different chromosomal regions, and the other contained an intragenic region (892 bp in the middle of a 4.3-kb open reading frame [ORF]) located more than 2.5 kb from the start codon of the next ORF. None of the remaining clones contained an intergenic region in the wrong orientation. Three identified fragments from clones isolated in three independent screenings had exactly the same sequence: only one clone was kept for further analysis. Twenty clones containing an intergenic region in the correct orientation and, thus, likely to contain a promoter region were finally selected and named cold-induced promoter (CIP) clones. The pHT304-I derivative (i.e., the pHT304-I plasmid containing a DNA fragment harboring a promoter) was extracted from each of these 20 CIP clones (Km resistant and Sp sensitive) and reintroduced into the original B. cereus R2SK strain (Km sensitive and Sp resistant). Transformants were then grown at 30 and 10°C in parallel to determine the resolution frequencies at these two temperatures. Resolution frequencies were determined as the ratio of the number of Km-resistant cells to the total number of cells (i.e., Em-resistant cells). Populations of Km-resistant cells grown at 30°C that were below the detection threshold (10 CFU/ml) were arbitrarily fixed at 9 CFU/ml to have an excessive estimate of the resolution frequency. Experiments were repeated twice, and the mean values are presented. Clones (n = 3) for which the resolution frequencies displayed less than a 1-log difference between 30 and 10°C in at least one of the two experiments were discarded.
In silico analysis.
Sequences were analyzed using the BLAST server (NCBI, NIH) (3). Protein domains were identified using SMART software (37, 42). The MEME program was used to align promoters of all the CIP regions identified (6).
RT-PCR experiments.
Total RNA was extracted from B. cereus ATCC 14579 (wild-type [WT]) cells grown at 10°C in LB at mid-exponential phase (OD600 = 0.7) by using TRI Reagent RNA extraction solution as recommended by the manufacturer (Ambion, Huntingdon, United Kingdom). cDNA synthesis from 0.5 μg of total RNA was performed using avian myeloblastosis virus reverse transcriptase polymerase from a Titan one-tube reverse transcription-PCR (RT-PCR) kit (Roche). Specific amplifications were performed with primer couples listed in Table 2, and the positions of amplified regions are shown in Fig. 1A. The RT prefix designates these primers. For instance, RT-5402-F and RT-5402-R were used to detect the mRNA transcript of the BC5402 gene; RT-5402-01-F and RT-5402-01-R were used to detect the mRNA corresponding to a region overlapping BC5402 and BC5401. The reverse transcription step was followed by 30 cycles of PCR amplification with Expand high-fidelity polymerase according to the instructions of the manufacturer (Roche Diagnostics).
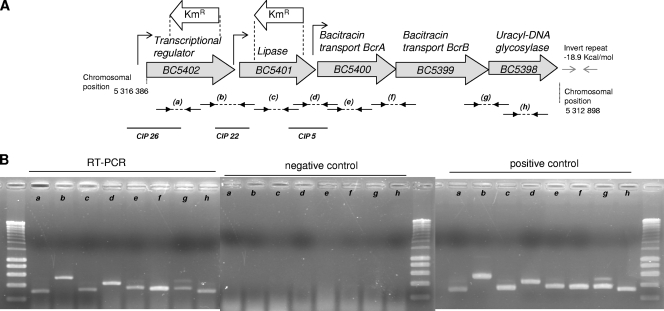
FIG. 1.
(A) Map of the B. cereus ATCC 14579 BC5398-BC5402 chromosomal region. Gray arrows represent ORFs. The positions of the kanamycin resistance gene integrated into the chromosomes of the mutants to disrupt BC5402 or BC5401 are indicated. Small arrows represent promoters. Functions or putative functions of gene products are indicated above the ORFs. Dashed lines with small black arrows annotated by letters in parentheses correspond to RT-PCR amplicons (see the text and Fig. 1B). The inverted repeat corresponding to a predicted stem-loop at the end of BC5398 is represented by two facing gray arrows. The solid black lines correspond to the IVET-identified fragments containing promoters specifically expressed at 10°C (the CIP clone numbers are indicated). (B) RT-PCR detection of BC5402 and downstream ORFs in B. cereus strain ATCC 14579. RT-PCR was performed with 500 ng of RNA. As a negative control, RT-PCR was performed with 500 ng of RNA and a heat-inactivated reverse transcriptase. As a positive control, PCR was performed with 100 ng of genomic DNA. Letters refer to the positions of the RT-PCR products relative to the locus, as represented in Fig. 1A. Experiments were performed with the Titan one-tube RT-PCR system (Roche). RNAs were extracted from strains grown at 10°C in LB and harvested in the exponential phase (OD600 = 0.7).
Relative quantification of gene expression by real-time PCR.
Real-time RT-PCR was performed with a LightCycler instrument (Roche) as described previously (9). Briefly, the QuantiFast SYBR green RT-PCR kit was used according to the instructions of the manufacturer (Qiagen) with 10 ng of total RNA as a template. RNAs were extracted from cells grown at 10 or 37°C and harvested during the middle of exponential phase, at the end of exponential phase, or at the beginning of stationary phase (at OD600s of 0.4, 1.0, and 2.0, respectively). Two independent cultures for each growth condition were performed. For each RNA sample, at least two independent measurements were performed. Altogether, presented results correspond to at least four measurements. The mRNA level changes for each gene were normalized to the RNA level for the ssu gene, encoding 16S RNA, and quantified by the 2−ΔΔCT method as described previously (38). The coefficient of variation of the ΔCT values (where ΔCT represents the differences in threshold cycle between the target and control genes) was <10%. Oligonucleotides listed in Table 2 with the LC prefix were used for real-time PCR.
Null mutant construction.
