Abstract
The viral metagenome within an activated sludge microbial assemblage was sampled using culture-dependent and culture-independent methods and compared to the diversity of activated sludge bacterial taxa. A total of 70 unique cultured bacterial isolates, 24 cultured bacteriophages, 829 bacterial metagenomic clones of 16S rRNA genes, and 1,161 viral metagenomic clones were subjected to a phylogenetic analysis.
Bacteriophages play an active role in the ecology of natural environments, influencing prokaryotic population dynamics (5, 15) and mediating lateral gene transfer between diverse bacterial species, for example. Activated sludge (AS) microbial assemblages in wastewater treatment plants have been shown to harbor great numbers of viruses with a wide range of genome sizes (7, 9, 10, 16). Historically, the focus of wastewater viral studies has been on specific host-virus interactions, the application of phages as tools in microbial source tracking, or the use of phages to improve the efficiency of the wastewater treatment process (e.g., foam and pathogen control) (2, 4, 8, 12, 17). Despite the interest in the wastewater viral community, a census of the activated sludge total viral community has not, to our knowledge, been investigated using both culture-based and metagenomic approaches.
Taxonomic assignment of viral metagenomic clone sequences.
Samples were collected from the AS aeration basin at the H. C. Morgan Water Pollution Control Facility in Auburn, AL, treated with 10% beef extract buffer (to separate viruses from aggregates), precipitated with polyethylene glycol, and subjected to cesium chloride gradient centrifugation. Viral DNA was then extracted by benzonase, proteinase K, and sodium dodecyl sulfate treatments and purified by phenol-chloroform extraction and isopropanol precipitation. An AS viral metagenomic library was constructed at the Lucigen Corporation (Middleton, WI) using a linker-amplified shotgun library approach (3, 18), and 1,161 cloned insert DNA sequences were determined using a single-vector primer. Trimmed viral insert sequences were classified as known (E value < 0.001; n = 694), unknown (n = 97), or novel (n = 370) as described previously (18). The known sequences were assigned to their taxonomic affiliations by BLASTx comparisons to the GenBank nonredundant nucleotide (nr/nt) database and selection of the top BLASTx hits for phylogenetic affiliation (Fig. 1).
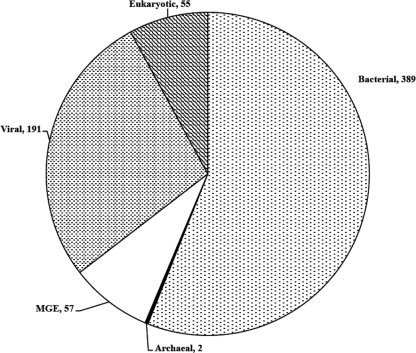
FIG. 1.
Taxonomic breakdown of the significant hits from the AS viral metagenomic libraries based on a BLASTx comparison against the GenBank nr/nt and env databases. Numbers indicate the number of sequences assigned to a particular phylogenetic category based on significant homology with known sequences in GenBank. MGE, mobile genetic element. The viral metagenomic DNA sequences were registered as part of the Wastewater Viral Metagenome project at GenBank (Trace Archives) and are under TI reference numbers 2251203077 to 2251204802.
The dominance of viral metagenomic sequences with significant homology to bacterial DNA (nearly 60% of the known sequences) is likely the consequence of the high frequency of prophage sequences within bacterial genomes deposited in GenBank databases (1). Considerable care was taken to prevent inclusion of prokaryotic (or eukaryotic) chromosomal DNA within the viral metagenomic library (i.e., CsCl purification and nuclease treatment were performed), and the high percentage of viral metagenomic sequences with homology to bacterial genomes has been observed in other environments (3). Of the 191 known sequences with homology to viral genomes, 95% were homologous to bacteriophage sequences and were within the viral families Myoviridae (40.3%), Siphoviridae (31.9%), or Podoviridae (25.6%) or considered unclassified phages (2.2%). The remaining sequences were classified as mobile genetic elements (9%), eukaryotic (8%), or archaeal (<1%) in nature. The taxonomic heterogeneity of sequences obtained in the viral metagenomic library underscores the diversity of the AS system, which receives an influx of microbiota from both human and environmental sources. In addition, this survey revealed a high percentage of novel predicted gene products (37%), illustrating the relative scarcity of wastewater viral DNA sequences in the GenBank nr/nt or environmental (env) databases.
