Abstract
The endolysin Lyb5, from Lactobacillus fermentum temperate bacteriophage φPYB5, showed a broad lytic spectrum against Gram-positive as well as Gram-negative bacteria. Sequence analysis revealed that the C terminus of the endolysin Lyb5 (Ly5C) contained three putative lysin motif (LysM) repeat regions, implying that Ly5C was involved in bacterial cell wall binding. To investigate the potential of Ly5C for surface display, green fluorescent protein (GFP) was fused to Ly5C at its N or C terminus and the resulting fusion proteins were expressed in Escherichia coli. After being mixed with various cells in vitro, GFP was successfully displayed on the surfaces of Lactococcus lactis, Lactobacillus casei, Lb. brevis, Lb. plantarum, Lb. fermentum, Lb. delbrueckii, Lb. helveticus, and Streptococcus thermophilus cells. Increases in the fluorescence intensities of chemically pretreated L. lactis and Lb. casei cells compared to those of nonpretreated cells suggested that the peptidoglycan was the binding ligand for Ly5C. Moreover, the pH and concentration of sodium chloride were optimized to enhance the binding capacity of GFP-Ly5C, and high-intensity fluorescence of cells was observed under optimal conditions. All results suggested that Ly5C was a novel anchor for constructing a surface display system for lactic acid bacteria (LAB). To demonstrate the applicability of the Ly5C-mediated surface display system, β-galactosidase (β-Gal) from Paenibacillus sp. strain K1, replacing GFP, was functionally displayed on the surfaces of LAB cells via Ly5C. The success in surface display of GFP and β-Gal opened up the feasibility of employing the cell wall anchor of bacteriophage endolysin for surface display in LAB.
Surface display of heterologous proteins or peptides on bacteria is potentially important in several areas of biotechnology, including development of live vaccine delivery systems, diagnostics, whole-cell absorbents, and novel biocatalysts (11). Lactic acid bacteria (LAB) have the status of being generally recognized as safe (GRAS), making them certainly more useful in food and medical applications than other bacterial species. The development of cell surface display systems for LAB has recently become one of the most active research areas. Most of the cell surface display systems for LAB reported thus far have made use of the C terminus of a cell wall-anchoring protein via an LPXTG motif (8, 12, 19, 24). This anchoring mechanism requires processing by a sortase for covalent anchoring of the protein to the cell wall peptidoglycan (15). Various anchoring proteins, such as membrane-spanning protein PgsA (16) and S-layer protein (3), have also been exploited for surface display. However, heterologous proteins have been anchored to the producer cells, and the use of genetically modified organisms is less desirable or at least still being debated. Surface display of heterologous proteins on genetically unmodified Gram-positive bacteria has been successfully carried out using the peptidoglycan binding lysin motif (LysM) domain of the major autolysin AcmA of Lactococcus lactis (1, 2, 4, 18, 28).
LysM was first discovered in the lysozyme of Bacillus phage φ29 as a C-terminal repeat composed of 44 amino acids separated by 7 amino acids (6). LysM is a common module found in more than 4,000 proteins of both prokaryotes and eukaryotes (6). Many bacterial proteins containing LysM are peptidoglycan hydrolases, such as p60 (20), Sep (26), LytF (31), AcmA (5), and Mur (7). The best-characterized LysM-containing protein is the N-acetylglucosaminidase AcmA of L. lactis subsp. cremoris MG1363. AcmA is the major autolysin and is required for cell separation and cell lysis during the stationary phase of L. lactis (5). It contains three domains: the N-terminal signal peptide, an active domain, and a C-terminal peptidoglycan anchor (cA) which consists of three LysM repeats (22). Several functional proteins, including malaria parasite surface antigen, β-lactamase, α-amylase, and viral capsid proteins, have been noncovalently bound to cell walls of AcmA-producing and non-AcmA-producing L. lactis as well as several other Gram-positive bacteria via cA (4, 17, 18, 23, 25).
