Abstract
Human enteric adenoviruses (HAdVs; serotypes 40 and 41) are important waterborne and food-borne pathogens. However, HAdVs are fastidious, are difficult to cultivate, and do not produce a clear cytopathic effect during cell culture within a reasonable time. Thus, we examined whether the viral transactivator proteins cytomegalovirus (CMV) IE1 and hepatitis B virus (HBV) X promoted the multiplication of HAdVs. Additionally, we constructed a new 293 cell line expressing CMV IE1 protein for cultivation assays. We analyzed the nucleic acid sequences of the promoter regions of both E1A and hexon genes, which are considered to be the most important regions for HAdV replication. Expression of either HBV X or CMV IE1 protein significantly increased the promoter activities of E1A and hexon genes of HAdVs by as much as 14-fold during cell cultivation. The promotion of HAdV expression was confirmed by increased levels of both adenoviral DNA and mRNA expression. Finally, the newly developed 293 cell line expressing CMV IE1 protein showed an increase in viral DNA ranging from 574% to 619% compared with the conventional 293 cell line. These results suggest that the newly constructed cell line could be useful for efficient cultivation and research of fastidious HAdVs.
Human enteric adenoviruses (HAdVs; serotypes 40 and 41) are among the most common etiological agents of gastroenteritis, particularly among children (1, 33). These viruses are transmitted by the fecal-oral route, via contaminated water and food. Although HAdVs are cultivable in several cell lines, including 293, A549, PLC/PRF5, and Caco-2 cells, they are fastidious and do not produce a clear and consistent cytopathic effect (CPE) within a reasonable time (6, 17-19, 20, 31). They are sensitive to type I interferon (IFN), and the HAdV E1A gene is deficient in its ability to transactivate its own genes (4, 23, 36, 39). These characteristics make cultural analyses of HAdVs difficult because of their low concentrations and the presence of other fast-culturing viral agents in environmental samples. However, the standard method of detecting viral pathogens in water samples uses replication in mammalian cell culture (13). Thus, better culture methods or other techniques are required for the rapid quantitative detection of infectious HAdVs in water.
One way to promote the replication of fastidious virus could be to apply other viral transactivator proteins. Viral transactivator proteins can activate and stimulate a variety of genes, including other viral genes, by (i) binding directly to specific DNA sequence motifs (sequence-dependent transcriptional regulation) and (ii) influencing transcription by interacting with other proteins (sequence-independent transcriptional regulation). Viral transactivator proteins can activate not only the viral genes but also many other genes by activating common transcription factors (e.g., AP1 and NF-κB) or signal transduction pathways (35). For example, simian virus T antigen (SV-T), hepatitis B virus (HBV) X, and cytomegalovirus (CMV) IE1 and CMV IE2 proteins can significantly activate a variety of genes, including viral and cellular genes (10, 22, 37). In addition, cellular transcription factors such as AP1 or NF-κB can be introduced into cells and can markedly increase gene transcription (35). These biological characteristics can be applied both to increase the levels of target mRNA and to promote the multiplication of fastidious HAdVs in cell culture.
The objectives of the present study were to determine whether viral transactivation proteins, including HBV X and CMV IE1, can activate the transcription of essential genes of HAdVs and subsequently promote the replication of HAdVs and to construct a new cell line that promotes the replication of fastidious HAdVs.
MATERIALS AND METHODS
Preparation of virus stocks.
HAdV serotype 41 (HAdV-41) was obtained from the American Type Culture Collection (ATCC VR-930). HAdVs were cultivated in 293 cells in minimum essential medium (MEM; Gibco, Grand Island, NY) containing 10% fetal bovine serum (FBS; Gibco). The viral DNA was estimated to be 3.3 × 106 viral genome copies/ml by serial dilution and subsequent PCR amplification. This stock had a titer of 1.6 × 105 50% tissue culture infective doses (TCID50)/ml, which was calculated by the Reed-Muench method using 293 cells (30). The stock was stored at −80°C until analysis. In addition to laboratory-adapted HAdV-41, clinical stool samples containing HAdV-40 (1 × 106 viral genome copies/ml) and HAdV-41 (3 × 105 viral genome copies/ml) were provided by the Korea Center for Disease Control and Prevention.
Subcloning of the promoters of the hexon and E1A genes of HAdV-41.