Comparison of bacterial and viral phylogenetic affiliations.
Because bacterial community structure is expected to be the primary determinant of bacteriophage diversity within an environment, the bacterial taxa from AS were surveyed using culture-dependent and -independent analyses. A survey of AS 16S rRNA genes was conducted with AS bacterial cultures (70 unique bacterial cultures) isolated on LB or a synthetic wastewater medium (14) using aerobic and anaerobic growth conditions. In addition, a bacterial culture-independent survey (n = 829) was conducted using as the template AS metagenomic DNA extracted from the same sample used for viral studies and PCR amplified using universal bacterial primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GYTACCTTGTTACGACTT-3′) (11). The 16S rRNA gene amplicons from the metagenomic DNA template were cloned into the TOPO-TA pCR2.1 vector (Invitrogen, Carlsbad, CA) and transformed into Escherichia coli cells. Each 16S rRNA gene clone sequence was trimmed for quality, compared to the GenBank nr/nt database using the BLASTn search algorithm, and classified according to bacterial division affiliation of the top BLASTn hit (Fig. 2).
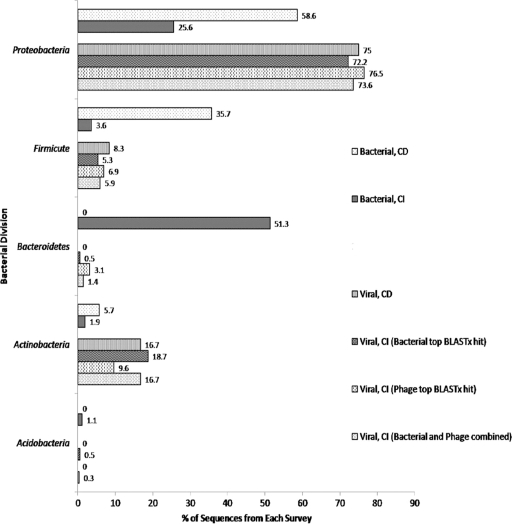
FIG. 2.
Relative dominance of bacterial divisions represented by sequences from the bacterial culture collection, 16S rRNA gene clone library, cultured bacteriophages, and viral metagenomic library. The percentages of sequences affiliated with each bacterial division are compared among the four phylogenetic surveys. CD, culture dependent; CI, culture independent. Other bacterial divisions with <2% representation include the Chlamydiae (0.1%; bacterial, CI), Nitrospira (0.2%; bacterial, CI), Planctomyces (0.7%; bacterial, CI), Cyanobacteria (1.6%; viral, CI), Spirochetes (0.7%; viral, CI), Chloroflexi (0.5%; viral CI), and Deinococcus (0.5%; viral, CI).
The culture-independent 16S rRNA gene clone library revealed more than a 3-fold increase in the number of represented bacterial divisions (n = 10) compared to that observed from the culture collection (n = 3), and a significant shift in relative dominance of each phylum was observed between the two surveys. Although Proteobacteria taxa dominated the culture-dependent survey (58% of analyzed sequences), the percentage of sequences representing this division in the culture-independent survey dropped to 25%. Similarly, Firmicutes representation was almost 10-fold lower in the culture-independent survey than in the culture-dependent study (4% versus 36%, respectively). In contrast, the Bacteroidetes taxa, which were only minimally represented in the culture-dependent survey, comprised more than half (51%) of the bacterial taxa identified in the 16S rRNA clone library. The minimal overlap between data obtained from these culture-dependent and -independent analyses reflects the strong cultivation bias that is common to virtually all environmental studies.