Endolysins from bacteriophages are cell wall hydrolases involved in cell lysis to release the progeny particles from the host cells (9, 30). Most endolysins lack a signal peptide and are translocated across the membrane by the aid of the holin protein. This protein typically contains an N-terminal catalytic domain and a C-terminal cell wall binding domain (33). The endolysins Ply118 and Ply500 of a Listeria monocytogenes phage share a unique C-terminal cell wall binding domain which establishes specific recognition of and high-affinity binding to bacterial cell wall carbohydrates (13). The temperate bacteriophage φPYB5, isolated from the Lactobacillus fermentum YB5 strain, has a hexagonal head, noncontractile tails, and several fibers and belongs to Bradley's group B as defined by the International Committee on Taxonomy of Viruses (32). The sequence of the endolysin gene lyb5 from the genome of φPYB5 has been deposited in GenBank under accession number EF531306, and the gene product has been successfully expressed in Escherichia coli and has shown a broad lytic spectrum (30).
Here, we generated a fusion of green fluorescent protein (GFP) to the C terminus of Lyb5 (Ly5C) to construct a surface display system for LAB. The GFP was bound to the surfaces of various LAB cells by the aid of Ly5C. Moreover, by using the system constructed, β-galactosidase (β-Gal) was functionally displayed on the surfaces of LAB cells and retained its activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli BL21(DE3) and E. coli Origami B(DE3) were used as the expression hosts, and E. coli DH5α was used as the cloning host. E. coli strains were grown aerobically in Luria-Bertani (LB) broth at 37°C. L. lactis and Streptococcus thermophilus were grown anaerobically in M17 broth (Oxoid, Basingstoke, United Kingdom) supplemented with 0.5% (wt/vol) glucose (hereinafter referred to as GM17) at 30 and 42°C, respectively. Lactobacillus strains were cultured in MRS broth (Oxoid, Basingstoke, United Kingdom) at 37°C without aeration. Antibiotics were added for E. coli as follows: 100 μg/ml ampicillin, 34 μg/ml chloramphenicol, and 12.5 μg/ml tetracycline.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers used in this study
| Strain, plasmid, or primer | Relevant characteristic(s), description, or sequencea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169 φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Novagen |
| E. coli BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| E. coli Origami B(DE3) | F−ompT hsdSB(rB− mB−) gal dcm lacY1 ahpC gor522::Tn10 (Tcr) trxB::Kan (DE3) | Novagen |
| L. lactis NZ9000 | MG1363 pepN::nisRK; most commonly used host for NICE system | O. P. Kuipers |
| Lb. plantarum K10 | Wild type | Our lab |
| Lb. brevis K31 | Wild type | Our lab |
| Lb. casei BL23 | Kind gift from S. Hazebrouck | 10 |
| S. thermophilus K11 | Wild type | Our lab |
| Lb. helveticus K17 | Wild type | Our lab |
| Lb. buchneri K43 | Wild type | Our lab |
| Lb. fermentum K37 | Wild type | Our lab |
| Lb. delbrueckii subsp. bulgaricus ATCC 11842 | Kind gift from M. van de Guchte | 27 |
| Plasmids | ||
| pBluescript-SK-lysin | pBluescript SK ligated to 1,257-bp PCR product containing lyb5 gene | 30 |
| pET-22b (+) | Apr; E. coli expression vector | Novagen |
| pWaldo-Control | pET-28b ligated to 725-bp GFPuv gene at NdeI and HindIII sites | 29 |
| pUC-bga | pUC ligated to 2,000-bp PCR product containing bga gene | 14 |
| pET-ly5C | Apr; pET-22b (+) carrying HindIII/XhoI-digested product expressing His6-tagged Ly5C | This study |
| pET-gfp | Apr; pET-22b (+) carrying NdeI/HindIII-digested product expressing His6-tagged GFP | This study |
| pET-gfp-ly5C | Apr; pET-22b (+) carrying NdeI/XhoI-digested product expressing His6-tagged GFP-Ly5C | This study |
| pET-gal | Apr; pET-22b (+) carrying NdeI/HindIII-digested product expressing His6-tagged β-Gal | This study |
| pET-gal-ly5C | Apr; pET-22b (+) carrying NdeI/XhoI-digested product expressing His6-tagged β-Gal-Ly5C | This study |