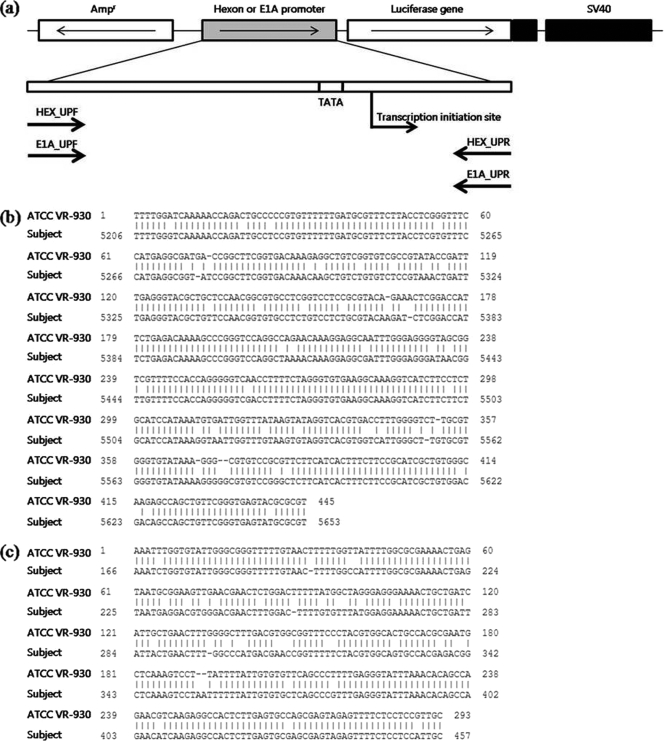
Multiple alignments of the hexon and E1A genes of HAdVs (GenBank accession numbers NC_001454, DQ315364, and L19443) were performed to design the PCR primer sets shown in Table 1 and Fig. 1. The primers (primer pair E1A_UPF and E1A_UPR and primer pair Hex_UPF and Hex_UPR) produced amplicons of 403 bp and 489 bp for E1A and hexon regions, respectively. The resulting E1A and hexon amplicons were purified for subcloning using a QIAquick PCR purification kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. After purification of the PCR amplicon, either the E1A or hexon upper promoter region was inserted into the pGL2-basic vector (Promega, Madison, WI) as shown in Fig. 1. The purified PCR amplicons were prepared for ligation by treatment with 5′ phosphorylase and T4 nucleotide kinase (New England Biolabs, Beverly, MA). The pGL2 vector DNA was digested with SmaI (New England Biolabs). The terminal phosphates were removed by treatment with Antarctic phosphatase (Invitrogen, Carlsbad, CA). The prepared insert and pGL2-basic vector DNA were then ligated using T4 DNA ligase (Invitrogen). The resulting ligation product was used to transform competent Escherichia coli cells. After transformation, colonies from each E1A and hexon ligation were picked and cultured in 3 ml of LB medium with 50 μg/ml ampicillin at 37°C with shaking overnight. Then, DNA was then purified from cultures using a QIAquick Spin miniprep kit (Qiagen) according to the manufacturer's instructions. Samples were quantified by spectrophotometric analysis and sequenced by a commercial company (Cosmo, Seoul, South Korea) to confirm the correct insert using a vector-specific primer. Samples that were identified by sequencing as having the insert in the correct orientation were used for large-scale culture and DNA purification for subsequent transient transfection assay. A large-scale culture (100 ml) was inoculated with 200 μl of the small preculture and analyzed using an EndoFree Plasmid maxi-kit (Qiagen) according to the manufacturer's instructions.
TABLE 1.
Summary of PCR primer sets for cloning both hexon and E1A promoter regions of HAdV-41
| Name | Target | Sequence (5′→3′) | Locationa |
|---|---|---|---|
| E1A_UPF (5′) | E1A promoter | GACTAGGGGTGGTGTAAGGTGACG | 60-83 |
| E1A_UPR (3′) | E1A promoter | CAACAGCAATGGAGGAGAAAAC | 467-488 |
| Hex_UPF (5′) | Hexon promoter | CACACGGTTTCGCACTCCACTAA | 5230-5252 |
| Hex_UPR (3′) | Hexon promoter | ACGCGCATACTCACCCGAACAG | 5697-5718 |
FIG. 1.
(a) Schematic diagram for subcloning the promoter regions of HAdV-41. Both hexon and E1A amplicons were cloned into the pGL2-basic vector (Promega) and directly controlled luciferase expression. Hexon (b) and E1A (c) genes of HAdV F group (ATCC VR-930) were compared with L19443 using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Transient transfection and subsequent assay of luciferase activity.
Both newly constructed recombinant plasmid DNA (pGL2 vectors containing either the E1A or hexon promoter) and transactivator protein expression vectors (either HBV X or CMV IE1) were cotransfected into 293 cells by Lipofectamine (Invitrogen) in accordance with the manufacturer's protocol. Both HBV X and CMV IE1 expression vectors (LNC IE) were described previously (16, 40). After incubation for 24 h, the luciferase activity in transfected cells was measured using a Dual-Luciferase Reporter Assay System (Promega) and a GloMax 20/20 luminometer (Promega) according to the manufacturer's instructions. Transactivation was determined by monitoring the level of firefly luciferase from the pGL2 vector. Renilla luciferase from cotransfected pRL-SV40 vector (Promega) was used as cotransfection control. The luciferase assay was performed by adding lysis buffer (500 μl) and harvesting the cells through centrifugation (at 13,000 rpm for 15 s). The supernatant was transferred to fresh tubes, and 20 μl of lysate was added to 100 μl of beetle luciferin solution in an assay tube. This substrate produced oxyluciferin formed by firefly luciferase, which was measured using a luminometer (Turner Biosystems, Sunnyvale, CA). Each experiment was repeated at least three times.
Expression of the CMV IE1 protein in 293 cells.
