Abstract
The acid tolerance response (ATR) is one of the major virulence traits of Streptococcus mutans. In this study, the role of GlnR in acid-mediated gene repression that affects the adaptive ATR in S. mutans was investigated. Using a whole-genome microarray and in silico analyses, we demonstrated that GlnR and the GlnR box (ATGTNAN7TNACAT) were involved in the transcriptional repression of clusters of genes encoding proteins involved in glutamine and glutamate metabolism under acidic challenge. Reverse transcription-PCR (RT-PCR) analysis revealed that the coordinated regulation of the GlnR regulon occurred 5 min after acid treatment and that prolonged acid exposure (30 min) resulted in further reduction in expression. A lower level but consistent reduction in response to acidic pH was also observed in chemostat-grown cells, confirming the negative regulation of GlnR. The repression by GlnR through the GlnR box in response to acidic pH was further confirmed in the citBZC operon, containing genes encoding the first three enzymes in the glutamine/glutamate biosynthesis pathway. The survival rate of the GlnR-deficient mutant at pH 2.8 was more than 10-fold lower than that in the wild-type strain 45 min after acid treatment, suggesting that the GlnR regulon participates in S. mutans ATR. It is hypothesized that downregulation of the synthesis of the amino acid precursors in response to acid challenge would promote citrate metabolism to pyruvate, with the consumption of H+ and potential ATP synthesis. Such regulation will ensure an optimal acid adaption in S. mutans.
Streptococcus mutans is the principal etiologic agent of human dental caries (40). The development of dental caries is closely associated with bacterial sugar metabolism, mainly acid production. The major virulence factors associated with the cariogenicity of S. mutans are its abilities to produce acid efficiently via carbohydrate metabolism (acidogenicity) and to tolerate acid stress in dental plaque (aciduricity) (35). The inherent tolerance of S. mutans and the inducible adaptive response upon exposure to sublethal acidic pHs are important characteristics of the acid tolerance response (ATR) in S. mutans (56). A number of mechanisms contributing to the aciduric response have been identified in S. mutans (35, 36). The membrane-bound F-ATPase (H+-translocating ATPase) is the primary factor or determinant in maintaining a cytoplasmic pH more alkaline than the extracellular environment (35). It has been suggested that the agmatine deiminase system (AgDS), an analog of the arginine deiminase system (ADS) in Streptococcus rattus and Streptococcus gordonii (11), can provide competitive fitness for S. mutans via the production of ammonia and ATP to increase the cytoplasmic pH (23, 24). Another factor contributing to acid tolerance is malolactic fermentation, in which the conversion of the dicarboxylic l-malate to the monocarboxylic lactic acid and CO2 increases the cytoplasmic pH (52). In addition to the above mechanisms, induction of stress regulons, alterations in the cell membrane composition, and activation of various two-component signal transduction systems (TCSs) and global regulators also participate in controlling the acid adaptation of S. mutans (21, 35, 36, 50). Specifically, a recent study by Gong et al. demonstrates that upregulation of five TCSs, including CiaHR, LevSR, ScnKR, HK/RR1037/1038, and ComDE, is essential for an optimal ATR in S. mutans (21), indicating that a complicated signal transduction network is required for acid adaption. On the other hand, inactivation of vicK, encoding the sensor kinase of the VicRKX TCS, results in impaired acid production and increased survival at pH 3.5, suggesting that the Vic system is essential for maintenance of intracellular pH homeostasis in S. mutans (50).
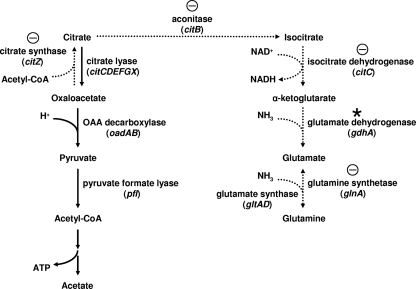
S. mutans can cope with acid stress by modulating its metabolic networks (14, 35, 36, 38). We have demonstrated previously that the expression of citBZC is repressed 10-fold after exposure to pH 5.5 for 30 min (14), suggesting that downregulation of the citBZC operon is part of the ATR. The enzymes encoded by citBZC, aconitase, citrate synthase, and isocitrate dehydrogenase, together with pyruvate dehydrogenase can drive the conversion of pyruvate to α-ketoglutarate (Fig. 1) (15), providing the de novo carbon skeleton for glutamate and glutamine. The link between metabolism and stress responses is well documented in Bacillus subtilis. Nitrogen assimilation in B. subtilis is controlled by GlnR and TnrA, both of which belong to the MerR family (20, 47). GlnR is a global regulator that represses the expression of glnRA (20), the urease operon (10, 64), and tnrA (20) under excess nitrogen conditions, whereas TnrA regulates the expression of its own gene and additional genes under nitrogen-limiting conditions (20). The GlnR regulon, but not the TnrA regulon, has been identified in S. mutans (17). Gram-positive bacteria that lack TnrA generally use GlnR to carry out some of the global roles that are assumed by TnrA in B. subtilis (53).
FIG. 1.
GlnR-repressed gene clusters and metabolic pathways at acidic pH. The reactions that were repressed at acidic pH are indicated by dotted arrows, and the genes that were negatively regulated by GlnR are indicated by the circled minus signs. The slight repression of gdhA at acidic pH was observed in the microarray (0.58- to 0.75-fold; data not shown) and is indicated by an asterisk. The genes encoding the enzymes are shown in parentheses. Abbreviations: acetyl-CoA, acetyl coenzyme A; OAA, oxaloacetate.