| Primers | ||
| LM1 | 5′-ACTAAGCTTTCTAGACACCAGGACACGCACAAC-3′ | This study |
| LM2 | 5′-GTGCTCGAGATAGTAGAGAGTTTGGCCG-3′ | This study |
| GFP-HindIII | 5′-GCGAAGCTTATGAGCAAAGGAGAAG-3′ | This study |
| GFP-XhoI | 5′-ACACTCGAGGTAGAGCTCATCCATG-3′ | This study |
| LM-NdeI | 5′-CGACATATGTCTAGACACCAGGACA-3′ | This study |
| LM-HindIII | 5′-GTGAAGCTTATAGTAGAGAGTTTGG-3′ | This study |
| Gal1 | 5′-CATCATATGATTAGCAGCAAATTACC-3′ | This study |
| Gal2 | 5′-AACAAGCTTCATCTCAAGCAGCTGAAC-3′ | This study |
The restriction sites in the primer sequences are underlined. Tcr, tetracycline resistance; Kan, kanamycin resistance; Apr, ampicillin resistance; NICE system, nisin-controlled gene expression system.
General molecular biology.
Molecular cloning techniques were performed essentially as described previously (21). All PCRs were carried out using Taq polymerase (TaKaRa, Tokyo, Japan), and the oligonucleotides used are listed in Table 1. Restriction enzymes and T4 DNA ligase were used according to the instructions of the manufacturer (TaKaRa, Tokyo, Japan).
The Ly5C gene was PCR amplified from the plasmid pBluescript-SK-lysin by using primers LM1 and LM2. The PCR products were digested with HindIII and XhoI and then cloned into similarly digested pET-22b (+) to generate pET-ly5C. The GFP gene was digested from the plasmid pWaldo-Control by using NdeI and HindIII and inserted into the corresponding sites of pET-22b (+) and pET-ly5C, yielding pET-gfp and pET-gfp-ly5C, respectively. The GFP gene was amplified by PCR from the plasmid pWaldo-Control with oligonucleotides GFP-HindIII and GFP-XhoI, and then HindIII/XhoI-digested PCR products were cloned into the corresponding sites of pET-22b (+) to generate pET-gfp2. The Ly5C gene was amplified by PCR with primers LM-NdeI and LM-HindIII and inserted into the NdeI and HindIII sites of the plasmid pET-gfp2, generating the plasmid pET-ly5C-gfp, which could express the fused protein Ly5C-GFP. The β-Gal gene (gal) from Paenibacillus sp. strain K1 (14) was PCR amplified with primers Gal1 and Gal2 using pUC-bga as the template. The PCR products were digested with NcoI and HindIII and ligated into the similarly digested pET-22b (+) and pET-ly5C. The resulting plasmids were designated pET-gal and pET-gal-ly5C. E. coli BL21(DE3) was transformed with the recombinant plasmids pET-gfp, pET-gfp-ly5C, and pET-ly5C-gfp by the standard CaCl2 heat shock protocol (21), and pET-gal and pET-gal-ly5C were introduced into E. coli Origami B(DE3).
Overexpression of Ly5C fusion proteins in E. coli.
E. coli strains harboring the recombinant plasmids constructed as described above were grown overnight at 37°C in LB medium supplemented with 100 μg/ml ampicillin. Subsequently, the overnight cultures were diluted 100-fold in 500 ml of fresh LB broth, and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the cultures reached an optical density at 600 nm (OD600) of 0.8. After further incubation for 4 h at 30°C, cells were harvested, washed, and resuspended in 100 ml of phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4). Then the cells were disrupted by sonication on ice at 300 W for two cycles (one cycle consists of 50 periods of sonication for 3 s with intermissions of 8 s), and the clear lysates were centrifuged at 12,000 rpm for 30 min and passed through a filter (0.22-μm pore size; Millipore) to remove the cell debris. Next, the clear lysates were applied to HisTrap FF crude columns (GE Healthcare) equilibrated with binding buffer (20 mM sodium phosphate, 500 mM NaCl, 25 mM imidazole, pH 7.4), the columns were washed with washing buffer (20 mM sodium phosphate, 500 mM NaCl, 50 mM imidazole, pH 7.4), and the His-tagged proteins were eluted in elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). Concentrations of purified proteins were determined using the Bradford protein assay. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to analyze protein expression.