The expression of CMV IE1 protein in the 293 cell line was checked by Western blotting. Briefly, each of the cell lysates was fractionated by SDS-PAGE. ProSieve color protein markers (Cambrex, Walkersville, MD) were used as the molecular weight standards in 8% SDS-PAGE. Samples were subjected to SDS-PAGE on two gels for staining with Coomassie brilliant blue (Sigma Chemical Co., St. Louis, MO) and for Western blotting, as described previously (26). For immunoblotting, protein samples were transferred onto polyvinylidene difluoride (PVDF) membranes (Amersham, Buckinghamshire, United Kingdom) using a Mini-PROTEAN 3 blot module (Bio-Rad Laboratories, Richmond, CA) and then blocked with 5% Difco skim milk (BD Biosciences, Mansfield, MA) in TBS (10 mM Tris-HCl, pH 8.0, 150 mM NaCl) for 1 h. Membranes were washed with TBS-0.1% Tween 20 (Sigma) three times for 10 min each time and then incubated for 1 h at room temperature with 6 ml of washing buffer and 4 ml of 5% skim milk containing a 1/1,000 dilution of anti-CMV IE monoclonal antibody (Chemicon, Temecula, CA). The membranes were washed with washing buffer three times for 10 min each time and then incubated for 1 h in 6 ml of washing buffer and 4 ml of 5% skim milk containing a 1/1,000 dilution of secondary goat anti-mouse IgG-horseradish peroxidase conjugate (Thermo Fisher Scientific, Pittsburgh, PA). The membranes were again washed with washing buffer three times for 10 min each time, detected with ECL Western blotting detection reagent (Amersham) for 1 min, and immediately exposed to X film (Agfa, Mortsel, Belgium) for 3 to 10 min in a darkroom. Exposed film was developed using an FPM 1200 medical film processor machine (Fuji, Tokyo, Japan).
Characterization of the promotion of viral replication by CMV IE1 protein.
To determine the activation of transcription and replication of HAdVs by CMV IE1, we extracted both viral mRNA and DNA from infected cells and quantified them. Briefly, 293 cells were prepared in six-well tissue culture plates (SPL Lifesciences, Pocheon, South Korea) 24 h before transfection. When they reached 90 to 95% confluence, the cells were transfected with 2 μg of LNC IE plasmid using Lipofectamine 2000 reagent (Invitrogen). CMV IE1 genes were inserted into 293 cells by formation of liposomes and expressed their proteins for 1 day. After 24 h, cells were infected with 5 × 103 genome copies of HAdV-41 suspended in 500 μl of phosphate-buffered saline (PBS) in each well. After 3 days of incubation, infected cells were harvested by trypsinization and centrifuged (100 × g for 3 min). The resulting cell pellets were subjected to mRNA extraction. Total RNA was extracted using an RNeasy Mini kit (Qiagen) according to the manufacturer's protocol. To avoid viral DNA contamination, we treated 8 μl of RNA with 1 μl of RQ1 RNase-Free DNase (Promega) and an equal volume of RQ1 RNase-Free DNase 10× reaction buffer (Promega) for 30 min at 37°C. DNase was inactivated by incubation at 65°C for 10 min with 1 μl of RQ1 DNase stop solution (Promega). To determine the activation of viral DNA replication, we harvested infected cells by freeze-thaw cycling. The resulting cell lysates were subjected to viral DNA extraction using a QIAamp viral DNA Mini kit (Qiagen). The extracted viral DNA and mRNA were quantified by real-time PCR using an ABI 7300 real-time PCR instrument (Applied Biosystems, Foster City, CA) as described previously (17).
Construction of the new cell line 293-CMV by electroporation.
The 293 cell line was permanently transformed with CMV IE1 expression vector by electroporation to construct the new cell line 293-CMV. Briefly, 293 cells were maintained in MEM (Gibco) with 10% FBS (Gibco). The cells were harvested at 80 to 95% confluence and resuspended in PBS adjusted to 5 × 106 to 5 × 107 cells/600 μl. The resulting cell suspensions and 3 μg of LNC IE vector were added to prechilled electroporation cuvettes (4 cm; Bio-Rad) and subjected to electroporation (960 V; 1,050 μF) using a Gene Pulser II unit (Bio-Rad). The same amount of pcDNA3 vector was used as control DNA. After electroporation, the cuvettes were transferred immediately to ice and stored for 5 min. Cell suspension was incubated in MEM with 10% FBS at 37°C in 5% CO2. After 48 h, neomycin-resistant cells were selected by culture in medium containing 1 mg/ml G418 (Gibco). To isolate single clones, we performed limiting serial dilution on 96-well plates (SPL) at least three times.