The response of genes in transcriptomes to acidification have been analyzed by microarray technology in pathogens, such as Helicobacter pylori (6), Mycobacterium tuberculosis (19), Streptococcus pneumoniae (41), and most recently, S. mutans (21). In all of these organisms, acid resistance is a critical factor in their pathogenesis. To date, microarrays have been used to analyze the global responses of S. mutans to various environmental challenges (3, 5, 7, 21, 44), whether in a biofilm life style (51) or with specific gene mutations (1, 2, 37, 42, 44, 58, 61). In this study, we analyzed the transcription profile of the adaptive ATR in S. mutans using a whole-genome microarray. In silico promoter analyses revealed a GlnR box, ATGTNAN7TNACAT (17), located in the 5′ untranslated regions (5′ UTRs) of several genes, specifically acid-repressed genes in clusters. The effects of GlnR and the GlnR box in pH regulation were evaluated further.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. mutans GS-5 and its derivatives were routinely grown in brain heart infusion (BHI) or Todd-Hewitt (TH) (Difco) broth at 37°C in a 5% CO2 atmosphere. Erythromycin (Em) at 10 μg ml−1, kanamycin (Km) at 500 μg ml−1, and spectinomycin (Spe) at 500 μg ml−1 were included, where indicated, in the culture media to maintain mutant strains. Recombinant Escherichia coli strains were grown in Luria-Bertani broth supplemented with ampicillin (Amp) at 100 μg ml−1, Km at 50 μg ml−1, and Spe at 100 μg ml−1 as needed. The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotypea | Description or relevant characteristic | Source |
|---|---|---|---|
| S. mutans strains | |||
| GS-5 | GlnR+ | Wild-type strain | H. K. Kuramitsu |
| ΔglnR mutant | Emr GlnR− | GS-5 strain lacking glnR | This study |
| CglnRΔglnR | Emr Sper GlnR+ | ΔglnR complementation strain | This study |
| Cit1-luc | Kmr GlnR+ | GS-5 chromosome harboring a single copy of pcit1-luc | This study |
| Cit2-luc | Kmr GlnR+ | GS-5 chromosome harboring a single copy of pcit2-luc | This study |
| Cit3-luc | Kmr GlnR+ | GS-5 chromosome harboring a single copy of pcit3-luc | This study |
| luc | Kmr GlnR+ | GS-5 chromosome harboring a single copy of the luc gene | This study |
| Plasmids | |||
| pDL277 | Sper | E. coli-Streptococcus shuttle vector | LeBlanc et al. (34) |
| pDL277-glnR | Sper | pDL277 harboring glnR | This study |
| pMC340B | Emr Kmr | Integration vector for S. mutans | Y.-Y. M. Chen |
| pMC340Bluc | Emr Kmr | pMC340B containing promoterless luc gene | This study |
| pcit1-luc | Emr Kmr | pMC340Bluc with 254-bp DNA fragment of the citB promoter | This study |
| pcit2-luc | Emr Kmr | pMC340Bluc with 101-bp DNA fragment of the citB promoter | This study |
| pcit3-luc | Emr Kmr | pMC340Bluc with 75-bp DNA fragment of the citB promoter | This study |
The r superscript indicates resistance to the drugs (erythromycin [Em], spectinomycin [Spe], and kanamycin [Km]).
The innate ATR in early-exponential-phase cells was evaluated by the method of Svensater et al. (56) with some modifications. Briefly, overnight cultures in BHI were diluted at 1:10 in fresh BHI (pH 7.5) and grown to an optical density at 600 nm (OD600) of 0.4. The cells were harvested by centrifugation and concentrated in 1/5 of the original culture volume in fresh BHI or BHI with HCl (BHI-HCl) (pH 5.5) and incubated for an additional 5 or 30 min for short and long acid challenges, respectively, prior to further manipulations (14, 57).
To evaluate pH-dependent regulation, continuous cultures were grown in a chemostat system (Sartorius) in tryptone-yeast extract (TY) (3% tryptone and 0.5% yeast extract) medium supplemented with 20 mM glucose at a dilution rate (D) of 0.3 h−1. The cultures were maintained at pH 7.5 or pH 5.5 by the addition of 2 N KOH. The cells were grown for a minimum of 10 generations to achieve the steady state at each condition. Steady-state cultures were collected and quickly frozen in liquid nitrogen.
General genetic manipulations.
DNA extraction, PCR, and reverse transcription-PCR (RT-PCR) analysis were performed as described previously (13, 14). Primers used in this study are listed in Table S1 in the supplemental material. Automated DNA sequencing (ABI) was performed by the Molecular Biology Facility of the National Taiwan University. All sequencing was performed in both directions. Total RNA was isolated as described previously (13) and further purified by using the RNeasy minikit (Qiagen).
The transcription initiation site (TIS) of citB was mapped by primer extension analysis. Briefly, 5 μg of total RNA was hybridized with [γ-32P]ATP-labeled primer citBR49 at 58°C for 20 min, and cDNA was generated at 42°C with avian myeloblastosis virus (AMV) reverse transcriptase (Promega). A DNA sequencing reaction mixture using the same primer was included on the gel as a marker to facilitate identification of the start sites.
Construction of glnR mutant and complementation strain.
All references to genomic loci are based on entries in the S. mutans UA159 genome database (http://www.oralgen.lanl.gov). The glnR gene in S. mutans GS-5 was deleted by PCR ligation mutagenesis (33). Briefly, a fragment of 704 bp 5′ to glnR and a fragment of 544 bp 3′ to glnR were PCR amplified from S. mutans GS-5 by using primer pairs IGR293F286-glnRR3 and glnRF352-glnAR484, respectively. The PCR products were digested with EcoRI and ligated with an erm cassette from Tn917 (48) without a promoter and a transcription terminator. The ligation mixture was used to transform strain GS-5 to generate a nonpolar mutation in glnR. The resulting isogenic strain (ΔglnR) harbored a deletion of the glnR locus with the first 3 bp and the last 21 bp of glnR remaining. The correct configuration of allelic replacement was verified by PCR and Southern blot hybridization. The expression of 3′ flanking gene (glnA) of glnR was examined by RT-PCR to confirm the nonpolar effect of the mutation.
A DNA fragment containing the 81-bp region immediately 5′ to the translation start codon of GlnR, the intact glnR gene, and the 39-bp region 3′ to the translation stop codon was generated by PCR using primers EcoRI-IGR294F1 and EcoRI-IGR295R39. The PCR product was digested with EcoRI and cloned into E. coli-Streptococcus shuttle vector pDL277 (34) to construct a glnR complementation construct. The recombinant plasmid (pDL277/glnR) was used to transform the ΔglnR mutant to generate the complementation strain CglnRΔglnR. The presence of pDL277/glnR in strain CglnRΔglnR was verified by PCR. The expression of glnR in strain CglnRΔglnR was further confirmed by RT-PCR.
Microarray hybridization and analysis.