The BC5401 gene, encoding a putative lipase, and the BC5402 gene, encoding a putative transcriptional regulator, were interrupted in B. cereus ATCC 14579 by allelic exchange with a cassette conferring kanamycin resistance (Kmr), as described previously (5). Briefly, DNA fragments of BC5401 upstream and downstream regions were PCR amplified using the primer couples 5Up-01_Bam/3Up-01_Pst and 5Dn-01_Xba/3Dn-01_Eag, respectively (Table 2). Similarly, the upstream and downstream regions of BC5402 were PCR amplified with the primer couples 5Up-02_Bam/3Up-02_Pst and 5Dn-02_Xba/3Dn-02_Eag, respectively (Table 2). PCR products were digested with BamHI/PstlI and XbaI/EagI using the primer-incorporated restriction sites (Table 2). In parallel, the Kmr cassette (a 1.5-kb fragment corresponding to the aphA3 kanamycin resistance gene with its own promoter) was digested from pHT304-Km (Table 1) with PstI/XbaI. pHT304-Km was obtained by cloning a 1.5-kb fragment carrying the aphA3 gene (Kmr) from the pDG783 plasmid (25). The three digested DNA fragments were purified, ligated into EagI/BamHI-digested pRN5101, and introduced by electroporation into E. coli ET12567. Unmethylated plasmids from E. coli ET12567 were then prepared, and B. cereus ATCC 14579 was transformed with the resulting recombinant plasmids pRN5101ΔBC5401 and pRN5101ΔBC5402. Transformants were then subjected to allelic exchange as described previously (5). Colonies that were resistant to Km and sensitive to Em arose through a double-crossover event in which the chromosomal WT copies of the BC5401 or BC5402 gene were deleted and replaced by the Kmr cassette. The chromosomal allelic exchange in the mutants was checked by PCR using the appropriate primer couples (Km5out/5Up-01_Bam and Km3out/3Dn-01_Eag or Km5out/5Up-02_Bam and Km3out/3Dn-02_Eag). PCR products were sequenced for confirmation.
Extraction and quantification of FA compounds by GC-MS.
FA profiles were determined by using approximately 40 mg (fresh weight) of WT or ΔBC5401 cells grown at 37°C (overnight) or at 12°C (for 21 days) on LB agar. FA methyl esters (FAMEs) were produced from total lipids by saponification (using NaOH in methanol at 100°C for 30 min) coupled with esterification (using HCl in methanol at 80°C for 10 min) as described previously (39, 40). Extraction of the FAMEs by CH2Cl2 was followed by a washing step with a 0.1 M sodium hydrogen carbonate solution. Samples were then injected into a gas chromatography-mass spectrometry (GC-MS) instrument (Shimadzu QP2010) equipped with an UBWAX column (length, 30 m; diameter, 0.25 mm; film thickness, 0.5 μm). The injection port temperature (in splitless mode) was set at 250°C. The carrier gas was helium, with a linear velocity of 37 cm/s. The oven temperature was held at 50°C for 1 min, increased to 190°C at a rate of 20°C/min, and further increased to a final temperature of 230°C at a rate of 2°C/min. For the MS, the ionization source temperature was 200°C. The mass spectra were recorded by electron impact ionization at 70 eV, and the acquisition of the total ion current was between 50 and 350 atomic mass units (2 scans/s). The FA compounds were identified by the equivalent chain length (ECL) method and/or by derivatization methods (using picolinyl derivates to determine the positions of the ramifications and DMOX [4,4-dimethyloxazoline] derivates to determine the positions of unsaturation) (18, 21). The ECL data for the identified compounds are listed in Table 3. The areas of the detected peaks (each peak corresponds to a distinct FA) were measured. The relative amount of one FA compound was expressed as the area of the individual peak divided by the sum of areas for all FAs detected (means ± standard errors of the means [SEM] of results for triplicate biological samples measured twice). Significant differences in mean FA peak areas were determined by one-way analysis of variance (ANOVA) and Tukey's honestly significant difference (HSD) test at the 1% level using SYSTAT version 9 (SPSS, Chicago, IL). Only data for the most abundant FAs (those with peak areas greater than 2% of the total peak area for at least one of the two strains grown at either 37 or 12°C) are presented.
TABLE 3.
ECLs of the identified FAMEs from B. cereus ATCC 14579
| Compound | Chemical name | Retention time (min)a | ECL |
|---|---|---|---|
| i13 | Methyl 11-methyldodecanoate | 10.535 | 12.48 |
| i14 | Methyl 12-methyltridecanoate | 11.721 | 13.48 |
| n14 | Methyl tetradecanoate | 12.380 | 14.00 |
| i15 | Methyl 13-methyltetradecanoate | 13.159 | 14.49 |
| a15 | Methyl 12-methyltetradecanoate | 13.408 | 14.65 |
| i16 | Methyl 14-methylpentadecanoate | 14.875 | 15.48 |
| i16:1(1) | Methyl 14-methyl-5-pentadecenoate | 15.213 | 15.64 |
| i16:1(2) | Methyl 14-methyl-10-pentadecenoate | 15.574 | 15.83 |
| n16 | Methyl hexadecanoate | 15.842 | 16.00 |
| C16:1n−11 | Methyl 5-hexadecenoate | 16.195 | 16.15 |
| C16:1n−6 | Methyl 10-hexadecenoate | 16.629 | 16.33 |
| i17 | Methyl 15-methylhexadecanoate | 16.932 | 16.48 |
| a17 | Methyl 14-ethylhexadecanoate | 17.308 | 16.65 |
| i17:1(1) | Methyl 15-methyl- 5-hexadecenoate | 17.300 | 16.65 |
| i17:1(2) | Methyl 15-methyl 10-hexadecenoate | 17.676 | 16.82 |
| n18 | Methyl stearate | 20.574 | 18.00 |
| C18:1n−9 | Methyl oleate | 21.206 | 18.21 |
| C18:2 | Methyl linoleate | 22.567 | 18.68 |
| Standard C19 | Methyl nonadecanoate | 23.422 | 19.00 |
The retention time was obtained with a GC-MS instrument (Shimadzu QP2010) equipped with an UBWAX column (see Materials and Methods).
RESULTS
Screening of transcriptional libraries for CIP genes.
Recombinase-based IVET was adapted to the investigation of gene activation during low-temperature growth of B. cereus ATCC 14579. From four independent screenings performed at 10°C, about 150 CIP clones for which a resolution event and a switch from a Km-sensitive, Sp-resistant phenotype to a Km-resistant, Sp-sensitive phenotype occurred were isolated. After the removal of potential replicates of the same clone based on the insert size and the removal of a clone containing an intragenic region, 20 distinct CIP clones were finally selected.
Determination of induction profiles of CIP genes at 10 and 30°C.