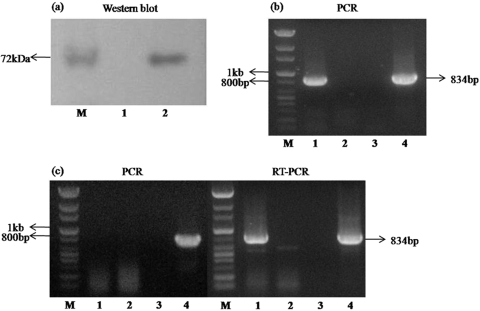
After clonal selection of transformed cells, incorporation into the chromosome and expression of CMV IE1 genes were checked by both PCR and reverse transcription-PCR (RT-PCR) assays (see Fig. 4). Transformed cells were cultivated in MEM containing G418. Cultured cells were harvested by trypsinization and centrifuged (at 100 × g for 3 min). Genomic DNA was extracted from the resulting cell pellets using a DNeasy Blood and Tissue kit (Qiagen). Incorporation of the LNC IE vector into the cell chromosome was checked by PCR amplification of genomic DNA. PCR amplification was performed in a mixture containing 2.5 μl of 10× reaction buffer (Cosmo), 1 μl of the deoxynucleoside triphosphates (dNTPs; 10 mM), 1.25 μl of LNCX-F primer (10 pmol/μl), 1.25 μl of LNCX-R primer (10 pmol/μl), 0.2 μl of G-Taq polymerase (Cosmo), and 2 μl of DNA template in a final volume of 25 μl. A forward (5′-GAT CCC CTC GCG AGT TGG TTC-3′) and reverse (5′-AAT GGG GCG GAG TTG TTA CGA C-3′) primer set was designed based on the pLNCX vector that would amplify a product of 834 bp. The PCR assay was performed under the following conditions: denaturation for 2 min at 94°C and then 40 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 30 s at 68°C. To check CMV IE1 mRNA expression, we extracted mRNA using an RNeasy Mini kit (Qiagen) from the cell pellets described above. To remove contaminating DNA, we treated aliquots of 8 μl of the final mRNA suspension with 1 μl of RQ1 RNase-Free DNase (Promega) in the same volume of RQ1 RNase-Free DNase 10× reaction buffer (Promega) for 30 min at 37°C. DNase was inactivated as described above. The RT reaction mixture contained 2 μl of 10× Buffer RT (Qiagen), 2 μl of the dNTPs (5 mM; Qiagen), 2 μl oligo(dT) primer (10 pmol/μl), 1 μl of RNase inhibitor (10 units/μl; TaKaRa, Shiga, Japan), 1 μl of Omniscript reverse transcriptase (Qiagen), and 5 μl of mRNA template in a final volume of 20 μl. RT was performed for 60 min at 37°C, and PCR was performed as described above.
FIG. 4.
Confirmation of protein expression, DNA incorporation, and mRNA expression of the CMV IE1 vector in the 293 cell line. (a) Protein expression of CMV IE1 from the transiently transfected 293 cell line. Lane M, size marker; lane 1, mock-treated 293 cells; lane 2, transiently transfected 293 cell line. (b) PCR amplification of genomic DNA of the new cell line. (c) PCR and RT-PCR amplification of the CMV IE1 gene using mRNA extraction solution from the new cell line. Lane M, 100-bp size marker; lane 1, 293 cell line stably expressing IE1; lane 2, 293 cell line; lane 3, negative control (distilled water); lane 4, positive control (CMV IE1 expression vector).
Test of the activation of HAdV replication in the new cell line.
Both 293 and 293-CMV cells were seeded into six-well tissue culture plates and incubated for 24 h before infection. After cells were inoculated with 5 × 103 genomic copies of HAdVs (laboratory-adapted HAdV-41; two clinical stool samples) in 500 μl, the appearance of CPE was checked daily for 1 week and compared to mock-infected cells. After 72 h postinfection (p.i.), both 293 and 293-CMV cells infected with HAdVs were harvested and fixed with 2.5% glutaraldehyde in 0.1 M phosphate (pH 7.4). Then, viral particles were observed with a transmission electron microscope (TEM; JEOL, Tokyo, Japan) at the Clinical Research Institute of Seoul National University Hospital. In addition, viral DNA was extracted after 3 days using a DNeasy Blood and Tissue kit (Qiagen), and viral genomic copy numbers were quantified by real-time TaqMan PCR as described above (17).
Statistical analysis.
Both transcript levels and copy numbers were analyzed statistically using a paired t test using SAS Enterprise Guide 4 (SAS Inc., Cary, NC).
RESULTS
Transactivation of the promoters of HAdVs by transient transfection.
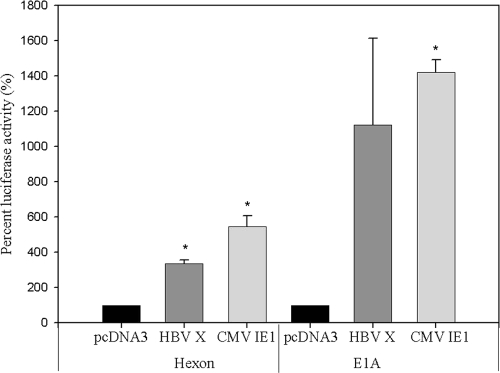
To determine the transcriptional levels of HAdV promoters by viral transactivator proteins (CMV IE1 protein), we performed a transient transfection assay using the constructed plasmids. The results are summarized in Fig. 2. In addition to the CMV IE1 expression vector, the HBV X gene was tested because it encodes another viral transactivator protein. Either the E1A or hexon promoter construct was cotransfected with the transactivator expression vector into 293 cells. Luciferase reporter gene expression was used to measure the contributions of the viral proteins to the transactivation of the HAdV genes (E1A and hexon). As shown in Fig. 2, both HBV X and CMV IE1 proteins were able to transactivate the HAdV hexon gene promoter by, on average, 338% and 547%, respectively (P = 0.007 and P = 0.018, respectively). E1A promoter showed 3-fold greater activation than the hexon promoter by the two transactivator proteins, with average values of 1,121% and 1,421%, respectively (P = 0.174 and P = 0.003, respectively). These results suggest that viral transactivators, including HBV X and CMV IE1 proteins, can increase the transcription of essential genes of HAdVs.