Viral enrichments performed on all of the bacterial isolates identified 24 bacterial hosts from which phages could be isolated. Seventy-five percent of the phages were isolated from Proteobacteria hosts, while the remainder of the phages were isolated from Firmicutes (8%) or Actinobacteria (17%) hosts. In the culture-independent viral metagenomic survey, the 389 sequences from the viral metagenomic library that were most similar to bacterial genomes were included in this taxonomic classification (i.e., viral, culture independent [top bacterial BLASTx hit]), as were the predicted hosts of the 187 viral sequences with significant homology to bacteriophage DNA (i.e., viral, culture independent [top phage BLASTx hit]) (Fig. 2). Although the two viral surveys demonstrate similar phylogenetic distributions, there are some striking differences observed between the analyses of the viral metagenomic library and the bacterial surveys. For example, only two sequences from the viral metagenomic library (0.5% of bacterial BLASTx hits and 3.1% of phage BLASTx hits) were highly similar to Bacteroidetes DNA despite the high relative abundance of Bacteroidetes taxa in the bacterial culture-independent survey (Fig. 2). Proteobacteria was the dominant phylum in every survey except for the bacterial culture-independent study, in which the Bacteroidetes division was the most highly represented (51%). In addition, four of the bacterial phyla predicted to be present within the viral metagenomic library (Deinococcus, Cyanobacteria, Chloroflexi, and Spirochetes) were absent from both of the bacterial phylogenetic surveys. Both of the Gram-positive phyla (Actinobacteria and Firmicutes) had slightly higher representation within the viral metagenomic survey than in the bacterial culture-independent survey. The phylogenetic distributions of the phage metagenomic sequences with the top BLASTx hits as bacterial versus the top BLASTx hits as viral were similar in every case, with the Proteobacteria, Firmicutes, and Actinobacteria taxa dominating the viral metagenomic survey. This study highlights the relative paucity of information within the GenBank databases concerning bacteriophages infecting Bacteroidetes taxa; presumably a very high percentage of the phage metagenomic sequences annotated as unknown or novel could be affiliated with the Bacteroidetes phylum.
Comparison of abundant viral metagenomic loci and cultured phage DNA.
Contig assembly of viral metagenomic sequences was performed using assembly criteria of 80% identity with at least a 20-bp overlap in order to assemble sequences from the most abundant viral types (3). The resulting contigs were compared to the nr/nt and env GenBank databases by BLASTx, and PCR primers were designed to target contigs with significant homology to bacterial hosts in the AS culture collection. Purified DNA from cultured phages was used as the template in a PCR with the metagenomic contig-specific primers, yielding three amplicons from the cultured phages. Predicted phylogenetic relationships between these amplicons, the metagenomic contigs, and their nearest neighbors in the nr/nt database were inferred using the maximum parsimony method. The resulting cladogram (>50% bootstrap support) indicated that every metagenomic contig was more closely related to a cultured phage amplicon than to any other sequence in GenBank databases (data not shown).
Metabolic profiling of viral metagenomic DNA sequences.
Sequences from the viral metagenomic library clones were analyzed by the MetaGene annotator, which identified 1,815 open reading frames (ORFs) within the library. The ORF sequences were then compared to those found in the SEED platform (http://www.theseed.org) and organized into predicted functional categories based on the subsystems approach described by Overbeek et al. (13).
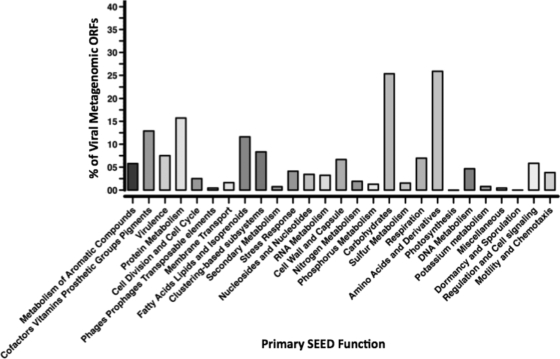
Metabolic profiles were predicted for the viral metagenomic sequences using the SEED platform (Fig. 3). The most dominant subsystems were those involved with macromolecule metabolism, although ORFs encoding virulence factors and pigment production collectively comprised approximately 20% of the sequences from the viral library. Several common subsystems evident in the AS viral metagenome (e.g., carbohydrate and amino acid metabolism) are highly represented in viral metagenomic surveys in other environments (6). Considering that this was not an exhaustive survey of AS viral metagenomes, no conclusions regarding changes in the relative dominance of each subsystem may be made, but overall the distribution of metabolic systems in an AS viral metagenome is similar to that of viral metagenomes from other environments (6). This is the first study to explore the functional genetic diversity present within an activated sludge viral community. Future studies will further this work by employing next-generation sequencing methods and expanding upon the viral sequence repository available from multiple natural environments.
FIG. 3.
Predicted metabolic profiles obtained from SEED analyses of viral metagenomic open reading frames (ORFs).
Nucleotide sequence accession numbers.