Binding of Ly5C fusion proteins to LAB.
LAB strains were grown as mentioned above, and cells from 1-ml aliquots of stationary-phase cultures were collected, washed twice, and resuspended in 100 μl of PBS. The cells were then incubated with 1-ml samples of the clear lysates containing excessive GFP, GFP-Ly5C, Ly5C-GFP, β-Gal, and β-Gal-Ly5C proteins for 30 to 60 min at 37°C. After binding, cells were collected by centrifugation at 10,000 rpm for 5 min and resuspended in 1 ml of PBS with vortex mixing. After being washed five times with PBS, cells were resuspended in 1 ml of PBS. The cells bound with Ly5C fusion proteins were assayed by whole-cell fluorescence measurement, fluorescence microscopy, and analysis of β-Gal activity.
The cell-associated fluorescence on an untreated black 96-well polystyrene test plate was measured by using an LS-50B spectrofluorometer (PerkinElmer) with excitation at 488 nm and emission at 511 nm. The background fluorescence of cells was subtracted to obtain the relative fluorescence units (RFU). The cell density at 600 nm was measured, and the whole-cell fluorescence per OD600 unit was calculated. The fluorescent cells were observed by fluorescence microscopy with an Eclipse TE2000-S instrument (Nikon, Japan). The β-Gal activity of the cells was assayed using o-nitrophenyl-β-d-galactopyranoside (ONPG) as a chromogenic substrate (14).
Cell pretreatments.
L. lactis and Lb. casei are the most widely used LAB, and here, they were chosen as representatives for Lactococcus and Lactobacillus strains for the subsequent experiments. Cells of L. lactis and Lb. casei in the stationary phase were harvested, resuspended in PBS, and adjusted to an OD600 of 1.0 before being treated with 0.2 volumes of different chemicals. The chemical treatments were as follows: 10% trichloroacetic acid (TCA), 5% TCA, 0.02% TCA, 0.01 M hydrochloric acid (HCl), 5.6 M acetic acid (HAc), 0.72 M lactic acid, and 10% SDS at 100°C for 10 min and 90% acetone at room temperature for 10 min. After the treatments, the cells were washed completely with PBS to remove any residual chemicals. The pretreated cells were subsequently used for Ly5C binding assays as described above.
Influence of cell growth phase, NaCl, pH, and binding time on Ly5C binding capacity.
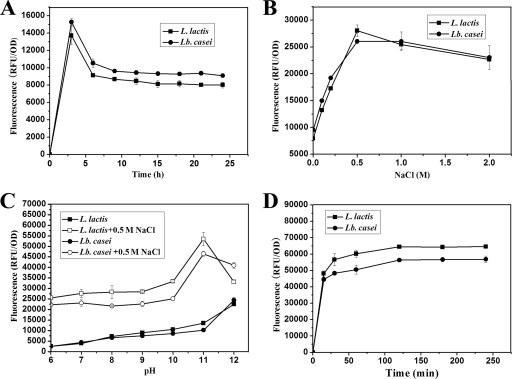
L. lactis and Lb. casei were grown in GM17 and MRS medium, respectively. At regular intervals, the cells were collected and washed twice with PBS and cell samples were diluted to an OD600 of 1.0. After being treated with a 0.2 volume of 5% TCA and washed with PBS, the cells from different time points were used for Ly5C binding assays.