FIG. 2.
Transcriptional activation by HBV X and CMV IE1 expression vector in 293 cells. Transcriptional levels of HAdV genes were monitored by luciferase enzyme activity. Data indicate the percent luciferase activity for HBV X and CMV IE1 compared with the pcDNA control vector. Both hexon and E1A promoter showed increased transcriptional levels relative to the control DNA (pcDNA 3 vector) by as much as 307 to 632% and 437 to 2,078%, respectively. The level of luciferase activity with control DNA was set at 100%, and others are shown relative to this level. Each experiment was repeated three times in duplicate. Error bars indicate the standard error. *, P < 0.05 compared to pcDNA vector control.
Increases in HAdV-41 mRNA and DNA levels by CMV IE1 in 293 cells.
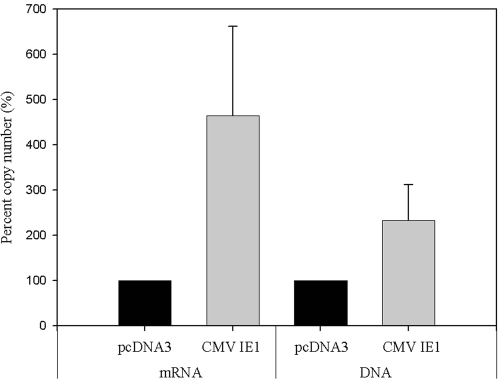
HAdV-41 was inoculated into 293 cells with and without transient transfection of the CMV IE1 expression vector. During the transient transfection assay, the expression of CMV IE1 protein was confirmed by Western blotting (see Fig. 4a). When the levels of viral mRNA and DNA were quantified by real-time TaqMan RT-PCR, the levels of viral mRNA in 293 cells transfected with CMV IE1 expression vector were more than four times higher than levels in control 293 cells (P = 0.206) (Fig. 3). Additionally, adenoviral DNA in 293 cells transfected with CMV IE1 expression vector was more than double that in control 293 cells (P = 0.341). These observations suggest that CMV IE1 protein increases both mRNA expression and DNA replication of HAdV in 293 cells.
FIG. 3.
Increments of viral RNA and DNA levels by CMV IE1 in 293 cells. The 293 cells were transiently transfected with either pcDNA3 (control) or CMV IE1 expression vector and then infected with HAdV-41. After 3 days of inoculation, 293 cells were harvested, and total RNA and DNA were extracted and quantified by real-time TaqMan RT-PCR assays. The copy number with control DNA was set at 100%, and others values are shown relative to this level. These mRNA and DNA experiments were repeated three and two times in duplicate, respectively. Error bars indicate the standard error.
Construction and confirmation of 293 cell line expressing CMV IE1.
As the vector expressing CMV IE1 contains the neomycin resistance gene, cells with this vector integrated into the chromosome were selected by cultivation in medium containing G418. To confirm the incorporation of the viral vector, we isolated the genomic DNA of cell clones and amplified it by PCR with primers for CMV IE1 (Fig. 4b). The neomycin-resistant 293 cells contained the vector in their chromosome. In addition, CMV IE1 mRNA expression was confirmed by RT-PCR using mRNA cell extracts (Fig. 4c). Viral DNA was not detected in mRNA extracts analyzed by PCR. These results indicate that the CMV IE1 expression vector was incorporated into the chromosome of 293 cells, and mRNA was expressed appropriately in these cells.
Cultivation and replication of HAdVs in 293-CMV cells.
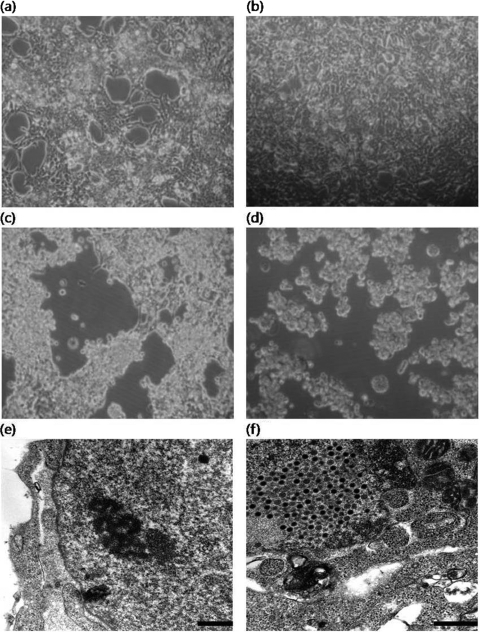
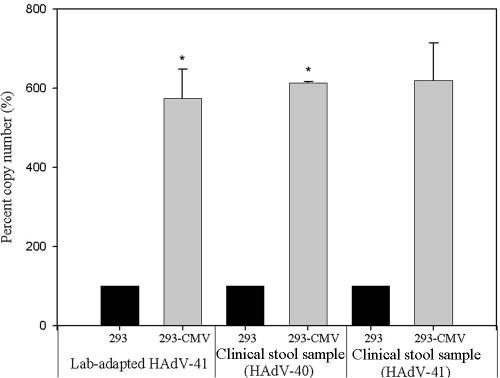
Figure 5 summarizes the cultivation of HAdV-41 in both 293 and 293-CMV cells. The 293 cells began to detach from the surface of the flask at 6 days p.i. Under the same experimental conditions, complete CPE was observed in 293-CMV cells at 6 days p.i. (Fig. 5d). At 72 h p.i., a number of viral particles were observed in the nucleus in 293-CMV cells (Fig. 5f). However, no clear sign of HAdV was observed in 293 cells (Fig. 5e). In addition to the appearance of CPE, three different HAdVs were inoculated into both the 293 and 293-CMV cell lines, and viral replication was measured by real-time TaqMan PCR assay. The viral DNA of laboratory-adapted HAdV-41 and clinical HAdV-40 and HAdV-41 in new 293-CMV cells increased by 574 to 619% in comparison with 293 cells (P = 0.023, P = 0.003, and P = 0.115, respectively) (Fig. 6). These data suggest that HAdVs show better replication in 293-CMV cells than in parental 293 cells.