All 16S rRNA gene sequences from the cultured isolates and bacterial 16s rRNA clones were submitted to GenBank under accession numbers GU002706 to GU003868. The viral metagenomic DNA sequences were registered as part of the Wastewater Viral Metagenome project at GenBank (Trace Archives) and are under TI reference numbers 2251203077 to 2251204802.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Angly, F., B. Rodriguez-Brito, D. Bangor, P. McNairnie, M. Breitbart, P. Salamon, B. Felts, J. Nulton, J. Mahaffy, and F. Rohwer. 2005. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aström, J., T. J. Pettersson, T. A. Stenstrom, and O. Bergstedt. 2009. Variability analysis of pathogen and indicator loads from urban sewer systems along a river. Water Sci. Technol. 59:203-212. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costán-Longares, A., M. Montemayor, A. Payan, J. Mendez, J. Jofre, R. Mujeriego, and F. Lucena. 2008. Microbial indicators and pathogens: removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 42:4439-4448. [DOI] [PubMed] [Google Scholar]
- 5.Debartolomeis, J., and V. J. Cabelli. 1991. Evaluation of an Escherichia coli host strain for enumeration of F male-specific bacteriophages. Appl. Environ. Microbiol. 57:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinsdale, E. A., R. A. Edwards, D. Hall, F. Angly, M. Breitbart, J. M. Brulc, M. Furlan, C. Desnues, M. Haynes, L. L. Li, L. McDaniel, M. A. Moran, K. E. Nelson, C. Nilsson, R. Olson, J. Paul, B. R. Brito, Y. J. Ruan, B. K. Swan, R. Stevens, D. L. Valentine, R. V. Thurber, L. Wegley, B. A. White, and F. Rohwer. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629-632. (Erratum, 455:830.) [DOI] [PubMed] [Google Scholar]
- 7.Hantula, J., A. Kurki, P. Vuoriranta, and D. H. Bamford. 1991. Ecology of bacteriophages infecting activated sludge bacteria. Appl. Environ. Microbiol. 57:2147-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamalludeen, N., Y. M. She, E. J. Lingohr, and M. Griffiths. 2009. Isolation and characterization of virulent bacteriophages against Escherichia coli serogroups O1, O2, and O78. Poult. Sci. 88:1694-1702. [DOI] [PubMed] [Google Scholar]
- 9.Khan, M. A., H. Satoh, H. Katayama, F. Kurisu, and T. Mino. 2002. Bacteriophages isolated from activated sludge processes and their polyvalency. Water Res. 36:3364-3370. [DOI] [PubMed] [Google Scholar]
- 10.Khan, M. A., H. Satoh, T. Mino, H. Katayama, F. Kurisu, and T. Matsuo. 2002. Bacteriophage-host interaction in the enhanced biological phosphate removing activated sludge system. Water Sci. Technol. 46:39-43. [PubMed] [Google Scholar]
- 11.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J. E., M. Y. Lim, S. Y. Kim, S. Lee, H. Lee, H. M. Oh, H.-G. Hur, and G. Ko. 2009. Molecular characterization of bacteriophages for microbial source tracking in Korea. Appl. Environ. Microbiol. 75:7107-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rezania, B., J. A. Oleszkiewicz, N. Cicek, and H. Mo. 2005. Hydrogen-dependent denitrification in an alternating anoxic-aerobic SBR membrane bioreactor. Water Sci. Technol. 51:403-409. [PubMed] [Google Scholar]
- 15.Shapiro, O. H., A. Kushmaro, and A. Brenner. 19 November 2009. Bacteriophage predation regulates microbial abundance and diversity in a full-scale bioreactor treating industrial wastewater. ISME J. [Epub ahead of print.] [DOI] [PubMed]
- 16.Song, J. S., J. H. Jeon, J. H. Lee, S. H. Jeong, B. C. Jeong, S. J. Kim, J. H. Lee, and S. H. Lee. 2005. Molecular characterization of TEM-type beta-lactamases identified in cold-seep sediments of Edison Seamount (south of Lihir Island, Papua New Guinea). J. Microbiol. 43:172-178. [PubMed] [Google Scholar]
- 17.Withey, S., E. Cartmell, L. M. Avery, and T. Stephenson. 2005. Bacteriophages—potential for application in wastewater treatment processes. Sci. Total Environ. 339:1-18. [DOI] [PubMed] [Google Scholar]
- 18.Wommack, K. E., S. R. Bench, J. Bhavsar, D. Mead, and T. Hanson. 2009. Isolation independent methods of characterizing phage communities 2: characterizing a metagenome. Methods Mol. Biol. 502:279-289. [DOI] [PubMed] [Google Scholar]