The genetic basis of the acid response was analyzed by using the whole-genome S. mutans UA159 microarrays obtained from the Pathogen Functional Genomics Resource Center (PFGRC) at The Institute for Genomic Research (TIGR) (http://pfgrc.tigr.org) (supported by the National Institute of Dental and Craniofacial Research). Analysis of global gene expression was performed using six independent array experiments (i.e., two fluorescent-dye reversal experiments with three independent S. mutans GS-5 cultures). In addition, each S. mutans UA159 microarray slide contained four sets of probes (i.e., four technical replicates) for each gene at different locations. This yielded a total of 24 expression measurements per gene for each pH condition. Experiments were performed by using protocols from PFGRC (http://pfgrc.tigr.org/protocols.shtml) with minor modifications. Briefly, 2 μg of total RNA isolated from early-exponential-phase cells grown in BHI or BHI-HCl was used to set up standard reverse transcription reactions using pd(N)6 random hexamer (Amersham) and SuperScript indirect cDNA labeling system (Invitrogen) to generate cDNA. The cDNA was purified and indirectly labeled with Cy3 or Cy5 monoreactive dye (Amersham). Reaction mixtures were combined, and the unincorporated dye was removed according to the manufacturer's specifications. The labeled cDNA was vacuum concentrated without heat before it was dissolved in 45 μl of hybridization buffer (50% formamide, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, and 300 μg sheared salmon sperm DNA). The probe was heated at 95°C for 5 min before it was placed on a prehybridized array enclosed in a prewarmed array hybridization chamber (Genetech). After an 18-h hybridization at 42°C, the array was washed once with 2× SSC plus 0.1% SDS for 10 min at 55°C, once with 0.1× SSC plus 0.1% SDS for 10 min at 25°C, and then twice with 0.1× SSC for 5 min each time at 25°C. The slide was rinsed with distilled water (dH2O) for 5 min and dried. The microarray images were scanned and analyzed using a flat scanner (PowerLook 3000; UMAX, Taiwan) and GenePix 3.0 software (Axon). The brightest spots on each array were used to establish a scan setting (80% of signal maximum) for each individual array. Photomultiplier tube gain and laser settings were varied from array to array to achieve this setting. The data were analyzed using the Spotfire Pro version 7.1 software program (trimmed method and trimmed value of 5%; SpotFire, Inc.). The actual signal intensity was calculated by subtracting the local mean background intensity of each spot from the mean signal intensity of each spot. Data were normalized per spot and per chip using Lowess transformation. Each spot represents the average of four experiments. The Student t test was then applied to the data which could find genes with highly reproducible gene expression. Differentially expressed genes were defined as genes in which the normalized ratios were greater than 2-fold (pH 7.5/pH 5.5 for acid-repressed genes or pH 5.5/pH 7.5 for acid-enhanced genes) with a P value of <0.01. Genes with a P value of >0.01 were excluded from consideration.
Searching for regulatory motifs in the upstream regions of transcriptionally altered genes.
The 200-nucleotide (nt) regions 5′ to the predicted start codon of the first gene of each operon were searched for potential regulatory motifs with the MEME tool (8). To assign genes to operons, the gene organization on the TIGR web page was analyzed, based on a previously published method (18) and using STRING (60). Two contiguous genes were considered part of the same operon if they had a confidence value over 75% according to the TIGR web page and/or a STRING score higher than 0.5. When genes were predicted to be part of the same operon, only the upstream region of the first gene of the operon was considered. All sequences that presented in at least two promoter regions of similar controlled genes with P ≤ 2 × 10−3 were analyzed as potential acid-dependent regulatory motifs.
Construction of the integratable, transcriptional luciferase fusion plasmids.
Plasmid pMC340B contains a pACYC184 replicon and an Ωkan insertion (45) flanked by the 5′ flanking region of S. mutans mtlA1 (SMU.1183c) and the 3′ flanking region of the mtlD gene (SMU.1182c). The mtlA1 gene, encoding enzyme II (EII) of the mannitol phosphotransferase system (PTS), and mtlD, encoding mannitol 1-phosphate 5-dehydrogenase, are the first and last genes of the mtl operon, respectively. The design of this integration vector allows the integration of foreign genes into the S. mutans chromosome at the mtl operon with the deletion of the entire mtl operon and concomitant acquisition of a Km-resistant (Kmr) phenotype. Note that Ωkan contains transcription terminators at both ends of the resistance gene, which will prevent any readthrough from the cassette.
Plasmid pMC340Bluc was derived from pMC340B. The promoterless firefly luciferase gene of pGL3 basic vector (Promega) was digested with KpnI and SalI and then ligated to pMC340B digested with KpnI and XhoI. The recombinant plasmid (pMC340Bluc) was used to clone various lengths of the citB upstream region. Briefly, three promoter regions, pcit1, pcit2, and pcit3, were amplified by PCR using primer pairs citBF228-citBR27, citBF75-citBR27, and citBF49-citBR27, respectively. The PCR products were first cloned into pDrive (Qiagen), and the sequences were verified by sequencing analysis. The correct promoter fragments were released from pDrive and cloned into pMC340Bluc at the KpnI site. The orientation of the promoter fragments on pMC340Bluc was verified by PCR, and the correct clones were used to transform S. mutans. Kmr isolates were selected, and the correct integration events in Kmr transformants were verified by colony PCR. The resulting recombinant strains in which promoter regions pcit1, pcit2, and pcit3 had been amplified by PCR were designated Cit1-luc, Cit2-luc, and Cit3-luc, respectively.
Luciferase assays.
Luciferase activity was measured as described previously (22). Briefly, the concentrated cell suspensions in lysis reagent (luciferase assay system; Promega) were subject to mechanical disruption with half of the volume of glass beads (0.1 mm in diameter) (Biospec Products, Bartlesville, OK) in a mini-Beadbeater homogenizer (FastPrep) at speed 4.5 for 25 s. The tubes were chilled on ice for 5 min prior to the second run of homogenization. The homogenizing/cooling cycle was repeated for a total of five times. The total cell lysate was then recovered by centrifugation. To perform luciferase assays, 20 μl of cell extract was added to 100 μl of luciferase assay reagent (Promega). After a 10-s delay, luminescence was quantified in a liquid scintillation counter (Spectra Max Gemini XS; Tekon Technologies) according to the manufacturer's recommendations and expressed as total counts integrated over the first 15-s interval. All reactions were carried out in three replicate experiments. Protein concentrations were determined using the Bio-Rad protein assay reagent with bovine serum albumin (BSA) as the standard. The specific activity was expressed as the counts per mg of total protein.