The influence of NaCl on the binding of Ly5C to the surfaces of L. lactis and Lb. casei cells was tested with PBS buffer supplemented with final concentrations of 0, 0.1, 0.2, 0.5, 1.0, and 2.0 M NaCl. Cells of both L. lactis and Lb. casei pretreated with 5% TCA were mixed with GFP-Ly5C. After incubation for 30 min at 37°C, the cell-associated fluorescence was measured as described above. The effect of pH was evaluated over a range from pH 5 to 12 with 0.5 M NaCl or no NaCl. The GFP-Ly5C binding assay was performed as described above.
The binding time course assay was carried out essentially as the standard GFP-Ly5C binding assay described above, using 5% TCA-pretreated L. lactis and Lb. casei cells. At different time points (15, 30, 60, 120, 180, and 240 min) after being mixed with GFP-Ly5C, cells were pelleted and washed with PBS and the fluorescence level per OD600 unit was measured.
RESULTS
Sequence analysis of endolysin Lyb5.
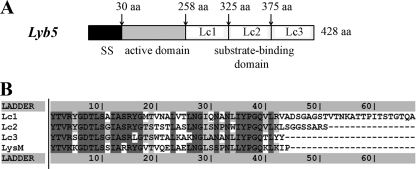
The endolysin gene lyb5 from Lb. fermentum temperate bacteriophage φPYB5 encodes a protein of 418 amino acids with a deduced molecular mass of 45 kDa (30). A signal peptide was predicted by using SOPMA software, and the cleavage site was located between amino acids 29 and 30 at the N terminus of Lyb5 (Fig. 1A). Based on data from reverse position-specific BLAST, two conserved domains in Lyb5 were identified. One, between amino acids 30 and 257, is the active domain and belongs to the GH25 muramidase superfamily, which can degrade cell walls by catalyzing the hydrolysis of 1,4-β-linkages between N-acetylmuramic acid and N-acetyl-d-glucosamine residues. The other is the substrate binding domain located in the C terminus (Ly5C) and consists of three putative LysM regions (Lc1, Lc2, and Lc3) between amino acids 258 and 418. Each of the Lc regions is composed of 41 amino acids, and the regions are separated by intervening sequences that vary in length and composition. The sequence between Lc1 and Lc2 consists of 26 amino acids, while the sequence between Lc2 and Lc3 consists of 10 amino acids. The overall level of similarity among the three repeated regions of Ly5C is approximately 70%. Moreover, the LysM repeated regions of Ly5C have about 63% similarity to the pfam01476 LysM domain (Fig. 1B). Comparing Ly5C with the C termini of AcmA (cA; containing three LysM repeats), Sep (containing one LysM repeat), Mur (containing four LysM repeats), and LAF_0486 from Lb. fermentum IFO 3956 revealed the levels of similarity to be 45, 46, 45, and 52%, respectively. The three LysM regions of Ly5C contain YG repeat units found in the carbohydrate or cell wall binding domains.
FIG. 1.
(A) Schematic illustration of Lyb5. SS indicates a signal peptide; the N-terminal catalytic domain and C-terminal repeated sequences are schematically indicated. aa, amino acids. (B) Alignment of the three putative LysM repeats of Lyb5, Lc1, Lc2, and Lc3, with the consensus LysM domain pfam01476.
Displaying GFP-Ly5C on the cell surfaces of LAB.