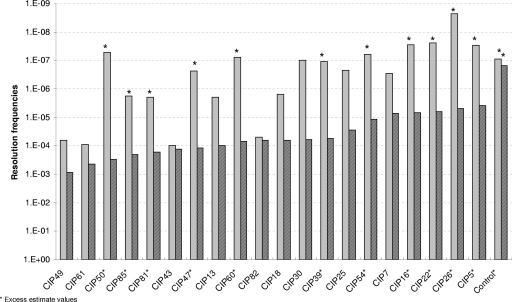
In the IVET system, the activity of a promoter is expressed by the frequency of resolution of the R2SK cassette in the B. cereus R2SK strain, leading to cells that switch from a Km-sensitive, Sp-resistant phenotype to a Km-resistant, Sp-sensitive phenotype. To confirm the promoter activity of the inserts present in the 20 CIP clones at low temperatures, their abilities to induce resolution in the B. cereus R2SK strain at 10 and 30°C were measured. For this purpose, the originally isolated pHT304-I plasmids containing the CIP-tnpI transcriptional fusions from the 20 CIP clones (expressing a Km-resistant, Sp-sensitive phenotype) were extracted and reintroduced into the unresolved B. cereus R2SK strain (Km sensitive and Sp resistant). These transformants, representing new clones of the original 20 CIPs, were selected on medium containing spectinomycin and erythromycin. They were analyzed again for resolution of the R2SK cassette, after growth in LB medium at both 10 and 30°C. The resolution frequencies induced by the 20 selected CIPs were calculated and are presented in Fig. 2. The greater the differences in the resolution frequencies obtained from clones at 10 and 30°C, the greater the differences in CIP expression between the two growth temperatures.
FIG. 2.
Resolution frequencies induced by the 20 CIPs at two temperatures. Transformants of the B. cereus R2SK strain harboring each of the 20 selected pHT304-I derivatives were grown at two temperatures. Resolution frequencies at 30°C (solid bars) and 10°C (striped bars) were calculated as the number of Kmr CFU per milliliter divided by the total population (the number of Emr CFU per milliliter). Experiments were repeated twice, and the mean values are presented (variability between results from duplicate experiments was below 15%). Asterisks indicate excessive estimate values: at 30°C, the population of Kmr CFU was below the detection threshold (10 CFU/ml) and was therefore artificially set at 9 CFU/ml. For the control (the B. cereus R2SK strain harboring pHT304-I), no Kmr CFU at either temperature tested was detectable.
Seventeen promoter regions were able to induce, reproducibly (i.e., in each of the two independent experiments), the transcription of the tnpI gene at frequencies at least 10-fold higher at 10°C than at 30°C. Three CIP clones (CIP43, CIP61, and CIP82) induced resolution frequencies with less than a 1-log difference between the two growth temperatures and were therefore excluded from further studies. For most of the remaining CIP clones (n = 11; those marked with an asterisk in Fig. 2), no expression of the promoter was detected at 30°C, as no Km-resistant colony arose in two independent experiments performed with cells grown at 30°C. Consequently, the resolution frequency was assigned by excessive estimation. For these 11 clones, this high (and underestimated) difference in resolution frequencies strongly suggests that the CIPs are specifically expressed during growth at 10°C. For the other 6 CIPs, the at least 10-fold-higher resolution frequencies they induced at 10°C than at 30°C suggest that these promoters were expressed at a higher level at 10°C than at 30°C. Thus, 17 selected promoters in B. cereus ATCC 14579 seemed to be true cold-induced fusions.
Analysis of DNA sequences fused to tnpI in the highly upregulated CIP clones.
Determination of the plasmid insert sizes in the remaining 17 chromosomal DNA fragments (CIP fragments) revealed a size range of 498 to 1,931 bp, with an average size of 1,070 bp (Table 4). By comparing the nucleotide sequence of each CIP fragment with that of the genome of B. cereus ATCC 14579 and by considering the promoter orientation relative to the tnpI reporter gene, analyses of these 17 CIP fragments inserted into the pHT304-I plasmids characterized 17 different putative promoter sequences, all in the 5′-3′ orientation, able to allow the initiation of tnpI transcription. In all 17 cases, at least one putative promoter region and its corresponding ORF were identified (Table 4). The genes corresponding to the 17 CIPs are randomly located within the genome. According to the proposed clusters of orthologous groups (COG) categories (44, 45), most of the identified genes (those corresponding to CIP60, CIP81, CIP30, CIP13, CIP47, CIP7, CIP18, CIP54, and CIP22) encode proteins involved in metabolism. Interestingly, all COG metabolism categories (energy production and conversion; transport and metabolism of carbohydrates, amino acids, nucleotides, coenzymes, lipids, and inorganic ions; and secondary metabolite biosynthesis, transport, and catabolism) were represented. Genes whose translation products are involved in information storage and processing (those corresponding to CIP26 and CIP39) and cellular processes and signaling (those corresponding to CIP16, CIP50, and CIP39) were also found. In addition, three genes with unknown functions (those corresponding to CIP49, CIP25, and CIP85) were identified. No highly conserved motif in the 17 promoter regions was identified (data not shown). This finding strongly suggests that not one but several transcriptional regulators may be implicated in the expression of those promoters at low temperatures.
TABLE 4.
