Abstract
Two types of endosymbiotic bacteria were identified in the gastrodermis of the marine invertebrate Xenoturbella bocki (Xenoturbellida, Bilateria). While previously described Chlamydia-like endosymbionts were rare, Gammaproteobacteria distantly related to other endosymbionts and pathogens were abundant. The endosymbionts should be considered when interpreting the poorly understood ecology and evolution of Xenoturbella.
Xenoturbella bocki is a benthic marine worm first described in 1949 and only found at a few geographical locations at apparently low population densities. It has a very simple body plan (Fig. 1A) with a blind gut (cul-de-sac) and lacks coelomic cavities, a brain, and reproductive and excretory organs (35). Xenoturbella was originally classified as a flatworm, but molecular analyses have placed it within its own animal phylum, with various affiliations (reviewed in reference 34). The most recent phylogenetic analyses using mitochondrial genome and phylogenomic data indicate a position either within Deuterostomia (5) or at the base of Bilateria (14). Thus, the evolutionary history of Xenoturbella remains elusive and it is unclear whether its simple body plan represents a plesiomorphic character or, alternatively, is the result of secondary loss (34). In addition, the ecology of Xenoturbella is not well understood, leaving, e.g., its food source unresolved (4, 18).
FIG. 1.
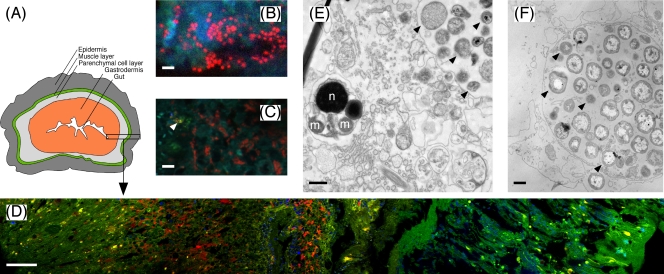
(A) Schematic drawing of the simple body plan of Xenoturbella (redrawn from reference 35). (B) Cells of the gammaproteobacterial endosymbiont (red) detected by probe XenoGam441-CY3 at high magnification (scale bar, 2 μm). (C) Double hybridization of gammaproteobacterial and chlamydial endosymbionts with probes XenoGam441-CY3 (red) and Cps-1353mod-CY5 (displayed in yellow for better contrast, indicated by the arrow); blue-green structures, autofluorescent tissue recorded in the fluorescein isothiocyanate channel (scale bar, 5 μm). (D) FISH detection of gammaproteobacterial endosymbionts with probe XenoGam441-CY3 (red) in a cross section of an X. bocki specimen. The image is composed of five consecutive image stacks, spanning from the gut (to the very left) to the epidermis (to the very right) (magnification, ×400; scale bar, 50 μm). Dense clusters of gammaproteobacterial endosymbionts (red) are confined to the gastrodermis; 4′,6-diamidino-2-phenylindole (DAPI)-stained cell nuclei and mitochondria of X. bocki are displayed in blue; the green and yellow structures are autofluorescent tissue recorded in the fluorescein isothiocyanate channel. (E and F) TEM images of bacterial cells in sections of germinal tissue from a spermatid cluster (E) and of gastric tissue (F) of X. bocki. Arrows point to examples of different putative bacterial morphotypes. Scale bars, 0.5 μm. n, spermatid nucleus; m, spermatid mitochondrion.
Symbiotic bacteria are found in a remarkable number of diverse marine invertebrates (11), where they affect both the ecology and the evolution of their hosts. The first indications of intracellular endosymbionts in Xenoturbella were published by Israelsson (17) and confirmed by electron microscopy of a Xenoturbella spermatid cluster (Fig. 1B); however, our first molecular analyses identified the putative endosymbionts as Gammaproteobacteria, whereas Israelsson (17) had reported a group of endosymbiotic Chlamydiae. The objectives of this study were therefore (i) to identify the putative endosymbionts by 16S rRNA gene analysis, (ii) to localize them inside the host by fluorescence in situ hybridization (FISH), and (iii) to specifically test for the prevalence of the gammaproteobacterial endosymbionts in Xenoturbella.
Sample collection and preparation.