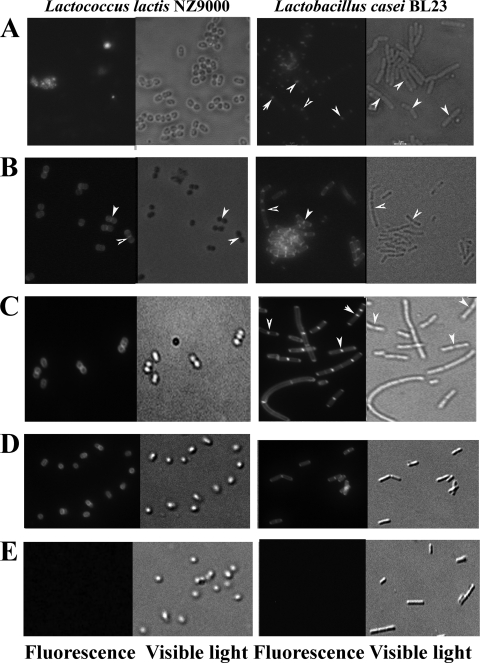
To investigate the potential of Ly5C for displaying heterologous proteins on the cell surfaces of LAB, the GFP which spontaneously emits green light at 508 nm when excited with blue light at 395 nm (GFPuv) was used as a reporter protein (29). The GFPuv gene was obtained from pWaldo-Control, and the gene product was fused to Ly5C at its N or C terminus. GFP-Ly5C and Ly5C-GFP (45 kDa) and GFP (28 kDa) were produced in E. coli (data not shown). After exogenous addition of cell lysates containing GFP-Ly5C, both L. lactis and Lb. casei cells were decorated with green fluorescence (Fig. 2A). As a control, no fluorescence was observed on cells incubated with GFP (Fig. 2E), suggesting that Ly5C was able to efficiently direct GFP to the surfaces of L. lactis and Lb. casei cells. Moreover, the Ly5C-GFP fusion protein was also displayed on the surfaces of L. lactis and Lb. casei cells (Fig. 2D), indicating that the location of GFP did not influence Ly5C binding. Interestingly, the binding of GFP was located at the specific loci of lactococcus and lactobacillus cells (arrowheads in Fig. 2) where the newly synthesized peptidoglycan was inserted into the cell wall.
FIG. 2.
Localization of GFP-Ly5C on L. lactis and Lb. casei cells. (A) Localization of the GFP-Ly5C fusion protein on nonpretreated L. lactis and Lb. casei cells. (B) Localization of the GFP-Ly5C fusion protein on TCA-pretreated L. lactis and Lb. casei cells. (C) Localization of the GFP-Ly5C fusion protein on TCA-pretreated L. lactis and Lb. casei cells at pH 11 in the presence of 0.5 M NaCl. (D) Localization of the Ly5C-GFP fusion protein on TCA-pretreated L. lactis and Lb. casei cells. (E) Control samples (L. lactis and Lb. casei cells mixed with GFP). Arrowheads indicate sites of GFP binding at specific loci where newly synthesized peptidoglycan is inserted into the cell wall.
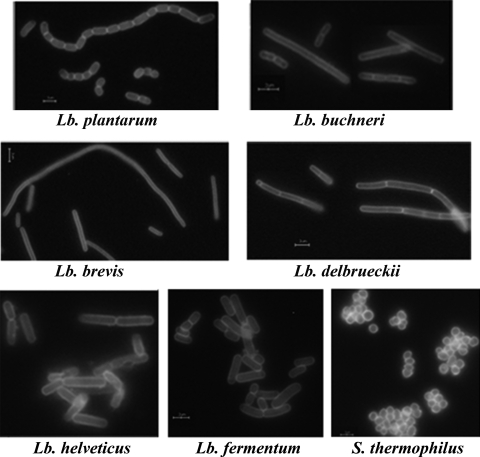
To further determine whether Ly5C can serve as an anchor protein with other LAB, TCA-pretreated cells of Lb. brevis, Lb. plantarum, Lb. delbrueckii, Lb. buchneri, Lb. helveticus, Lb. fermentum, and S. thermophilus were tested. As shown in Fig. 3, GFP was presented on the entirety of the surfaces of LAB cells by the direction of Ly5C. This result demonstrated that Ly5C was necessary and sufficient as an anchor for displaying proteins on the cell surfaces of LAB.
FIG. 3.
Localization of GFP-Ly5C fusion protein on various TCA-pretreated LAB cells at pH 11 in the presence of 0.5 M NaCl. Scale bars, 10 μm.
GFP-Ly5C binds to peptidoglycan.