B. cereus genes identified in this study as being specifically expressed during growth at low temperatures
| Clone no.a | Insert size (bp) | No. of potential promoters | Downstream ORFb |
COG category(ies)c | Broad COG categoryd | Assigned functional description of gene producte | |
|---|---|---|---|---|---|---|---|
| NCBI no. | ERGO no. | ||||||
| CIP60 | 719 | 1 | BC0544 (+3) | RZC04345 | C | Metabolism | Iron-sulfur cluster-binding protein |
| CIP81 | 711 | 1 | BC0749 (+6) | RZC02927 | H | Metabolism | Thiazole biosynthesis protein ThiG |
| CIP16 | 1,473 | 2 | BC0795 | RZC01484 | O | Cellular processes and signaling | Molybdopterin biosynthesis MoeB protein |
| BC0796 | RZC01487 | P | Metabolism | Rhodanese-related sulfurtransferase | |||
| CIP30 | 558 | 1 | BC1235 (+6) | RZC03344 | E | Metabolism | Indole-3-glycerol phosphate synthase |
| CIP50 | 1,083 | 1 | BC1647 (+8) | RZC03052 | NU | Cellular processes and signaling | Flagellar biosynthesis/type III secretory pathway ATPase |
| CIP13 | 1,931 | 1 | BC2489 | RZC06907 | I | Metabolism | Acyl-CoA synthetase/AMP-(fatty) acid ligase |
| CIP47 | 1,536 | 2 | BC3197 | RZC04150 | I | Metabolism | Permease of the major facilitator superfamily |
| BC3196 | RZC07700 | GEPR | Metabolism | Biotin carboxylase | |||
| CIP49 | 1,155 | 3 | BC4246 | RZC02388 | * | Poorly characterized | Hypothetical protein |
| BC4247 | RZC02389 | * | Poorly characterized | Hypothetical protein | |||
| BC4248 | RZC02393 | * | Poorly characterized | Hypothetical protein | |||
| CIP25 | 1,315 | 1 | BC4634 | RZC01131 | * | Poorly characterized | Hypothetical protein |
| CIP7 | 498 | 1 | BC4851 | RZC00481 | IQ | Metabolism | o-Succinylbenzoic acid-CoA ligase |
| CIP18 | 1,166 | 1 | BC4866 | RZC03491 | G | Metabolism | Glc-1-P adenyltransferase |
| CIP85 | 641 | 1 | BC4943 | RZC07012 | * | Poorly characterized | Hypothetical protein |
| CIP54 | 963 | 1 | BC4977 (+2) | RZC02376 | F | Metabolism | 5′-Nucleotidase/2′,3′-cyclic phosphodiesterase or related esterase |
| CIP5 | 702 | 1 | BC5400 (+2) | RZC04931 | V | Cellular processes and signaling | Bacitracin transport ATP-binding protein BcrA |
| CIP22 | 858 | 1 | BC5401 (+3) | RZC04935 | E | Metabolism | Lipase/acylhydrolase with GDSL-like motif |
| CIP26 | 1,691 | 1 | BC5402 (+4) | RZC04938 | K | Information storage and processing | Transcriptional regulator in the LacI family |
| CIP39, CIP2, CIP1 | 1,187 | 1 | BC5440 | RZC06561 | KT | Information storage and processing | Autolysin response regulator |
The listing of three numbers together indicates that the same clone was identified in three independent screenings.
Numbers in parentheses correspond to the number of genes organized as an operon found downstream of the CIP. ERGO numbers are according to the database for the ERGO bioinformatics system.
Data are from Tatusov et al. (44). *, no COG.
Broad functional COG category according to the NCBI website (ftp://ftp.ncbi.nih.gov/pub/COG/COG/fun.txt).
Assigned functional description according to the B. cereus genome databases (http://www.integratedgenomics.com and http://www.ncbi.nlm.nih.gov). Boldface indicates products of genes on which this study focused further.
Analysis of three CIP clones mapping at the same locus.
Among the 17 validated fusions, CIP5, CIP22, and CIP26 sequences mapped at the same chromosomal region (Table 4). Further study focused on this chromosomal region. All 3 were among the 11 selected CIP clones for which no resolution event occurred at 30°C (Fig. 2). The downstream genes corresponding to CIP26, CIP22, and CIP5 sequences are BC5402, BC5401, and BC5400, respectively. BC5402 encodes a putative transcriptional regulator, as revealed by the helix-turn-helix (HTH) domain found in the N-terminal region of the gene product (data not shown). BC5401 encodes a member of the GDSL (Gly-Asp-Ser-Leu) lipase family, as suggested by the sequence homology of the gene product to other GDSL lipases and by the GDSL amino acid signature motif found at positions 63 to 66. Additional conserved amino acids (some of which have been shown to play key roles in the catalytic function of the enzymes) are present in the GDSL lipase family proteins. These amino acids (N at position 98, G at position 103 and 134, ND at positions 136 and 137, D at position 242, and HP at positions 245 and 246 in the predicted product of BC5401) were also found (2). BC5400 was predicted to encode a bacitracin transport ATP-binding protein, BcrA. BlastN alignment indicated that BC5401 and BC5400 are found in strains of all B. cereus genetic groups and that BC5402 is found in all but the most thermotolerant strain, NVH391-98 (data not shown).
RT-PCR and characterization of the BC5402-BC5398 transcriptional unit.
The chromosomal locus of BC5402, BC5401, and BC5400 displays a putative operon structure, including the two additional genes BC5399 and BC5398 in the same orientation (Fig. 1A). All of these five genes except for BC5402, which is absent in strain NVH391-98, as stated above, are present with conserved synteny in strains of all B. cereus genetic groups. Using RNA extracted from exponential-phase cells (OD600 = 0.7) grown at 10°C, RT-PCR experiments revealed the presence of mRNA molecules overlapping the adjacent genes BC5402 and BC5401, BC5401 and BC5400, BC5400 and BC5399, and BC5399 and BC5398 (amplicons b, d, f, and g in Fig. 1), suggesting that the five genes of the locus were cotranscribed. An inverted repeat with an energy level of −18.9 kcal/mol was found downstream of BC5398 and may account for the termination of transcription (Fig. 1A). Interestingly, transcription of the BC5401 gene can be initiated both by the promoter upstream from BC5402 when the two genes BC5402 and BC5401 are cotranscribed (as revealed by the RT-PCR-detected mRNA overlapping BC5402 and BC5401) and by its own promoter (as revealed by the identified CIP22 fragment). Similarly, transcription of the BC5400 gene can be initiated either by the promoter upstream from BC5402 or by the promoter upstream from BC5401 [as revealed by the detected mRNAs overlapping (i) BC5402 and BC5401 and (ii) BC5401 and BC5400] and also by its own promoter (as revealed by the identified CIP5 fragment). Thus, the identified promoters located in this five-gene operon (BC5402 to BC5398) may also be responsible for the transcription of a four-gene operon (BC5401 to BC5398) or a three-gene operon (BC5400 to BC5398). The selection of three promoters in the same operon after the random screening with the IVET system at 10°C suggests that an important role is played by this locus during B. cereus growth at low temperatures.
Quantification of the expression of the BC5401 and BC5402 genes.
The levels of transcription of the first two genes of this operon at various time points during the kinetics of growth were analyzed by quantitative RT-PCR. Results presented in Table 5 confirmed that the levels of expression of the studied genes were significantly higher (showing >2-fold change) when cells were grown at 10°C than when cells were grown at 37°C. For BC5402, this finding was true for the three tested time points in the kinetics of growth, while for BC5401, this pattern was observed only during the exponential phase of growth.