Seven specimens of X. bocki were collected at two different sites on six different dates in 2005 (GPS position: 58°16′N, 11°26′E) and 2006 (GPS position: 58°17′N, 11°31′E) by dredging and sieving of surface sediment at a 100-m water depth in the Gullmar Fjord, Sweden (4). Whole animals were fixed in 4% (wt/vol) paraformaldehyde in sterile seawater and stored in 70% ethanol. Two spermatid clusters (i.e., aggregations of male gametes at the stage of maturation just before becoming fully mature spermatozoa) could be dissected from two whole animals and were preserved in 98% ethanol; since these dissected animals could not be utilized, a total of five whole animals and two spermatid clusters were available for further analysis. The identity of the X. bocki specimens and spermatid cluster was confirmed by cloning and sequencing cytochrome c oxidase I (cox1) gene fragments (3, 13). One animal and one spermatid cluster were used for transmission electron microscopy (TEM) as previously described (26). DNA was extracted from 2- to 4-mm-wide cross sections of the remaining four whole animals (Qiagen DNeasy tissue kit; Qiagen GmbH, Hilden, Germany). The second spermatid cluster was digested with 2 U proteinase K (Qiagen) in 10 μl of 0.1 M Tris-EDTA buffer for 12 h at 55°C, followed by inactivation for 15 min at 95°C. The resultant digest was used directly as the template for PCR.
PCR, cloning, and sequence analyses.
Nearly full-length 16S and 23S rRNA gene fragments were PCR amplified using general bacterial primer pairs 26F-1492R (15, 20) and 129F-2241R (16, 19) at an annealing temperature of 57°C. Short 16S rRNA gene fragments of the X. bocki endosymbionts were amplified with primers 16SigF and 16SigR (annealing temperature: 60°C), which are specific for Chlamydiales (12), and primer 64F, which was designed to target the gammaproteobacterial endosymbionts (Table 1), in combination with bacterial primer 518R (25) at an annealing temperature of 54°C. All PCRs were performed as 25-μl reaction mixtures for 28 cycles using HotStarTaq Master Mix (Qiagen). PCR products were gel purified (E.Z.N.A. gel extraction kit; Omega Bio-Tek, Doraville, GA), cloned (pGEM-T Easy vector system; Promega, Madison, WI), and sequenced (Macrogen, Seoul, Korea).
TABLE 1.
PCR primers and FISH probes developed in this study
| Primer or probe | Sequence (5′-3′) | Target, specificity | Temp (°C)a | % FAb |
|---|---|---|---|---|
| Primers | ||||
| 64F | GGA CGG TAA CAT TGT GGT GC | 16S rRNA gene, X. bocki gammaproteobacterial endosymbiont | 54 | |
| 1417F | CGA AGG AGA GAC GGA GAA GGT TAG | 23S rRNA gene, X. bocki gammaproteobacterial endosymbionts (sequencing primer) | 59 | |
| Probes | ||||
| Cps-1353modc | GAC GTT ATT GCT GAC ACG | 16S rRNA, X. bocki chlamydial endosymbiont and relatives | 15 | |
| XenoGam441 | ATA GCC TTC CTC AGT GAT | 16S rRNA, X. bocki gammaproteobacterial endosymbiont | 15 | |
| XenoGam666 | CGG AAA TTC CTC TAC CC | 16S rRNA, X. bocki gammaproteobacterial endosymbiont and relatives | 15 | |
| XenoGam727 | TCC AGG TAG ACG CCT TC | 16S rRNA, X. bocki gammaproteobacterial endosymbiont and relatives | 15 |
Temp, applied primer annealing temperature for PCR.
FA, applied formamide concentration in the hybridization buffer for FISH.
Modified from reference 28.
The retrieved rRNA gene sequences were aligned with the Silva SSURef and LSURef version 100 databases (30), and their phylogenetic position was inferred by Bayesian analysis (31). Tree searches (3,000,000 generations) were performed with program default priors on model parameters and default settings for the MCMC runs using the GTR+Γ+I evolutionary model, which, according to Modeltest version 3.7 (29), best fit the sequence data sets among 56 evaluated models. The input files of log likelihood scores for the various models were generated in PAUP* (33). The displayed trees (Fig. 2) have been cropped for better readability; for full trees, see the supplemental material.