Acid killing.
The ability of the wild-type and GlnR-deficient strains to resist acid challenge was evaluated by the acid killing assay of Wen and Burne (62). Briefly, overnight cultures of the wild-type, ΔglnR, and CglnRΔglnR strains in TH broth were diluted 1:20 in fresh TH broth and grown to an OD600 of 0.4. The cells were harvested by centrifugation, washed once with 0.1 M glycine buffer (pH 7), and then concentrated in 1/10 of the original culture volume in 0.1 M glycine buffer (pH 2.8). The viability of the culture in pH 2.8 at 15, 30, and 45 min was determined by serial dilution and plating. The survival rate was calculated as a percentage of the viable cells at each time point compared to the number of viable cells prior to acid treatment. For each strain, at least three independent experiments were performed, and all plating was done in three replicate samples.
RESULTS
ATR of S. mutans.
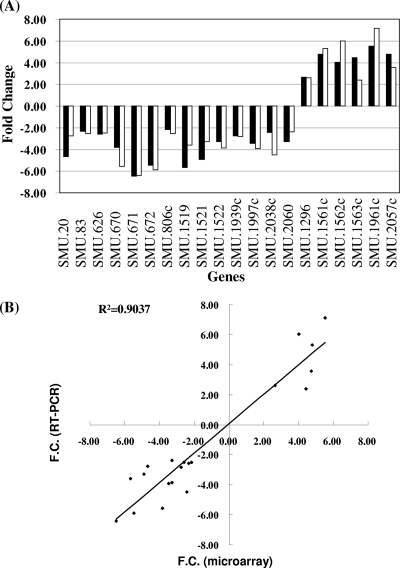
We used a whole-genome microarray to globally examine the alterations in gene expression in S. mutans under acidic stress in this study. Among genes with altered expression levels, 6 upregulated genes and 14 downregulated genes were randomly selected for validation by RT-PCR. The microarray and RT-PCR results were compared (Fig. 2A), and a correlation coefficient of 0.90 between the two sets of data was observed (Fig. 2B), confirming the accuracy of microarray analysis. Among genes altered significantly (P < 0.01) by acid challenge, 43 genes (∼2.1% of total open reading frames [ORFs]) and 55 genes (∼2.8% of total ORFs) were up- and downregulated (see Tables S2 and S3 in the supplemental material), respectively. This result confirmed that acidic pH had a pleiotropic effect on gene expression. Many genes that were known to be upregulated by acid stress were also observed in our transcriptome analysis. These genes included atpABCDEFGH (31, 38, 39, 43, 46), dexB (63), glgD (38), ahpF (63), gloA (38, 63), msmK (63), gtfD (13), hipO (14, 38), and butA (38, 63) (see Table S2 in the supplemental material). Microarray analysis (see Table S3 in the supplemental material) also revealed reduced (0.30- to 0.43-fold) expression of glnA, adhE, manL, and dnaJ, which was in agreement with translation analyses reported previously (38, 39, 63). In agreement with our previous study (14), the levels of expression of citB, citZ, and citC were significantly reduced in bacteria grown at low pH (15 to 26% of the levels of expression in bacteria grown at neutral pH). Note that a general downregulation of several genes encoding proteins involved in amino acid biosynthesis was detected by microarray analysis (see Table S3 in the supplemental material).
FIG. 2.
Comparison of microarray and RT-PCR results. (A) Twenty genes were randomly selected on the basis of microarray analysis, and the levels of expression of each selected gene at neutral and acidic pH values were determined by RT-PCR. The bars represent the change in expression (fold change for expression of the gene in bacteria grown at pH 5.5 compared to expression of the gene in bacteria grown at pH 7.5) analyzed by microarrays (black bars) or RT-PCR (white bars). The numerical ORF designation of each selected gene is indicated. SMU.670, SMU.671, and SMU.672 are citB, citZ, and citC, respectively. (B) Correlation of microarray and RT-PCR assay data. The best-fit line is shown. The two data sets showed a correlation coefficient (R2) of 0.9. F.C., fold change.
Identification of a regulatory motif among acid-repressed genes.
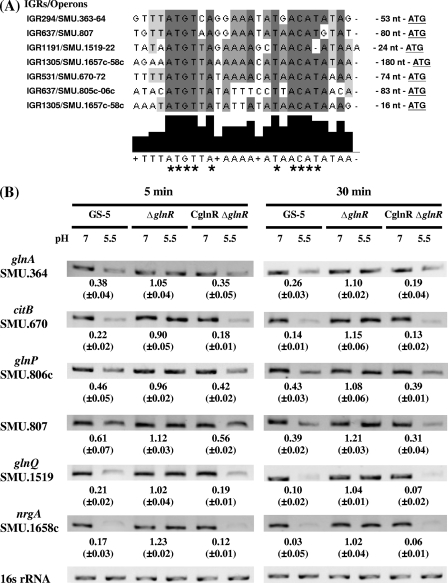
Most of the downregulated loci identified above belonged to the transport and binding proteins of the amino acid biosynthesis and energy metabolism functional classes (Operon Database [http://odb.kuicr.kyoto-u.ac.jp]). A conserved GlnR box sequence with 6-base inverted repeat (IR) spaced by 7 nt (ATGTNAN7TNACAT) (17) was found in the 5′ UTRs of six operons (Fig. 3A). Among these six operons, citBZC and glnRA encode proteins involved in the α-ketoglutarate and glutamate biosynthesis pathway (Fig. 1). glnA (SMU.364) of the glnRA operon encodes glutamine synthetase (GS), which forms glutamine out of glutamate and ammonium at the expense of ATP. The nrgA-glnB(SMU.1658c-SMU.1657c) operon encodes an ammonium transporter and a nitrogen regulatory protein PII homolog, respectively. Of note, two copies of the GlnR box sequence, 180 and 16 bases 5′ to the ATG start codon of nrgA, respectively, were present in IGR1305 (IGR stands for intergenic region). Both glnP-SMU.805c and glnQHMP encode amino acid ABC transporters. SMU.807 is a single locus encoding a putative membrane protein. The GlnR-dependent repression through the conserved GlnR box sequence has been demonstrated in S. pneumoniae (28). To test the hypothesis that the downregulation of these 6 operons in response to acidic pH was mediated by GlnR, the expression of all 6 operons in the wild-type, ΔglnR, and ΔglnR complementation (CglnRΔglnR) strains was examined under both neutral and acidic pHs by RT-PCR. Short (5-min) and long (30-min) acid challenges were performed to determine the kinetics of the response (Fig. 3B). In agreement with the microarray data, the expression of all 6 operons in the wild-type strain, GS-5, was downregulated upon exposure to acid stress, and the repression was more pronounced in cells with 30-min acid treatment. It is noteworthy that the downregulation of genes in the wild-type strain at pH 5.5 was most pronounced for citB, glnQ, and nrgA (Fig. 3B). The repression was abolished in the GlnR-deficient strain (ΔglnR) under both acid exposure treatments. The wild-type levels of expression were restored in the trans-complemented strain (CglnRΔglnR), confirming the negative regulation of GlnR at acidic growth conditions. In addition, the expression of glnA, citB, and SMU.807 was significantly enhanced (P < 0.05) in the ΔglnR strain even at neutral pH (data not shown), suggesting that a negative regulation of these three genes via GlnR also occurred at neutral pH.