TABLE 5.
Analysis by real-time RT-PCR of the levels of expression of BC5402 and BC5401 at various time points in the kinetics of growtha
| Gene | Fold change in expression at 10°C relative to expression at 37°C during: |
||
|---|---|---|---|
| Middle of exponential phase | End of exponential phase | Early stationary phase | |
| BC5402 | 9.46 | 2.69 | 4.42 |
| BC5401 | 3.41 | 3.22 | 1.53 |
See Materials and Methods for details.
Construction and characterization of the BC5401 and BC5402 knockout mutants.
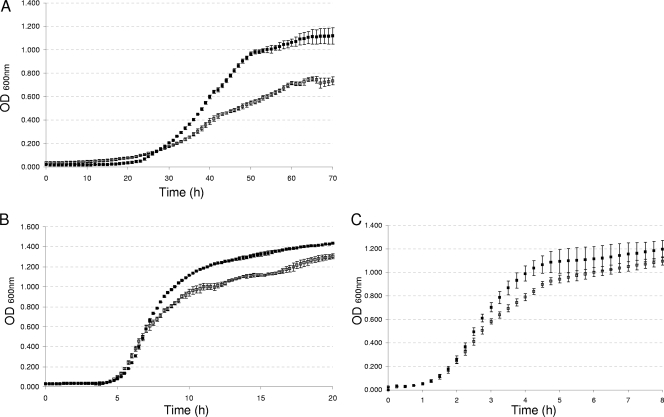
The first two genes of this operon specifically expressed at low temperatures were selected for mutagenesis to further investigate their contributions to B. cereus growth at low temperatures. Isogenic mutants of the BC5402 and BC5401 genes of B. cereus ATCC 14579 were constructed using pRN5101 for insertional inactivation via double-crossover integration. The two knockout strains (ΔBC5402 and ΔBC5401 mutants) and the WT strain were analyzed for the ability to grow at low temperatures in comparison with growth at 30 and 45°C. The growth of the ΔBC5402 strain at 12°C was similar to that of the WT (data not shown). In contrast, the ΔBC5401 mutant displayed impaired growth ability at the three tested temperatures, only slightly at 30 and 45°C, but particularly clearly at 12°C (Fig. 3). At this low temperature, a growth delay was observed from the beginning of the exponential phase. A final OD600 lower than that obtained for the WT was also observed. In contrast, at 30 and 45°C, the growth of the ΔBC5401 strain was impaired only slightly at the end of the exponential phase and this strain reached the stationary phase with a final OD600 slightly lower than (at 30°C) or similar to (at 45°C) that of the WT.
FIG. 3.
Growth curves of B. cereus WT and ΔBC5401 strains at various temperatures. The WT (black squares) and the BC5401 mutant (white squares) were grown at 12°C (A), 30°C (B), or 45°C (C). The cultures were incubated in an automated turbidometer with vigorous constant shaking, and the OD600 was measured at 15-min intervals (at 30 or 45°C) or 1-h intervals (at 12°C). Results shown are the means and standard deviations of data from triplicate experiments.
Quantitative analysis of FAs in the BC5401 mutant and WT strains.
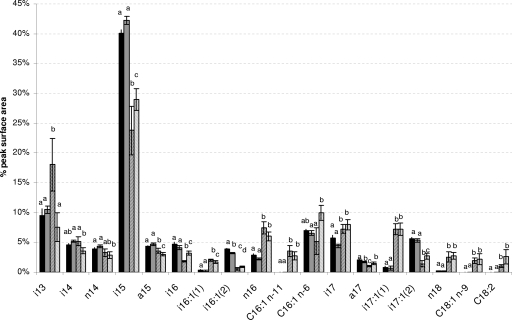
Disruption of one of the identified cold-induced genes (BC5401), encoding a putative lipase, led to a cold-impaired growth phenotype. We therefore determined the FA profiles of both WT and ΔBC5401 cells grown at either an optimal or low temperature (37 or 12°C, respectively) (Fig. 4). At 37°C, only slight differences between the WT and the ΔBC5401 strain were observed. The only significant differences (P < 0.01; Tukey's HSD test) were the slightly smaller amounts of two compounds [i16:1(2) and a17] in the mutant.
FIG. 4.
FA profiles of B. cereus WT and ΔBC5401 strains at two temperatures. The WT (black bars) and the ΔBC5401 strain (gray bars) were grown at 37°C (solid bars) or 12°C (striped bars) on LB agar. The profiles of FAMEs were determined by GC-MS as described previously (39). The areas of the detected peaks (each corresponding to a distinct FA) were measured. Each bar represents the relative amount of one FA compound, expressed as the area of the individual peak divided by the sum of the areas of all the peaks on the chromatogram (mean ± SEM of results for triplicate biological samples measured twice). Only data for major FA compounds (i.e., FAs for which the area of the individual peak was >2% of the total area at either 37 or 12°C) are presented. For a given FA, the presence of the same letters above the bars indicates that no significant difference between results for the two temperatures and/or strains was detected (P > 0.01; Tukey's HSD test).
Several changes in the FA profiles of WT cells grown at 12°C instead of 37°C were observed. FAs of cells grown at 12°C were changed in particular by decreased amounts of i15 and i17:1(2) and increased amounts of i13, n16, i17, i17:1(1), and n18 (differences between profiles at the two temperatures were significant, with a P value of <0.01). Three compounds (C16:1 n−11, C18:1, and C18:2, where n−11 indicates double bonds at position n−11) were found specifically in cells grown at 12°C. The ratio of unsaturated to saturated FAs increased for cells grown at the low temperature: the ratio was 0.24 for cells grown at 37°C and 0.32 for cells grown at 12°C.
Changes similar to those in WT cells were observed in the ΔBC5401 strain grown at 12°C instead of 37°C, with one major exception. The amount of i13 was not significantly different (P > 0.01) between cells of the ΔBC5401 strain grown at the two temperatures. In addition, this mutant displayed some differences (P < 0.01) in i14 and C16:1 n−6 compounds between cells grown at the two temperatures that were not observed in WT cells. The ratio of unsaturated to saturated FAs also increased when this strain was grown at the low temperature (ratios were 0.21 and 0.47 for cells grown at 37 and 12°C, respectively). Thus, during growth at the low temperature, a major increase of i13 was found for the WT, which was not observed for the BC5401 mutant.