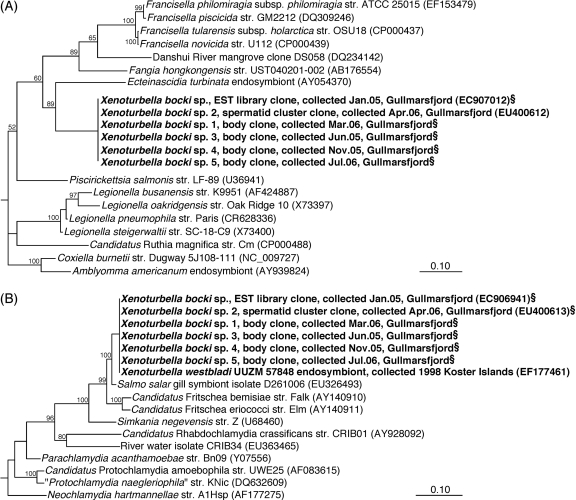
FIG. 2.
Phylogeny of the Xenoturbella endosymbionts (bold) inferred from Bayesian analysis of 16S rRNA gene sequences. Scale bars, 10% sequence divergence. (A) Position of the gammaproteobacterial endosymbiont among members of the class Gammaproteobacteria based on 1,301 sequence positions between E. coli positions 58 and 1,461. (B) Position of the chlamydial endosymbiont among members of the class Chlamydiae based on 1,296 sequence positions between E. coli positions 53 and 1,466. Values are neighbor-joining-based bootstrap percentages. §, short sequence added to the tree by the parsimony method. For complete versions of the trees, see Fig. S2 in the supplemental material.
FISH.
Three 16S rRNA-targeted probes, termed XenoGam, were designed specifically for the X. bocki gammaproteobacterial endosymbionts using the ARB program package (21). A previously published probe was modified to match the chlamydial endosymbiont sequences (Table 1), and probe specificity was evaluated in silico using ARB and the Ribosomal Database Project Probe Match tool (6). All XenoGam probes had seven or more mismatches with the chlamydial endosymbiont. Probe XenoGam441 had one or more mismatches with any other sequence in the database, whereas probes XenoGam666 and XenoGam727 had perfect matches with five or six other 16S rRNA gene sequences mostly affiliating with the endosymbiont sequence. Hybridization conditions were optimized for the CY3-labeled probe XenoGam441 by Clone-FISH (32) on Escherichia coli JM109(DE3)/pGEM-T clones (Promega) expressing the target 16S rRNA; untransformed E. coli JM109 (Promega) and Desulfovibrio strain I8 (GenBank accession number FJ655907) served as negative controls with six and five mismatches with the probe, respectively. FISH was performed at various formamide concentrations (5 to 60%) in the hybridization buffer according to standard protocols (8), signal intensities were quantified with the image analysis software DAIME version 1.1 (9), and sigmoidal best fits of the fluorescence intensity data were made with Prism v. 5.0b (GraphPad Software Inc., La Jolla, CA). The resulting probe dissociation curve showed rapid disappearance of probe-conferred signal from the perfect match target at formamide concentrations of >15% (see Fig. S1 in the supplemental material); no signal was obtained with the negative control strains under any of the conditions tested. Similar results were obtained with probes XenoGam666 and XenoGam727 in hybridization series of increasing formamide concentrations of X. bocki cryosections (8; data not shown). Therefore, 15% formamide was selected as the optimal stringency for all XenoGam probes.
One-half of an X. bocki specimen was embedded in OCT compound (Leica Microsystems, Wetzlar, Germany), frozen at −35°C, and sectioned at −15°C into 10- to 20-μm-thick slices on a CM1800 cryostat (Leica). Sections were immobilized on gelatin-coated slides, dehydrated, and hybridized with CY3- and CY5-labeled probes according to standard protocols (8). Besides the endosymbiont-specific probes, EUBI-III (7) and NON (2) were used as positive and negative control probes, respectively. All hybridizations were evaluated on an Axiovert 200M epifluorescence microscope equipped with an ApoTome module for optical sectioning (Carl Zeiss, Jena, Germany).
A novel putative endosymbiont in Xenoturbella.
In initial 16S and 23S rRNA clone libraries prepared from a single X. bocki spermatid cluster with general bacterial primers, 23 out of 24 sequenced 16S rRNA gene clones and all 5 sequenced 23S rRNA gene clones represented a single sequence type distantly related to symbiotic and pathogenic Gammaproteobacteria known for an intracellular lifestyle, e.g., Coxiella, Legionella, and Francisella (Fig. 2A; see Fig. S2A and B in the supplemental material). This 16S rRNA gene sequence showed <90% identity with its closest relatives and formed a monophyletic group with an endosymbiont of the tunicate Ecteinascidia turbinata (22), the marine isolate Fangia hongkongensis, and the genera Francisella and Piscirickettsia (Fig. 2A). A congruent phylogenetic position was inferred for the 23S rRNA gene sequence (see Fig. S2B in the supplemental material). The chlamydial endosymbionts reported by Israelsson (17) were not detected in these general clone libraries. Therefore, a Chlamydiales-specific PCR was employed which finally retrieved a 16S rRNA gene fragment identical to that of the chlamydial endosymbiont. This sequence clustered with insect endosymbionts of the candidate genus Fritschea and showed 98% identity with a salmon gill-associated sequence (Fig. 2B).