FIG. 3.
GlnR was involved in the acid repression of clusters of genes encoding proteins involved in amino acid metabolism in S. mutans. (A) Alignment of the putative GlnR box in the 5′ UTRs of acid-repressed genes encoding proteins involved in amino acid metabolism. The nucleotide sequences of the 5′ UTRs of downregulated genes were aligned using the ClustalW2 multialignment sequence analysis program (http://www.ebi.ac.uk/Tools/clustalw2). Nucleotides that are identical in all sequences are shown on dark gray background in the sequence alignment and indicated by tall black bars below the sequence alignment and by asterisks. Nucleotides that show 50% identity in the nucleotide sequences are shown on light gray background and shorter black bars below the sequence alignment. The intergenic regions (IGRs) and operons are shown to the left of the sequence alignment. The distance (in nucleotides) between the GlnR box and the translational start sites is listed to the right of the sequence alignment. (B) The expression of each gene in the wild-type, ΔglnR, and CglnRΔglnR strains after 5 and 30 min of acid adaptation. Specific primers were used to amplify each gene by RT-PCR. The mean values of the change in expression (fold change for expression of the gene in bacteria grown at pH 5.5 compared to expression of the gene in bacteria grown at pH 7.5) from three independent experiments with three replicate samples in each experiment (n = 9) are depicted below the blots after the intensities of 16S rRNA signal were normalized. When necessary, a higher dilution of cDNA was used in PCRs to avoid the saturation effect. The standard deviations are shown in parentheses.
pH-dependent regulation of GlnR in chemostat cultures.
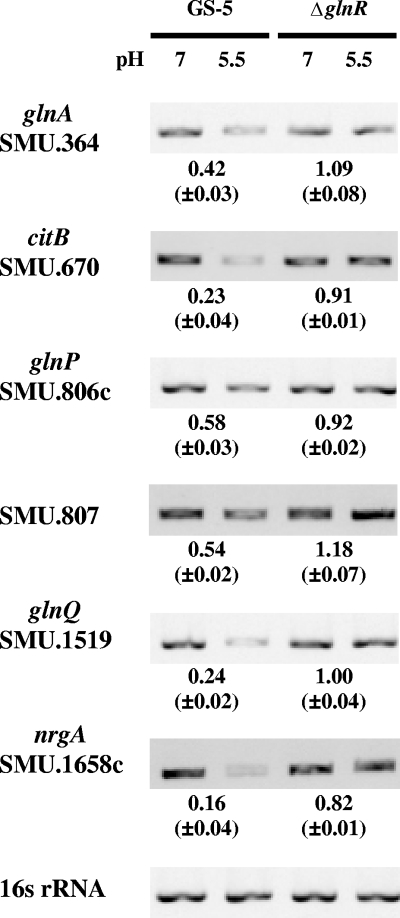
To eliminate the possible effect caused by growth arrest or slow growth under acidic pH, continuous chemostat cultures of wild-type and ΔglnR strains were prepared to evaluate the regulatory function of GlnR at low pH. Similar to the results obtained with batch cultures, the expression of all six operons was downregulated at pH 5.5 in the wild-type strain, and the repression of citB, glnQ and nrgA was most significant as seen in batch cultures (Fig. 4). The levels of repression at acidic pH in chemostat-grown cells were less than those in cells exposed to low-pH challenge (Fig. 3B and 4), indicating that prolonged adaption in acidic pH would result in a different balance in the expression of these genes. The derepression was consistently observed with all six operons in the ΔglnR strain, confirming that the biosynthesis and uptake of amino acids were synchronized by acidic pH.
FIG. 4.
Levels of expression of various clusters of genes encoding proteins involved in amino acid metabolism in chemostat cultures of wild-type S. mutans (GS-5) and ΔglnR mutant. Specific primers for each gene were used in the RT-PCR. The means and standard deviations of the change in expression (fold change for expression of the gene in bacteria grown at pH 5.5 compared to expression of the gene in bacteria grown at pH 7.5) were calculated and listed below the blots as described in the legend to Fig. 3.
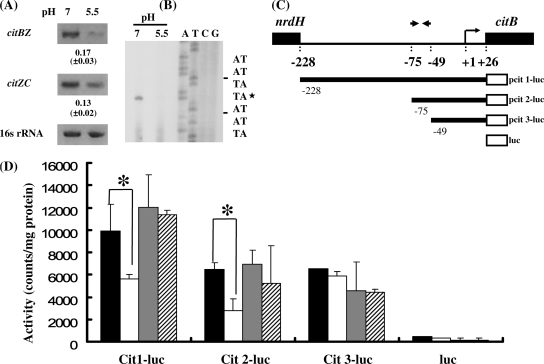
The palindromic sequence is involved in regulation of citBZC at acidic pH.
The expression of the citBZC operon, containing genes encoding the first three enzymes in the glutamine/glutamate biosynthesis pathway (15), was analyzed to further define the possible role of the GlnR box in expression of this pathway (Fig. 1). In agreement with our previous study (14) and microarray analysis (see Table S3 in the supplemental material), RT-PCR confirmed that citBZC was markedly repressed by acid challenge (Fig. 2A). Furthermore, contiguous transcripts were detected between cit genes, suggesting that all three genes were cotranscribed as a polycistronic message (Fig. 5A). The TIS of this operon was mapped to a T residue located 26 nt 5′ of the citB translation start codon by primer extension (Fig. 5B). The intensity of the signal in RNA isolated from cells grown at neutral pH was significantly higher than that from cells exposed to pH 5.5, suggesting that expression of citBZC was predominantly regulated at the transcriptional level.