DISCUSSION
Molecular microbial ecology is often hampered by the difficulty of unraveling how the environment shapes bacterial physiology and enables microorganisms to respond to environmental stresses. In the aim to identify genes specifically expressed at 10°C and thus putatively involved in B. cereus low-temperature adaptation, we used an IVET approach for B. cereus and identified 17 clones with elevated expression at 10°C relative to that at 30°C, which demonstrated induced expression during growth at low temperatures. To our knowledge, this is the first study that demonstrates the feasibility of this approach for identifying CIPs or cold-induced genes in a food-borne pathogen. The screening was performed with cells undergoing cold acclimation by using an approach allowing the identification of genes specifically expressed (even transiently) during growth at cold temperatures at any time during the lag phase or the exponential growth phase. In comparison, a conventional microarray analysis might have the limitation that not all stages in the kinetics of growth would be represented in samples, thus permitting the possibility of failure to isolate transiently expressed genes. The 17 CIP clones could be functionally divided into four categories based on the COG classification of the annotated downstream gene in the captured DNA of each clone. Although most of the promoters (11 of 17) identified here were specifically expressed at low temperatures, 6 of the identified promoters were still expressed at 30°C. In this case, the promoters were expressed at a much lower level at 30°C (CFU for which a resolution event had occurred were isolated at least 10 times less frequently at 30°C than at 10°C). This finding suggests that our growth steps at 30°C did not remove all promoters expressed at a low level. The list of genes identified here is of course not exhaustive, as illustrated by a low degree of overlap of identified clones among the four independent screenings. This outcome may be due to the relatively high stringency conditions we used during the screening steps. In addition, the probable multifactorial nature of the entire cold adaptation process implies that it is not governed by a few essential activated genes but rather by a large number of genes. Thus, many additional genes specifically expressed at 10°C probably remain to be discovered. However, the genes identified in this screening enable us to outline a first picture of the different strategies B. cereus uses to grow at low temperatures.
One strategy may be to activate alternative metabolic pathways. Preferential metabolic pathways in B. subtilis (32) or in B. cereus (13, 14) are modified during growth at low temperatures. Most of the genes identified in this study seem to be involved in various metabolic reactions. Among them, BC0749 is predicted to code for ThiG, a protein required for the biosynthesis of the thiazole moiety of thiamine (vitamin B1). Derived from thiamine, thiamine pyrophosphate is an essential cofactor involved in central metabolism and amino acid biosynthesis (35). In B. cereus ATCC 14579, the expression of BC0749 is under the control (though probably indirect) of the σB general stress response adaptation regulator (48). However, no other members of the σB regulon in B. cereus have been identified during this study, in contrast to the several σB-regulated genes in B. subtilis (8, 10) and in the food-borne pathogen Listeria monocytogenes (11) observed previously to be induced during growth at cold temperatures. This finding indicates that distinct roles may be played by σB in B. cereus and these other two bacterial species during growth at low temperatures. This hypothesis is in agreement with the findings of a recent study showing that the σB activation pathway in the B. cereus group is highly different from those in other low-GC-content Gram-positive bacteria (17).
Low-temperature adaptation often requires genes under the control of regulators able to sense and coordinate metabolic functions during cold stress. For example, in B. subtilis, the expression of cold-inducible genes responsible for membrane unsaturation is controlled by DesKR, a specific two-component system (1). Among the genes identified in this study, one (BC5440) was found to encode a protein of a two-component regulatory system that may function to sense and perceive the environment more precisely and respond to it. The BC5440 gene product does not belong to the same response regulator family as B. subtilis DesR (16). BC5440 encodes a response regulator probably involved in the regulation of enzymes with murein hydrolase activity (like regulators in the LytR/AlgR family) (24). The expression of murein hydrolase enzymes may have to be tightly controlled to allow optimal peptidoglycan turnover when bacteria are grown at low temperatures.
Modifications in the cell wall and/or membrane composition (e.g., changes in the ratio of saturated to unsaturated FAs) are also important for bacteria to adapt to the cold (29). Induced expression of the products of some of the identified genes (BC1235, BC2489, and BC4851) presumably targets precursors for FA metabolism. Their roles in FA metabolism may account for the modifications to the FA profiles observed when cells are grown at low temperatures. This hypothesis is supported by the identification in B. subtilis of an ortholog of BC4851, encoding an o-succinylbenzoic acid-coenzyme A (CoA) ligase, which is also induced during growth at cold temperatures (10).
One of the identified genes (BC3197) encodes a putative permease of the major facilitator superfamily (MFS). This family includes transporters of small solutes (15, 34), and some members also act as osmoprotectors. In L. monocytogenes, osmoprotectants permit better growth at low temperatures (52). In addition, several of the identified genes (BC4246, BC4634, and BC4943) were predicted to encode hypothetical proteins. A large number of genes with unknown functions have also been shown previously to be induced in B. subtilis during growth at low temperatures (10).
Finally, three promoters from the same locus (BC5402 to BC5398) were identified in distinct screenings. The lack of detection of resolution events in cells carrying the three corresponding CIP clones during growth at 30°C suggests specific expression of this locus when B. cereus was grown at 10°C. The BC5402 gene was predicted to encode a regulator in the LacI family. Five regulators from this family in Erwinia species have been studied previously and are involved in the repression of genes adjacent to the regulator genes (47). Gene BC5402 is the first of five genes occurring in an operon strongly expressed at low temperatures. However, the BC5402 gene, when mutated singly, does not affect the growth of B. cereus at low temperatures. This finding seems to indicate that this gene is not coding for an essential function by itself; however, we cannot assert that this gene therefore plays no role or only a very minor one in the acclimation of the bacterium to cold or in the regulation of adjacent genes. Many traits contributing to ecological performance result in only subtle or difficult-to-score phenotypic changes upon inactivation and are likely to be overlooked. Moreover, it is more and more frequently accepted that many regulators act in a partially or fully redundant manner and that mutation of one such gene is thus unlikely to fully abolish the activity of all the genes under its control. In the ATCC 14579 genome, two genes encoding putative lipase/acylhydrolase members of the GDSL family (BC5401 and BC4123) are located in operon structures that include putative transcriptional regulator-encoding genes (BC5402 and BC4124). A third gene (BC2449) also displayed sequence homology to BC5401, but the GDSL motif of the product was degenerated into GDSF, and this gene was not adjacent to a gene encoding a putative transcriptional regulator. These genes may have similar functions, and we cannot rule out the possibility that disruption of BC5402 is complemented by the expression of its paralog. A search for other possible redundant factors should accordingly be pursued.