A second general bacterial 16S rRNA gene clone library was generated from a section of a whole animal, thus also including the gut and gut content. In contrast to the spermatid library, none of the 32 sequences analyzed from the whole-body library affiliated with the gammaproteobacterial or chlamydial endosymbiont sequences described above. Instead, they were highly (95 to 100%) similar to one of six phylotypes representing various Alpha-, Beta-, and Gammaproteobacteria, probably originating from contaminating gut content of the worm (see Table S1 in the supplemental material). However, 16S rRNA gene sequences of both putative endosymbionts were detected by specific PCR also in the whole-animal preparation (Fig. 2; see Fig. S2A and C in the supplemental material).
Tissue-specific localization and abundance.
FISH analysis of sections from the same whole-animal specimen as used for the clone libraries showed that the gammaproteobacterial endosymbionts were highly abundant in the gastrodermis of X. bocki (Fig. 1B and D), whereas chlamydial cells were extremely rare (Fig. 1C). The gammaproteobacterial cells were coccoid, with a diameter of 0.5 to 1.0 μm, and often occurred in groups (Fig. 1B and D), an arrangement typical for cells inside bacteriocytes (Fig. 1F). No bacterial cells were detected outside the gastrodermis, i.e., in the parenchyma, muscles, or epidermis (Fig. 1A and D); the gut content, which presumably contains abundant bacteria, had been lost during preparation. Comparable results were obtained with all three XenoGam probes (Table 1) labeled with CY3 or CY5, and comparison to the general bacterial probe EUB, as well as simultaneous hybridizations with probes XenoGam441-CY3 and EUB-CY5 and probes XenoGam441-CY3 and Cps-1353mod-CY5, respectively, indicated that almost all of the Bacteria detectable by FISH belonged to the gammaproteobacterial endosymbionts, while chlamydial cells were estimated to account for <1% of the cells (Fig. 1C and results not shown).
It should be noted, however, that during their life cycle, Chlamydiae form metabolically inactive elementary bodies that may be difficult to detect by FISH (28); but even if 90% of the Chlamydiae present were elementary bodies, their total number would still be much lower than that of the Gammaproteobacteria. Israelsson (17) reported several different intracellular bacterial morphotypes in the gastrodermis of Xenoturbella, with cell diameters of 0.3 to 1 μm; these results agree well with our TEM analysis of both spermatid clusters and gastrodermal cells (Fig. 1E and F). Even though the FISH analysis does not directly prove the intracellular localization of either gammaproteobacterial or chlamydial endosymbionts, the combined results of TEM, FISH, and sequence analysis suggest that the different morphotypes represent the reticulate and elementary bodies of the chlamydial cells, as well as the gammaproteobacterial endosymbionts, and thus that both types of endosymbionts occur within the same host cells.
General occurrence of both types of endosymbionts in Xenoturbella.
Three additional individual animals from different samplings were analyzed for both endosymbionts by specific PCR; all three tested positive for both sequence types (Fig. 2; see Fig. S2A and C in the supplemental material). Further evidence of the ubiquitous presence of both endosymbionts in Xenoturbella was derived by blastn searches (1) of an X. bocki expressed sequence tag (EST) library; two sequences of the library (GenBank accession numbers EC907012 and EC907192) apparently originated from bacterial rRNA contamination and showed >99% identity with the 16S rRNA gene of the gammaproteobacterial endosymbionts (Fig. 2A). The same library also contained the 16S and 23S rRNA gene sequences of the chlamydial endosymbionts (17). The chlamydial endosymbionts were originally described in samples collected already in 1998, and the detection of at least three different bacterial morphotypes by TEM in these specimens (17) suggests that also the gammaproteobacterial endosymbionts have been consistently associated with Xenoturbella. We realize that the number of specimens analyzed is still small, which is mainly due to the difficulty of finding the animal; however, the consistent detection of both endosymbionts in six or seven independent samples from different years or months that were analyzed in different laboratories is, in our view, a strong indication of a specific and stable symbiotic association between Xenoturbella and the two bacterial types.