FIG. 5.
HAdV multiplication in both 293 and 293-CMV cell lines. Cell morphology by inverted microscope is shown as follows (magnification, ×100): 293 cell line without infection (a), 293-CMV cell line without infection (b), 293 cell line at 6 days p.i. with HAdV-41 (c), and 293-CMV cell line at 6 days p.i. with HAdV-41 (d). Transmission electronic microscope sections are shown as follows: 293 cell line at 3 days p.i. with HAdV-41 (magnification, ×40,000; bar, 500 nm) (e) and 293-CMV cell line at 3 days p.i. with HAdV-41 (magnification, ×50,000; bar, 500 nm) (f).
FIG. 6.
Confirmation of increased viral replication in the newly constructed cell line. Three different HAdVs were inoculated into both 293 cells and the new cell line. Viral DNA was amplified using real-time TaqMan PCR at 3 days p.i. The experiments were repeated three times in duplicate for laboratory-adapted HAdV and twice in duplicate for clinical stool samples. Error bars indicate the standard error. *, P < 0.05 compared to the 293 control cell line.
DISCUSSION
The results of the present study indicate that viral transactivators such as CMV IE1 protein can activate the expression of essential genes of HAdVs and promote the replication of HAdVs. In addition, our newly constructed cell line, 293-CMV, may be useful for culturing and analyzing fastidious HAdVs. The fastidious characteristics of HAdVs are thought to be due to the antiviral effect of IFN, the inability of the E1A gene product to transactivate other essential genes, and a block in the release of progeny viruses (4, 32, 36). A recent study by Sherwood et al. demonstrated that the replication of HAdVs can be promoted by reducing the IFN response by the expression of simian virus 5 (SV5) T antigen (32). Cells with SV5 T antigen showed an increase in the titer of HAdV-40 by 1 log10 compared to conventional 293 cells. Our study demonstrates that the addition of viral transactivators such as CMV IE1 can activate the promoters of E1A and hexon genes and promote replication to a greater degree than blocking IFN. The increased DNA replication of HAdVs can overcome the shortcoming of cultivation of HAdVs by reducing the lag time required.
A recent study also indicated that among the cell lines in common use, the 293 cell line is the most suitable for culturing HAdVs (12). Because of its expression of E1A from HAdV-5, the 293 cell line has been used for the propagation of HAdVs (14). However, HAdV-40 and HAdV-41 replicate very slowly or do not show clear CPE on many other cell lines, such as BGMK. Because of the suitability of 293 cells for detecting HAdVs, 293 cells were also used in previous studies with SV5 T antigen. Other cell lines, such as A549 and PLC/PRF5, could be tested. PLC/PRF5 is a human hepatoma cell line transformed by hepatitis B virus (9). These cells express HBV antigen. Our study also demonstrated that some HBV transactivating proteins, such as HBV X, can transactivate the promoters of HAdV genes. HBV X protein in PLC/PRF5 cells is likely to help the replication of HAdVs. In addition to PLC/PRF5 cells, a plaque assay using A549 cells, a human lung carcinoma cell line, has been developed recently (7, 11). It is believed that the use of this newly developed cell line will improve the cultivation of fastidious HAdVs.
Initially, both HBV X and CMV IE1 proteins were examined by transient transfection assays. Both of these proteins can activate the transcription of HAdVs. As CMV protein has a greater capacity for the activation of HAdVs, 293 cells were permanently transformed with the CMV IE1 expression vector. CMV major IE1 and IE2 proteins are divided into several coding strategies (IEP72 and IEP86), and these proteins are capable of transcriptional control (22). Many studies have indicated the transactivating function of human CMV (HCMV) through binding to specific TATA elements and specific binding sites (2, 3, 15, 29). These proteins can activate multiple viral and cellular genes (28, 29), express cytokines and chemokines (34), and increase cellular DNA synthesis, including induction of various enzymes (25). Generally, HCMV inhibits major histocompatibility complex class II (MHC-II) expression and IFN regulatory factor 1 regulation in human endothelial cells and fibroblasts (24). HCMV IE1 protein can inhibit STAT1/STAT2 signaling (27). In fact, inhibition of STAT1 activity improves the replication of HAdVs (21). In addition, CMV IE1 protein antagonizes IFN-stimulated antiviral cellular gene expression (27). Reduction of the antiviral response is crucial for productive viral replication, including that of CMV (27). These characteristics can provide newly constructed 293-CMV cell lines capable of high-efficiency multiplication of fastidious HAdVs.