In contrast to the BC5402 deletion mutant, the BC5401 deletion mutant was shown to be significantly impaired in its ability to grow at low temperatures. This finding indicates that in the ΔBC5402 strain, transcription of BC5401, encoding a member of the GDSL lipase family, and possibly of the three remaining genes located downstream of BC5401 could still proceed adequately, probably from the promoter found upstream from BC5401. Despite the cold-induced IVET-identified promoter upstream from BC5400, a possible polar effect influencing the downstream genes in the BC5401 deletion mutant cannot be discounted. Thus, the possible roles of BC5400, BC5399, and BC5398 during growth at low temperatures remain to be clarified.
Although the lipases of the GDSL family are widely distributed in bacteria, most of the members of this family described previously are found in plants. Expression of a GDSL-like lipase is upregulated in rice overexpressing a gene involved in various types of stress tolerance, including cold stress (30). In bacteria, lipases of this family are generally described as lipolytic enzymes, but their physiological role is not well understood (2). Lipases (triacylglycerol acylhydrolases; EC 3.1.1.3) generally govern the turnover of lipids and the biogenesis of membranes in bacteria and are by definition carboxylesterases, whose major substrates are long-chain (≥10-carbon) triacylglycerols, releasing FAs and glycerol (31). They generally act at the water-lipid interface and display broad substrate specificity, a property that lends the microorganisms access to diverse carbon sources during plant cell wall degradation or during the recycling of lipid-containing nutrients (27, 28). It has also been suggested previously that some of these enzymes may play a role in the turnover of membrane lipids and lipid-anchored proteins by altering cell membrane composition to change cell membrane functions or adapt cell membranes to environmental changes (41, 46). The main factor controlling the expression of lipase activity is therefore the presence of lipid sources, although lipase production is also significantly influenced by other carbon sources such as sugars, polysaccharides, amino acids, and other complex compounds. In addition, lipase synthesis is influenced by other physicochemical factors such as temperature, pH, the presence of inorganic salts, agitation, and oxygen concentration (28). In the present study, the role of BC5401 in the FA profile of B. cereus was investigated. As already described for many other bacteria and other B. cereus strains (29), both the ATCC 14579 WT strain and the BC5401 mutant can adapt the fluidity of their membranes at lower growth temperatures by increasing the proportion of unsaturated FAs. Interestingly, compared with growth at 37°C, growth at a low temperature led to a major increase in i13 in the WT. This is a 13-carbon FA branched at position n−2. No such increase was observed for the BC5401 mutant. Although the role of this FA compound at low temperatures is not known, a similar increase in Shewanella piezotolerans grown at 4°C versus 20°C was observed previously (50). However, the exact role of the BC5401 product in i13 biosynthesis during bacterial growth at low temperatures remains to be investigated.
In conclusion, we have shown the applicability of the IVET approach to gaining a fuller understanding of the gene machinery specifically activated during the growth of B. cereus at low temperatures. This study gives insight into the properties required for the ecological adaptation of B. cereus to cold environments and provides an initial background for the investigation of the adaptation of gene expression mechanisms at low temperatures. The different genes identified in this study certainly reflect the complexity of the processes involved in B. cereus low-temperature adaptation (acclimation).
Acknowledgments
This work was supported by INRA (Institut National de la Recherche Agronomique) and by a grant from the Agence Nationale de la Recherche (ANR; France) as part of an ANR-05-PNRA-013 B. cereus contract.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Aguilar, P. S., A. M. Hernandez-Arriaga, L. E. Cybulski, A. C. Erazo, and D. de Mendoza. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akoh, C. C., G. C. Lee, Y. C. Liaw, T. H. Huang, and J. F. Shaw. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534-552. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 5.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 7.Bouillaut, L., N. Ramarao, C. Buisson, N. Gilois, M. Gohar, D. Lereclus, and C. Nielsen-Leroux. 2005. FlhA influences Bacillus thuringiensis PlcR-regulated gene transcription, protein production, and virulence. Appl. Environ. Microbiol. 71:8903-8910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigulla, M., T. Hoffmann, A. Krisp, A. Volker, E. Bremer, and U. Volker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brillard, J., K. Susanna, C. Michaud, C. Dargaignaratz, M. Gohar, C. Nielsen-Leroux, N. Ramarao, A. B. Kolsto, C. Nguyen-the, D. Lereclus, and V. Broussolle. 2008. The YvfTU two-component system is involved in plcR expression in Bacillus cereus. BMC Microbiol. 8:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budde, I., L. Steil, C. Scharf, U. Volker, and E. Bremer. 2006. Adaptation of Bacillus subtilis to growth at low temperature: a combined transcriptomic and proteomic appraisal. Microbiology 152:831-853. [DOI] [PubMed] [Google Scholar]
- 11.Chan, Y. C., K. J. Boor, and M. Wiedmann. 2007. σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl. Environ. Microbiol. 73:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, Y. C., and M. Wiedmann. 2009. Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 49:237-253. [DOI] [PubMed] [Google Scholar]
- 13.Choma, C., M. H. Guinebretiere, F. Carlin, P. Schmitt, P. Velge, P. E. Granum, and C. Nguyen-The. 2000. Prevalence, characterization and growth of Bacillus cereus in commercial cooked chilled foods containing vegetables. J. Appl. Microbiol. 88:617-625. [DOI] [PubMed] [Google Scholar]
- 14.Chung, B. H., R. Y. Cannon, and R. C. Smith. 1976. Influence of growth temperature on glucose metabolism of a psychotrophic strain of Bacillus cereus. Appl. Environ. Microbiol. 31:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culham, D. E., T. Romantsov, and J. M. Wood. 2008. Roles of K+, H+, H2O, and ΔΨ in solute transport mediated by major facilitator superfamily members ProP and LacY. Biochemistry 47:8176-8185. [DOI] [PubMed] [Google Scholar]