FIG. 5.
The palindromic sequence was involved in the regulation of citBZC at acidic pH. (A) Expression of the citBZC operon under neutral and acidic pH values. The polycistronic messages of citBZ and citZC were analyzed by RT-PCR with primer pairs SMU.670F-citZR78 and citZF1013-citCR122, respectively. The mean values of the change in expression (fold change for expression of the gene in bacteria grown at pH 5.5 compared to expression of the gene in bacteria grown at pH 7.5) were calculated and listed below the blots as described in the legend to Fig. 3. (B) Primer extension analysis of the citBZC operon. Total RNA was isolated from the wild-type strain (GS-5) exposed to pH 7.5 and pH 5.5 for 30 min. Primer citBR49, 49 bp 3′ to the ATG codon, was used in the cDNA extension and sequence analysis. The transcription initiation site (TIS) (+1) is marked by an asterisk. (C) Diagram of series of amplicons containing various lengths of the upstream region of citB (the chromosomal Cit1-luc to Cit3-luc strains). The relative positions of each promoter-luc fusion, the inverted repeat, +1 site, and ATG start site of citB are indicated. (D) The luciferase activities in the promoter-luc fusion strains. Luciferase activities from GS-5 cells not exposed to pH 5.5 (unadapted) (black bars), GS-5 cells exposed to pH 5.5 for 30 min (white bars), unadapted ΔglnR cells (gray bars), and ΔglnR cells exposed to pH 5.5 for 30 min (hatched bars) are shown. The means plus standard deviations (error bars) for three independent assays are shown. Statistically significant differences (P < 0.05, Student's t test) are indicated by asterisks.
To determine whether the GlnR box located upstream of the citB gene is involved in the regulation of the citBZC operon in response to acidic pH, promoter fragments of various lengths (from various 5′ ends to +27 generated by PCR with the specific primers) were fused with a promoterless firefly luciferase gene to monitor promoter activities in wild-type and ΔglnR strains (Fig. 5C). Both Cit1-luc and Cit2-luc recombinant strains responded to acidic pH repression in the wild-type background (Fig. 5D). However, the repression was completely abolished in the GlnR-deficient host, confirming the negative regulation of GlnR. Furthermore, the promoter activity of strain Cit1-luc was higher than strain Cit2-luc, suggesting that sequence further upstream was required for optimal promoter activity. The promoter activity of strain Cit1-luc was higher in the ΔglnR strain than in the wild-type strain at neutral pH, confirming that the transcriptional expression of citB was enhanced in the GlnR-deficient (ΔglnR) background. When the dyadic sequence of 27 bp (−75 to −49) was deleted (strain Cit3-luc), a comparable level of activity was observed in both the wild-type and GlnR-deficient hosts regardless of the growth pH, indicating that the dyadic sequence is essential for the repression of citBZC operon by GlnR in response to acidic pH.
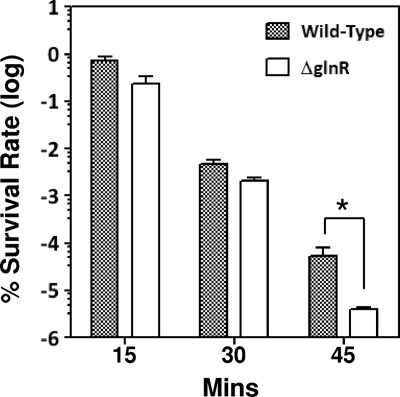
GlnR is required for optimal ATR.
When the survival rates at pH 2.8 were examined, it was found that the viability of the ΔglnR mutant declined at a higher rate than that of the wild-type strain, and a difference of more than 10-fold was detected 45 min after acid treatment (Fig. 6), indicating that GlnR was required for optimal ATR in S. mutans. The survival rate of the complementation strain recovered to the rate of the wild-type strain. Preincubation in medium that has been adjusted to pH 5.5 for 2 h could enhance the survival rates of both strains in pH 2.8 at a similar rate (data not shown).
FIG. 6.
Acid killing assay. Survival rates were determined by plating the cells on BHI agar plates, and results are expressed as percent survival rate. The means plus standard deviations (error bars) for three independent samples are shown. The statistically significant difference (P < 0.10, Student's t test) is indicated by an asterisk.
DISCUSSION
S. mutans can grow on the components of saliva as the sole carbon and nitrogen source within a mixed plaque community, suggesting that the fine balance between nitrogen and carbohydrate metabolism is essential for its survival (12, 16, 54, 59). The downregulation of the mannose-specific enzyme II (EIIMan) and glucose-specific enzyme II (EIIGlc) activity of the PTS in S. mutans by growth at acidic pH and with excess carbohydrate in the chemostat culture system clearly indicates that specific regulation of sugar uptake is required for optimal growth under each growth condition (27). Our array analysis also revealed GlnR-independent downregulation in several genes encoding proteins involved in sugar transport or energy generation at pH 5.5. Thus, regulatory mechanisms involved in both carbohydrate metabolism and nitrogen metabolism are activated upon acid challenge in S. mutans. The citrate metabolic pathway in S. mutans catalyzes the conversion of acetyl coenzyme A and oxalacetate to α-ketoglutarate, the precursor for glutamine synthesis (15). In addition to citBZC, orthologs of the citrate metabolic genes, including genes encoding citrate lyase and oxaloacetate decarboxylase, are located in the S. mutans genome (Fig. 1) (4), and the alternative catabolism of citrate has been demonstrated by Korithoski et al. (29). Thus, it is likely that citrate can either serve as the precursor of α-ketoglutarate or be processed by citrate lyase and oxaloacetate decarboxylase to produce pyruvate. The results of microarray and RT-PCR analyses revealed that citBZC, glnQHMP, and nrgA are highly sensitive to pH regulation (Fig. 3) (see Table S3 in the supplemental material), suggesting tight regulation in both ammonia and amino acid uptake and citrate metabolism. Such regulation will reduce energy spending in transportation and promote citrate fermentation to pyruvate at acidic pH, which would lead to the consumption of H+ by oxaloacetate decarboxylase and ATP synthesis in the acetate kinase reaction to further support cells to resist acid stress (9). A recent study by Krastel et al. also reveals a downregulation of glnQHMP in cells grown at pH 5.5 compared to cells grown at neutral pH. Furthermore, increased survival at the lethal pH of 3.5 is detected in a mutant lacking glnQHMP, confirming that the repression of glnQHMP by acidic growth pH is part of the ATR of S. mutans (30).