A number of methods for detecting HAdVs have been developed to date, including analytical methods for infectious HAdVs, such as the rapid culture centrifugation method (8), integrated cell culture-PCR (5, 6), mRNA conventional (18) or real-time RT-PCR (17), and the rapid shell vial culture technique (38). These methods are not capable of detecting infectious virus rapidly because susceptible cells are not available, and secondary confirmation tests are typically required. The rapid culture centrifugation method shows only a slight difference (1%) from conventional culture, and additional serotyping or a genus-specific enzyme-linked immunosorbent assay (ELISA) is still required. The mRNA RT-PCR method was targeted on mRNA as an index of infectious HAdVs (17, 18). These previous studies were able to rapidly detect infectious HAdV by targeting viral mRNA that was produced by infectious virus during multiplication. It can be useful because the mRNA of HAdVs could be detected at 6 h p.i. for high levels of virus (106 infectivity units) and as soon as 3 days p.i. for low levels of virus (5 infectious units). By combining our new cell line with previous mRNA RT-PCR assays, the sensitivity would likely be improved, and the analysis time would be reduced for the detection of low concentrations of infectious HAdV. Further study is warranted.
In the present study, we used stool specimens from diarrheal patients. The isolates of HAdV-40 and HAdV-41 in the present study grew approximately six times better in 293-CMV cells than in 293 cells. These observations suggest that our cell line is a robust host system for detecting various HAdVs. One of the limitations of this study is that only high levels (5 × 103 genomic copies) of HAdVs were tested. Low levels (<10 genomic copies) of viral inoculation should be tested in the future because HAdV is commonly present at low concentrations in most environmental samples. In conclusion, we have demonstrated that viral transactivators can significantly promote the replication of HAdVs. Our new cell line could be applicable in other HAdV-related research, such as studies regarding HAdV-based recombinant vectors.
Acknowledgments
This study was partially supported by a grant from American Water Works Research Foundation (AWWARF) and National Research Foundation of Korea funded by the Korean Government (MEST) (2009-0080246).
We thank Sunyoung Kim at Seoul National University and Shinko Takada at University of Texas MD Anderson Cancer Center for kindly providing CMV IE1 and HBV X expression vectors, respectively. We also thank Mark D. Sobsey at the University of North Carolina at Chapel Hill for helpful advice on this study.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Albert, M. J. 1986. Enteric adenoviruses. Arch. Virol. 88:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry, P. A., E. Pratt-Lowe, B. M. Peterlin, and P. A. Luciw. 1990. Cytomegalovirus activates transcription directed by the long terminal repeat of human immunodeficiency virus type 1. J. Virol. 64:2932-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biegalke, B. J., and A. P. Geballe. 1991. Sequence requirements for activation of the HIV-1 LTR by human cytomegalovirus. Virology 183:381-385. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M., H. L. Wilson-Friesen, and F. Doane. 1992. A block in release of progeny virus and a high particle-to-infectious unit ratio contribute to poor growth of enteric adenovirus types 40 and 41 in cell culture. J. Virol. 66:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo, Y. J., and S. J. Kim. 2006. Detection of human adenoviruses and enteroviruses in Korean oysters using cell culture, integrated cell culture-PCR, and direct PCR. J. Microbiol. 44:162-170. [PubMed] [Google Scholar]
- 7.Cromeans, T. L., X. Lu, D. D. Erdman, C. D. Humphrey, and V. R. Hill. 2008. Development of plaque assays for adenoviruses 40 and 41. J. Virol. Methods 151:140-145. [DOI] [PubMed] [Google Scholar]
- 8.Durepaire, N., S. Ranger-Rogez, and F. Denis. 1996. Evaluation of rapid culture centrifugation method for adenovirus detection in stools. Diagn. Microbiol. Infect. Dis. 24:25-29. [DOI] [PubMed] [Google Scholar]
- 9.Grabow, W. O. K., D. L. Puttergill, and A. Bosch. 1992. Propagation of adenovirus type 40 and 41 in the PLC/PRF/5 primary liver carcinoma cell line. J. Virol. Methods 37:201-208. [DOI] [PubMed] [Google Scholar]