- 16.de Been, M., C. Francke, R. Moezelaar, T. Abee, and R. J. Siezen. 2006. Comparative analysis of two-component signal transduction systems of Bacillus cereus, Bacillus thuringiensis and Bacillus anthracis. Microbiology 152:3035-3048. [DOI] [PubMed] [Google Scholar]
- 17.de Been, M., M. H. Tempelaars, W. van Schaik, R. Moezelaar, R. J. Siezen, and T. Abee. 2 December 2009. A novel hybrid kinase is essential for regulating the σB-mediated stress response of Bacillus cereus. Environ. Microbiol. doi: 10.1111/j.1462-2920.2009.02116.x. [DOI] [PubMed]
- 18.Destaillats, F., and P. Angers. 2002. One-step methodology for the synthesis of FA picolinyl esters from intact lipids. J. Am. Oil Chem. Soc. 79:253-256. [Google Scholar]
- 19.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Food Safety Authority. 2007. Request for updating the former SCVPH opinion on Listeria monocytogenes risk related to ready-to-eat foods and scientific advice on different levels of Listeria monocytogenes in ready-to-eat foods and the related risk for human illness. Scientific opinion of the panel on biological hazards. EFSA J. 599:1-42. [Google Scholar]
- 21.Fay, L., and U. Richli. 1991. Location of double bonds in polyunsaturated fatty acids by gas chromatography-mass spectrometry after 4,4-dimethyloxazoline derivatization. J. Chromatogr. 541:89-98. [Google Scholar]
- 22.Fedhila, S., N. Daou, D. Lereclus, and C. Nielsen-LeRoux. 2006. Identification of Bacillus cereus internalin and other candidate virulence genes specifically induced during oral infection in insects. Mol. Microbiol. 62:339-355. [DOI] [PubMed] [Google Scholar]
- 23.Francis, K. P., R. Mayr, F. von Stetten, G. S. Stewart, and S. Scherer. 1998. Discrimination of psychrotrophic and mesophilic strains of the Bacillus cereus group by PCR targeting of major cold shock protein genes. Appl. Environ. Microbiol. 64:3525-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galperin, M. Y. 2008. Telling bacteria: do not LytTR. Structure 16:657-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 26.Guinebretiere, M. H., F. L. Thompson, A. Sorokin, P. Normand, P. Dawyndt, M. Ehling-Schulz, B. Svensson, V. Sanchis, C. Nguyen-The, M. Heyndrickx, and P. De Vos. 2008. Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 10:851-865. [DOI] [PubMed] [Google Scholar]
- 27.Gunstone, F. D. 1999. Enzymes as biocatalysts in the modification of natural lipids. J. Sci. Food Agric. 79:1535-1549. [Google Scholar]
- 28.Gupta, R., N. Gupta, and P. Rathi. 2004. Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64:763-781. [DOI] [PubMed] [Google Scholar]
- 29.Haque, M. A., and N. J. Russell. 2004. Strains of Bacillus cereus vary in the phenotypic adaptation of their membrane lipid composition in response to low water activity, reduced temperature and growth in rice starch. Microbiology 150:1397-1404. [DOI] [PubMed] [Google Scholar]
- 30.Hu, H., J. You, Y. Fang, X. Zhu, Z. Qi, and L. Xiong. 2008. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 67:169-181. [DOI] [PubMed] [Google Scholar]
- 31.Jaeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 32.Kaan, T., G. Homuth, U. Mader, J. Bandow, and T. Schweder. 2002. Genome-wide transcriptional profiling of the Bacillus subtilis cold-shock response. Microbiology 148:3441-3455. [DOI] [PubMed] [Google Scholar]
- 33.Kaneda, T. 1972. Positional preference of fatty acids in phospholipids of Bacillus cereus and its relation to growth temperature. Biochim. Biophys. Acta 280:297-305. [DOI] [PubMed] [Google Scholar]
- 34.Law, C. J., P. C. Maloney, and D. N. Wang. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62:289-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leonardi, R., S. A. Fairhurst, M. Kriek, D. J. Lowe, and P. L. Roach. 2003. Thiamine biosynthesis in Escherichia coli: isolation and initial characterisation of the ThiGH complex. FEBS Lett. 539:95-99. [DOI] [PubMed] [Google Scholar]
- 36.Lereclus, D., O. Arantes, J. Chaufaux, and M. Lecadet. 1989. Transformation and expression of a cloned delta-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol. Lett. 51:211-217. [DOI] [PubMed] [Google Scholar]
- 37.Letunic, I., R. R. Copley, S. Schmidt, F. D. Ciccarelli, T. Doerks, J. Schultz, C. P. Ponting, and P. Bork. 2004. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32:D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasser, M. 1990. Technical note no. 101. Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). MIDI Labs, Inc., Newark, DE.
- 40.Sasser, M., C. Kunitsky, and G. Jackoway. 2005. Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J. AOAC Int. 88:178-181. [PubMed] [Google Scholar]
- 41.Schmid, R., and R. Verger. 1998. Lipases: interfacial enzymes with attractive applications. Angew. Chem. Int. Ed. Engl. 37:1608-1633. [DOI] [PubMed] [Google Scholar]
- 42.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumann, W. 2009. Temperature sensors of eubacteria. Adv. Appl. Microbiol. 67:213-256. [DOI] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 46.Titball, R. W. 1998. Bacterial phospholipases. Symp. Ser. Soc. Appl. Microbiol. 27:127S-137S. [PubMed] [Google Scholar]
- 47.Van Gijsegem, F., A. Wlodarczyk, A. Cornu, S. Reverchon, and N. Hugouvieux-Cotte-Pattat. 2008. Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Mol. Plant Microbe Interact. 21:1471-1481. [DOI] [PubMed] [Google Scholar]
- 48.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villafane, R., D. H. Bechhofer, C. S. Narayanan, and D. Dubnau. 1987. Replication control genes of plasmid pE194. J. Bacteriol. 169:4822-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, F., X. Xiao, H. Y. Ou, Y. Gai, and F. Wang. 2009. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J. Bacteriol. 191:2574-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber, M. H., and M. A. Marahiel. 2002. Coping with the cold: the cold shock response in the Gram-positive soil bacterium Bacillus subtilis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357:895-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood, J. M., D. E. Culham, A. Hillar, Y. I. Vernikovska, F. Liu, J. M. Boggs, and R. A. Keates. 2005. A structural model for the osmosensor, transporter, and osmoregulator ProP of Escherichia coli. Biochemistry 44:5634-5646. [DOI] [PubMed] [Google Scholar]