The endosymbionts were also detected in the spermatid clusters (Fig. 1E and 2), which may be indicative of vertical symbiont transmission (10). However, there is no TEM evidence that the endosymbionts also occur inside the sperm cells (Fig. 1E). In Xenoturbella, which is hermaphroditic, eggs and spermatids develop within the parenchymal cell layer (Fig. 1A) and are emptied into the gut via the gastrodermis (27). Therefore, spermatid clusters likely contain gastrodermal cells that harbor endosymbionts. In the absence of eggs and larvae for further analysis, the question of vertical transmission therefore remains unresolved.
Proposal of “Candidatus Endoxenoturbella lovénii.”
The gammaproteobacterial 16S rRNA gene sequence type was detected in a spermatid cluster, four adult worms, and an EST library, which all represent independent X. bocki individuals; FISH and TEM revealed a tissue-specific localization and a high abundance of the Gammaproteobacteria without apparent tissue necrosis. Together, these results suggest that the Gammaproteobacteria form a stable and specific association with Xenoturbella and may be provisionally classified as “Candidatus Endoxenoturbella lovénii” (24).
Description of the candidate genus Endoxenoturbella.
“Candidatus Endoxenoturbella” (En.do.xe.no.tur.bel′.la, pref. endo from G. endon, within; Xenoturbella, a marine worm; Endoxenoturbella, inside the worm Xenoturbella); small, coccoid bacteria (0.5 to 1.0 μm in diameter), members of the class Gammaproteobacteria, highly abundant and presumably endemic in the gastrodermis of Xenoturbella species.
Description of the candidate species Endoxenoturbella lovénii.
The description of “Candidatus Endoxenoturbella lovénii” [lo.ve′.ni.i, N.L. masc. adj., referring to the Sven Lovén Centre for Marine Sciences, University of Gothenburg (the union of the Kristineberg Marine Research Station and the Tjärnö Marine Biological Laboratory), to appreciate its importance as a base for research in marine biology and in particular on Xenoturbella] is the same as that of the genus. This species colonizes the gastrodermis of Xenoturbella bocki. The basis of its assignment was the 16S rRNA gene sequence (GenBank accession number EU400612) and hybridization with a 16S rRNA-targeted oligonucleotide probe (5′-ATAGCCTTCCTCAGTGAT-3′). The source was the gastrodermis of Xenoturbella bocki Westblad; so far, it has not been cultured.
Evolutionary and functional implications.
Without understanding the ecology and evolution of the host, at present we can only speculate about the function of the endosymbionts. Most intracellular bacterium-animal symbioses are related to nutrient provision or waste recycling, in particular of nitrogenous compounds (23), and some marine invertebrates with endosymbiotic bacteria show a reduced body plan, e.g., gutless oligochaetes or pogonophoran tube worms (11). If Xenoturbella is a derived deuterostome (5) that feeds on (protein-rich) mollusks (3, 4), nitrogen detoxification by symbionts may have facilitated a secondary loss of excretion organs. If it is an ancestral primitive worm (14) living on dissolved organic matter (18), it may be more likely that the symbionts supply growth factors or chemical defense to their host. Though still hypothetical, the possibility that the endosymbionts provide essential functions to Xenoturbella and may have contributed to its secondary simplification should be considered when interpreting the ecology and evolution of Xenoturbella.
Nucleotide sequence accession numbers.
The sequences reported here have been deposited in GenBank under accession numbers EU400612 to EU400614 and GU129934 to GU129939.
Supplementary Material
Acknowledgments
We are grateful to Britta Poulsen for expert technical assistance, to the crew of Oscar von Sydow and Arne Tiselius at the Sven Lovén Centre for Marine Sciences—Kristineberg for help with sampling, and to Sarah Bourlat, Thomas Stach, and Olle Israelsson for instructions on sampling methods.