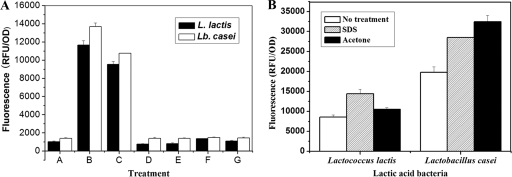
TCA is known to remove mainly (lipo)teichoic acids from cell walls, which can prevent the C terminus of AcmA from binding to the cell surface (4, 23). To improve the binding capacity of Ly5C, TCA, HCl, HAc, and lactic acid were used to pretreat L. lactis and Lb. casei cells. Compared with nonpretreated cells of L. lactis and Lb. casei, TCA-pretreated cells demonstrated a 10-fold increase in fluorescence intensity, indicating that TCA was effective in enhancing Ly5C binding capacity. Moreover, 10% TCA and 5% TCA had the same effect on the binding capacity of GFP-Ly5C, while 0.2% TCA had no impact (Fig. 4A). In addition, fluorescence microscopic observation showed that the 5% TCA treatment left the cellular morphology largely intact and made the binding of GFP-Ly5C occur on the entire cell surface (Fig. 2B).
FIG. 4.
Effects of different chemical pretreatments on the capacities of Ly5C to bind to L. lactis and Lb. casei cells. (A) Effects of pretreatment with different acids on Ly5C binding capacity. Bars: A, no treatment; B, 10% TCA; C, 5% TCA; D, 0.2% TCA; E, 0.01 M HCl; F, 5.6 M HAc; and G, 0.72 M lactic acid. (B) Effects of SDS and acetone on Ly5C binding at pH 11 in the presence of 0.5 M NaCl. The data presented are the averages and standard deviations of results from four independent experiments.
To identify to which cell wall component Ly5C binds, equal amounts of cells of L. lactis and Lb. casei were pretreated with SDS or acetone to remove cell wall-associated proteins or lipids, respectively. The results in Fig. 4B showed that GFP-Ly5C could bind to both SDS- and acetone-pretreated cells and that the fluorescence intensity of pretreated cells was improved. These results demonstrated that cell wall-associated proteins or lipids were not the Ly5C binding components. Considering the cell wall components of Gram-positive bacteria, the peptidoglycan was determined to be the binding ligand for Ly5C.
Factors influencing binding of GFP-Ly5C to lactococcus and lactobacillus cells.
To enhance the binding capacity of GFP-Ly5C, some factors that influence GFP-Ly5C binding, such as the cell growth phase, pH, NaCl concentration, and binding time, were tested. As shown in Fig. 5A, L. lactis and Lb. casei cells incubated for 3 h had the highest fluorescence intensity. The fluorescence intensity then decreased slightly with increasing incubation times. To harvest more cells, both L. lactis and Lb. casei cells incubated for 12 h were collected and used for the subsequent binding experiments. NaCl could significantly enhance the binding capacity, and the optimal concentration was 0.5 M, indicating that ionic interaction by way of charge was the molecular basis for the binding of Ly5C to the cells (Fig. 5B). The cell-associated fluorescence intensity obtained when binding was carried out at pH 11 was increased 2-fold over that obtained at lower pHs (Fig. 5C). In addition, the fluorescence intensity of cells reached a high level when the cells were mixed with GFP-Ly5C for 30 min, and no obvious increase was observed with prolonged exposure to GFP (Fig. 5D). Fluorescence microscopic observation also showed that both L. lactis and Lb. casei cells exhibited high-intensity green fluorescent decoration when binding was carried out at pH 11 in the presence of 0.5 M NaCl (Fig. 2C and 3). Furthermore, for the nonpretreated cells of L. lactis and Lb. casei, an 8-fold increase in fluorescence intensity at pH 11 in the presence of NaCl compared to the intensity under different conditions was also achieved (Fig. 4B). All the results suggested that changing the NaCl concentration or pH was an effective method to improve the capacity of GFP-Ly5C binding to LAB cells.
FIG. 5.