- 10.Gruda, M. C., J. M. Zabolotny, J. H. Xiao, I. Davidson, and J. C. Alwine. 1993. Transcriptional activation by simian virus 40 large T antigen: interactions with multiple components of the transcription complex. Mol. Cell. Biol. 13:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto, S., N. Sakakibara, H. Kumai, M. Nakai, S. Sakuma, S. Chiba, and K. Fujinaga. 1991. Fastidious human adenovirus type 40 can propagate efficiently and produce plaques on a human cell line, A549, derived from lung carcinoma. J. Virol. 65:2429-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, J. W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaykus, L. A. 1997. Epidemiology and detection as options for control of viral and parasitic foodborne disease. Emerg. Infect. Dis. 3:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, S. C. 2006. Human adenoviruses in water: occurrence and health implications: a critical review. Environ. Sci. Technol. 40:7132-7140. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. M., Y. Hong, K. T. Jeang, and S. Kim. 2000. Transactivation activity of the human cytomegalovirus IE2 protein occurs at steps subsequent to TATA box-binding protein recruitment. J. Gen. Virol. 81:37-46. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. M., Y. Hong, S. Kim, M. H. Cho, M. Yoshida, K. T. Jeang, and W. Burns. 1999. Sequences downstream of the RNA initiation site of the HTLV type I long terminal repeat are sufficient for trans-activation by human cytomegalovirus immediate-early proteins. AIDS Res. Hum. Retroviruses 15:545-550. [DOI] [PubMed] [Google Scholar]
- 17.Ko, G., N. Jothikumar, V. R. Hill, and M. D. Sobsey. 2005. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods 127:148-153. [DOI] [PubMed] [Google Scholar]
- 18.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2003. Detection of infectious adenovirus in cell culture by mRNA reverse transcription-PCR. Appl. Environ. Microbiol. 69:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, H. K., and Y. S. Jeong. 2004. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl. Environ. Microbiol. 70:3632-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locas, A., C. Barthe, A. B. Margolin, and P. Payment. 2008. Groundwater microbiological quality in Canadian drinking water municipal wells. Can. J. Microbiol. 54:472-478. [DOI] [PubMed] [Google Scholar]
- 21.Look, D. C., W. T. Roswit, A. G. Frick, Y. Gris-Alevy, D. M. Dickhaus, M. J. Walter, and M. J. Holtzman. 1998. Direct suppression of Stat1 function during adenoviral infection. Immunity 9:871-880. [DOI] [PubMed] [Google Scholar]
- 22.Lukac, D. M., J. R. Manuppello, and J. C. Alwine. 1994. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J. Virol. 68:5184-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mautner, V., A. Bailey, V. Steinthorsdottir, R. Ullah, and A. Rinaldi. 1999. Properties of the adenovirus type 40 E1B promoter that contribute to its low transcriptional activity. Virology 265:10-19. [DOI] [PubMed] [Google Scholar]
- 24.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, Y. B., Y. H. Cho, Y. M. Jee, and G. Ko. 2008. Immunomagnetic separation combined with real-time reverse transcriptase PCR assays for detection of norovirus in contaminated food. Appl. Environ. Microbiol. 74:4226-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. U. S. A. 103:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 80:3872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2007. The autoregulatory and transactivating functions of the human cytomegalovirus IE86 protein use independent mechanisms for promoter binding. J. Virol. 81:5807-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed, L. J., and H. A. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 27:493-497. [Google Scholar]
- 31.Rodriguez, R. A., P. M. Gundy, and C. P. Gerba. 2008. Comparison of BGM and PLC/PRC/5 cell lines for total culturable viral assay of treated sewage. Appl. Environ. Microbiol. 74:2583-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherwood, V., H. G. Burgert, Y. H. Chen, S. Sanghera, S. Katafigiotis, R. E. Randall, I. Connerton, and K. H. Mellits. 2007. Improved growth of enteric adenovirus type 40 in a modified cell line that can no longer respond to interferon stimulation. J. Gen. Virol. 88:71-76. [DOI] [PubMed] [Google Scholar]
- 33.Shinozaki, T., K. Araki, Y. Fujita, M. Kobayashi, T. Tajima, and T. Abe. 1991. Epidemiology of enteric adenoviruses 40 and 41 in acute gastroenteritis in infants and young children in the Tokyo area. Scand. J. Infect. Dis. 23:543-547. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, R. S., M. J. Tymms, L. H. McKinlay, M. F. Shannon, A. Seth, and I. Kola. 1997. ETS1, NF-κB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene 14:2845-2855. [DOI] [PubMed] [Google Scholar]
- 36.Tiemessen, C. T., and A. H. Kidd. 1993. Sensitivity of subgroup F adenoviruses to interferon. Arch. Virol. 128:1-13. [DOI] [PubMed] [Google Scholar]
- 37.Twu, J. S., and W. S. Robinson. 1989. Hepatitis B virus X gene can transactivate heterologous viral sequences. Proc. Natl. Acad. Sci. U. S. A. 86:2046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Doornum, G. J. J., and J. C. De Jong. 1998. Rapid shell vial culture technique for detection of enteroviruses and adenoviruses in fecal specimens: comparison with conventional virus isolation method. J. Clin. Microbiol. 36:2865-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Loon, A. E., P. Gilardi, M. Perricaudet, T. H. Rozijn, and J. S. Sussenbach. 1987. Transcriptional activation by the E1a regions of adenovirus types 40 and 41. Virology 160:305-307. [DOI] [PubMed] [Google Scholar]
- 40.Yaginuma, K., I. Nakamura, S. Takada, and K. Koike. 1993. A transcription initiation site for the hepatitis B virus X gene is directed by the promoter-binding protein. J. Virol. 67:2559-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]