The GlnR regulon (glnRA, nrgA, glnQHMP, and gdhA) is responsible for nitrogen assimilation in Gram-positive bacteria, and the regulation via the interaction between GlnR and GlnR box is highly conserved (17). Moreover, GlnR represses expression of the zwf-glnPQ operon in S. pneumoniae (28) and the amtB-glnK operon in Lactococcus lactis (32). In S. mutans, intracellular pH has been shown to fluctuate consistently with the external pH (25, 26, 56); thus, rapid responses to the alterations are required for optimal adaptation. It is likely that the quick response to acid challenge in clusters of genes encoding proteins involved in amino acid metabolism is mediated by GlnR and/or a cofactor(s) readily present in the cell. It is conceivable that GlnR acquires DNA-binding affinity through conformational changes under low pH, as shown for L. lactis GadR (49). Furthermore, the arrangement of glnRA and gltAD in S. mutans is identical to the glnA-nitR and gltAB genes of Clostridium saccharobutylicum and Clostridium beijerinckii (55). However, this arrangement is not found in other streptococci. The clustering and coordinated regulation of glnA and gltAB in C. saccharobutylicum and C. beijerinckii appear to be a simple and efficient mean of ensuring that glutamine synthetase (GS) and glutamate synthase are available at the same time (55). Our microarray data show that the expression of gltD was slightly reduced (0.41-fold) at acidic pH (see Table S3 in the supplemental material), suggesting that the expression of glnA and gltAD genes was regulated coordinately in S. mutans.
Unlike S. pneumoniae, a tnrA homolog (SMU.1287) was identified in the genome of S. mutans UA159. The presence of this locus in S. mutans GS-5 was confirmed by PCR and sequence analysis. In B. subtilis, both GlnR and TnrA regulate the expression of gene products involved in nitrogen metabolism, and both recognize very similar DNA-binding consensus sequences (20). Although the N-terminal DNA-binding domains in these two proteins are highly conserved, GlnR and TnrA exert regulatory functions under different nutritional conditions in B. subtilis. Furthermore, the expression of tnrA is negatively regulated by GlnR under excess nitrogen conditions, whereas TnrA can repress glnR expression under nitrogen-limiting conditions (20). Whether the identified TnrA homolog plays a role in the regulation of genes involved in S. mutans nitrogen metabolism and ATR and whether TnrA and GlnR can modulate each other's expression under specific growth conditions are not known. However, the repression of glnA, citB, and SMU.807 by GlnR at neutral pH strongly suggested that the regulation of GlnR is modulated by growth pH and an additional factor, presumably nitrogen availability. Experiments are currently under way to further dissect the regulation circuit and define the function of the TnrA homolog in S. mutans gene regulation.
One of the limitations of using batch cultures for transcriptome analysis is the lack of control of growth rate and nutrient concentrations. To verify the microarray result, we examined the expression of all genes in the GlnR regulon in the continuous chemostat culture system, where a constant growth rate and nutrient concentration were maintained throughout the study. A general downregulation at pH 5.5 was detected for all genes, confirming the GlnR-mediated repression. The slight differences in the degrees of reduction in the genes examined in the two culture systems also suggested that additional factors, such as nutrient availability, may also play a role in the GlnR regulation.
In conclusion, the roles of GlnR and GlnR box identified through microarray and bioinformatics were confirmed for the citBZC operon, and the impact of the GlnR regulon in the ATR was demonstrated in this study. The repression by GlnR is also found in other clusters of genes encoding proteins involved in glutamine and glutamate metabolism (glnRA, nrgA-glnB, citBZC, glnP-SMU.805c, glnQHMP, and SMU.807) in response to acid stress. Collectively, these results suggest that glutamine and glutamate biosynthesis and transport are stringently controlled during acid adaptation. It is likely that the GlnR-mediated acid responses are present in other oral streptococci.
Supplementary Material
Acknowledgments
This work was supported in part by the National Science Council (grants NSC-952320-B002-086 and NSC-963112-B002-031) and Chang Gung Memorial Hospital (CMRPD170172).
We thank H. K. Kuramitsu and P. Fives-Taylor for critical review of the manuscript and H. Hsieh for technical assistance. We thank NIDCR and TIGR for providing S. mutans microarray slides.