This study was supported by the Danish Natural Science Research Council (FNU grant to A.S., K.U.K., and P.F.). M.O. was funded by the Royal Swedish Academy, and H.N. was supported by the Human Frontier Science Program Long-Term Fellowship and the Swedish Research Council.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourlat, S. J., C. Nielsen, A. E. Lockyer, D. T. Littlewood, and M. J. Telford. 2003. Xenoturbella is a deuterostome that eats molluscs. Nature 424:925-928. [DOI] [PubMed] [Google Scholar]
- 4.Bourlat, S. J., H. Nakano, M. Åkerman, M. J. Telford, M. C. Thorndyke, and M. Obst. 2008. Feeding ecology of Xenoturbella bocki (phylum Xenoturbellida) revealed by genetic barcoding. Mol. Ecol. Resources 8:18-22. [DOI] [PubMed] [Google Scholar]
- 5.Bourlat, S. J., O. Rota-Stabelli, R. Lanfear, and M. J. Telford. 2009. The mitochondrial genome structure of Xenoturbella bocki (phylum Xenoturbellida) is ancestral within the deuterostomes. BMC Evol. Biol. 9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Daims, H., K. Stöcker, and M. Wagner. 2005. Fluorescence in situ hybridization for the detection of prokaryotes, p. 213-239. In A. M. Osborn and C. J. Smith (ed.), Advanced methods in molecular microbial ecology. BIOS Scientific Publishers, Abingdon, United Kingdom.
- 9.Daims, H., S. Lücker, and M. Wagner. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200-213. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, A. E. 1995. The ecology of symbiotic microorganisms. Adv. Ecol. Res. 26:69-103. [Google Scholar]
- 11.Dubilier, N., C. Bergin, and C. Lott. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6:725-740. [DOI] [PubMed] [Google Scholar]
- 12.Everett, K. D. E., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 13.Folmer, O., M. Black, W. Hoeh, R. Lutz, and R. Vrijenhoek. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3:294-299. [PubMed] [Google Scholar]
- 14.Hejnol, A., M. Obst, A. Stamatakis, M. Ott, G. W. Rouse, G. D. Edgecombe, P. Martinez, J. Baguñà, X. Bailly, U. Jondelius, M. Wiens, W. E. Müller, E. Seaver, W. C. Wheeler, M. Q. Martindale, G. Giribet, and C. W. Dunn. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc. Biol. Sci. 276:4261-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks, R. E., R. I. Amann, and D. A. Stahl. 1992. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl. Environ. Microbiol. 58:2158-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt, D. E., V. Klepac-Ceraj, S. G. Acinas, C. Gautier, S. Bertilsson, and M. F. Polz. 2006. Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl. Environ. Microbiol. 72:2221-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Israelsson, O. 2007. Chlamydial symbionts in the enigmatic Xenoturbella (Deuterostomia). J. Invertebr. Pathol. 96:213-220. [DOI] [PubMed] [Google Scholar]
- 18.Israelsson, O. 2008. Xenoturbella (Deuterostomia) probably feeds on dissolved organic matter. Mar. Biol. Res. 4:384-391. [Google Scholar]
- 19.Lane, D. J. 1991. 16/23S rRNA sequencing, p. 113-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 20.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss, C., D. H. Green, B. Pérez, A. Velasco, R. Henríquez, and J. D. McKenzie. 2003. Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: phylogenetic and in situ hybridisation analysis. Mar. Biol. 143:99-110. [Google Scholar]
- 23.Moya, A., J. Peretó, R. Gil, and A. Latorre. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218-229. [DOI] [PubMed] [Google Scholar]
- 24.Murray, R. G. E., and E. Stackebrandt. 1995. Implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 45:186-187. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obst, M., H. Nakano, S. J. Bourlat, M. C. Thorndyke, M. J. Telford, J. R. Nyengaard, and P. Funch. The spermatozoan ultrastructure of Xenotubella bocki Westblad 1949. Acta Zool, in press.
- 27.Pedersen, K. J., and L. R. Pedersen. 1986. Fine structural observations on the extracellular matrix (ECM) of Xenoturbella bocki Westblad, 1949. Acta Zool. (Stockh.) 67:103-113. [Google Scholar]
- 28.Poppert, S., A. Essig, R. Marre, M. Wagner, and M. Horn. 2002. Detection and differentiation of chlamydiae by fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:4081-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 30.Pruesse, E., C. Quast, K. Knittel, B. Fuchs, W. Ludwig, J. Peplies, and F. O. Glöckner. 2007. Silva: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 32.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 34.Telford, M. J. 2008. Xenoturbellida: the fourth deuterostome phylum and the diet of the worms. Genesis 46:580-586. [DOI] [PubMed] [Google Scholar]
- 35.Westblad, E. 1949. Xenoturbella bocki n.g., n.sp., a peculiar, primitive turbellarian type. Ark. Zool. 1:11-29. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.