Footnotes
Published ahead of print on 19 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abranches, J., M. M. Nascimento, L. Zeng, C. M. Browngardt, Z. T. Wen, M. F. Rivera, and R. A. Burne. 2008. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 190:2340-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, S. J., Z. T. Wen, and R. A. Burne. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189:8519-8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ajdic, D., and V. T. Pham. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189:5049-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang, S., C. Z. Lee, K. Peck, M. Sindici, U. Matrubutham, M. A. Gleeson, and J. T. Wang. 2001. Acid-induced gene expression in Helicobacter pylori: study in genomic scale by microarray. Infect. Immun. 69:1679-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arirachakaran, P., E. Benjavongkulchai, S. Luengpailin, D. Ajdic, and J. A. Banas. 2007. Manganese affects Streptococcus mutans virulence gene expression. Caries Res. 41:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 9.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78-88. [PubMed] [Google Scholar]
- 10.Brandenburg, J. L., L. V. Wray, Jr., L. Beier, H. Jarmer, H. H. Saxild, and S. H. Fisher. 2002. Roles of PucR, GlnR, and TnrA in regulating expression of the Bacillus subtilis ure P3 promoter. J. Bacteriol. 184:6060-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 12.Carlsson, J. 1970. Nutritional requirements of Streptococcus mutans. Caries Res. 4:305-320. [DOI] [PubMed] [Google Scholar]
- 13.Chen, P. M., J. Y. Chen, and J. S. Chia. 2006. Differential regulation of Streptococcus mutans gtfBCD genes in response to copper ions. Arch. Microbiol. 185:127-135. [DOI] [PubMed] [Google Scholar]
- 14.Chia, J. S., Y. Y. Lee, P. T. Huang, and J. Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cvitkovitch, D. G., J. A. Gutierrez, and A. S. Bleiweis. 1997. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J. Bacteriol. 179:650-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jong, M. H., and J. S. Van der Hoeven. 1987. The growth of oral bacteria on saliva. J. Dent. Res. 66:498-505. [DOI] [PubMed] [Google Scholar]
- 17.Doroshchuk, N. A., M. S. Gel'fand, and D. A. Rodionov. 2006. Regulation of nitrogen metabolism in gram-positive bacteria. Mol. Biol. (Mosk.) 40:919-926. (In Russian.) [PubMed] [Google Scholar]
- 18.Ermolaeva, M. D., O. White, and S. L. Salzberg. 2001. Prediction of operons in microbial genomes. Nucleic Acids Res. 29:1216-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223-232. [DOI] [PubMed] [Google Scholar]
- 21.Gong, Y., X. L. Tian, T. Sutherland, G. Sisson, J. Mai, J. Ling, and Y. H. Li. 2009. Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155:3322-3332. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, S. D., and Q. Gao. 1999. Firefly luciferase as a reporter to study gene expression in Streptococcus mutans. Plasmid 42:154-157. [DOI] [PubMed] [Google Scholar]
- 23.Griswold, A. R., Y. Y. Chen, and R. A. Burne. 2004. Analysis of an agmatine deiminase gene cluster in Streptococcus mutans UA159. J. Bacteriol. 186:1902-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griswold, A. R., M. Jameson-Lee, and R. A. Burne. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton, I. R. 1990. Maintenance of proton motive force by Streptococcus mutans and Streptococcus sobrinus during growth in continuous culture. Oral Microbiol. Immunol. 5:280-287. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton, I. R., and N. D. Buckley. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6:65-71. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton, I. R., L. Gauthier, B. Desjardins, and C. Vadeboncoeur. 1989. Concentration-dependent repression of the soluble and membrane components of the Streptococcus mutans phosphoenolpyruvate:sugar phosphotransferase system by glucose. J. Bacteriol. 171:2942-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 29.Korithoski, B., K. Krastel, and D. G. Cvitkovitch. 2005. Transport and metabolism of citrate by Streptococcus mutans. J. Bacteriol. 187:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krastel, K., D. B. Senadheera, R. Mair, J. S. Downey, S. D. Goodman, and D. G. Cvitkovitch. 2010. Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance. J. Bacteriol. 192:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhnert, W. L., G. Zheng, R. C. Faustoferri, and R. G. Quivey, Jr. 2004. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186:8524-8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen, R., T. G. Kloosterman, J. Kok, and O. P. Kuipers. 2006. GlnR-mediated regulation of nitrogen metabolism in Lactococcus lactis. J. Bacteriol. 188:4978-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 34.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 35.Lemos, J. A., J. Abranches, and R. A. Burne. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95-107. [PubMed] [Google Scholar]
- 36.Lemos, J. A., and R. A. Burne. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemos, J. A., M. M. Nascimento, V. K. Lin, J. Abranches, and R. A. Burne. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353-1366. [DOI] [PubMed] [Google Scholar]
- 39.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339-1351. [DOI] [PubMed] [Google Scholar]
- 40.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Galiano, A. J., K. Overweg, M. J. Ferrandiz, M. Reuter, J. M. Wells, and A. G. de la Campa. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935-3946. [DOI] [PubMed] [Google Scholar]
- 42.Merritt, J., J. Kreth, W. Shi, and F. Qi. 2005. LuxS controls bacteriocin production in Streptococcus mutans through a novel regulatory component. Mol. Microbiol. 57:960-969. [DOI] [PubMed] [Google Scholar]
- 43.Nascimento, M. M., J. A. Lemos, J. Abranches, R. B. Goncalves, and R. A. Burne. 2004. Adaptive acid tolerance response of Streptococcus sobrinus. J. Bacteriol. 186:6383-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nascimento, M. M., J. A. Lemos, J. Abranches, V. K. Lin, and R. A. Burne. 2008. Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 190:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 47.Robichon, D., M. Arnaud, R. Gardan, Z. Pragai, M. O'Reilly, G. Rapoport, and M. Debarbouille. 2000. Expression of a new operon from Bacillus subtilis, ykzB-ykoL, under the control of the TnrA and PhoP-PhoR global regulators. J. Bacteriol. 182:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubens, C. E., and L. M. Heggen. 1988. Tn916 delta E: a Tn916 transposon derivative expressing erythromycin resistance. Plasmid 20:137-142. [DOI] [PubMed] [Google Scholar]
- 49.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 50.Senadheera, D., K. Krastel, R. Mair, A. Persadmehr, J. Abranches, R. A. Burne, and D. G. Cvitkovitch. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shemesh, M., A. Tam, and D. Steinberg. 2007. Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiology 153:1307-1317. [DOI] [PubMed] [Google Scholar]
- 52.Sheng, J., and R. E. Marquis. 2007. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 272:196-201. [DOI] [PubMed] [Google Scholar]
- 53.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917-927. [DOI] [PubMed] [Google Scholar]
- 54.St. Martin, E. J., and C. L. Wittenberger. 1980. Regulation and function of ammonia-assimilating enzymes in Streptococcus mutans. Infect. Immun. 28:220-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stutz, H. E., K. W. Quixley, L. D. McMaster, and S. J. Reid. 2007. Co-regulation of the nitrogen-assimilatory gene cluster in Clostridium saccharobutylicum. Microbiology 153:3081-3090. [DOI] [PubMed] [Google Scholar]
- 56.Svensater, G., U. B. Larsson, E. C. Greif, D. G. Cvitkovitch, and I. R. Hamilton. 1997. Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12:266-273. [DOI] [PubMed] [Google Scholar]
- 57.Svensater, G., B. Sjogreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 58.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Dobler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terleckyj, B., and G. D. Shockman. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Mering, C., L. J. Jensen, B. Snel, S. D. Hooper, M. Krupp, M. Foglierini, N. Jouffre, M. A. Huynen, and P. Bork. 2005. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33:D433-D437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen, Z. T., H. V. Baker, and R. A. Burne. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wen, Z. T., and R. A. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.