Binding of GFP-Ly5C to cells of L. lactis and Lb. casei under different conditions with the variables of growth phase (A), NaCl concentration (B), pH (C), and binding time (D). The data points represent the averages and standard deviations of results from four independent experiments.
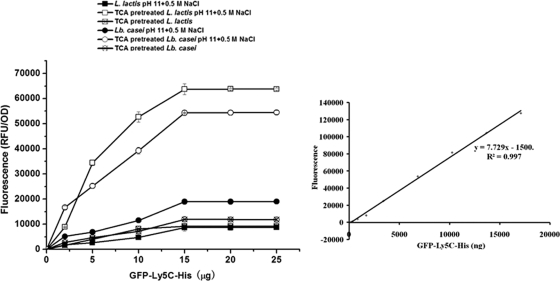
To determine the maximum binding capacity, different amounts of purified GFP-Ly5C fusion protein were mixed with cells of L. lactis and Lb. casei at an OD of 1 under various conditions. As shown in Fig. 6, cells of L. lactis and Lb. casei at an OD of 1 (approximately 108 CFU/ml) could bind approximately 35 to 40 μg of GFP-Ly5C fusion protein (45 kDa) under the optimal conditions. Thus, each cell could bind approximately 106 GFP-Ly5C molecules.
FIG. 6.
(Left) Quantificational detection of surface-displayed GFP-Ly5C fusion protein on cells of L. lactis and Lb. casei under different conditions; (right) standard curve of the fluorescence intensity and the concentration of GFP-Ly5C fusion protein. Different amounts of purified GFP-Ly5C were added to equal amounts of cells for 1 h, and then the cells were collected, washed, and resuspended in 1 ml PBS. Samples of 200 μl were added to the test plate to measure the fluorescence and cell density at 600 nm. The data points represent the averages and standard deviations of results from four independent experiments.
Targeting galactosidase on the surfaces of LAB cells.
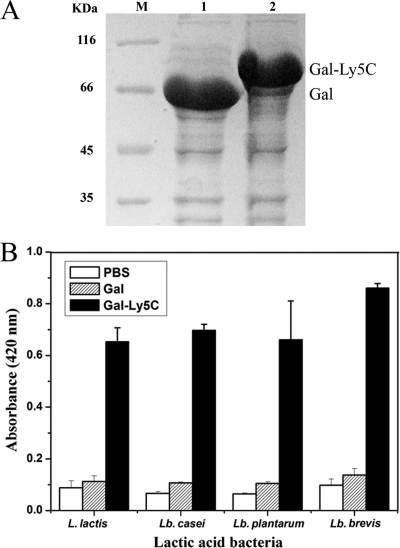
To confirm the application of the surface display system constructed by using Ly5C, β-Gal was fused to Ly5C at its N terminus and E. coli Origami B(DE3) was transformed with the recombinant plasmid expressing the fusion protein. As shown in Fig. 7A, the obvious bands of β-Gal (66 kDa) (lane 1) and β-Gal-Ly5C (85 kDa) (lane 2) were observed at the estimated sizes. The clear lysates containing active β-Gal-Ly5C were added to pretreated cells of L. lactis, Lb. brevis, Lb. casei, and Lb. plantarum for 30 to 60 min. β-Gal activity on the surfaces of cells was detected, indicating that β-Gal-Ly5C was associated with the surfaces of LAB cells and retained β-Gal activity (Fig. 7B). As a control, no galactosidase activity from the cells incubated with β-Gal or PBS was detected.
FIG. 7.
Functional display of β-Gal on the surfaces of LAB cells. (A) Analysis of the expression of β-Gal and β-Gal-Ly5C fusion protein by SDS-PAGE. M, molecular mass markers. (B) Absorbances (at 420 nm) of cells mixed with PBS, β-Gal, and β-Gal-Ly5C. TCA-pretreated cells were mixed with PBS and excessive β-Gal and β-Gal-Ly5C fusion protein. After being mixed for 1 h, cells were collected, washed, and resuspended in the same volume of PBS. The galactosidase activity of 100 μl of bound cells was then measured. The data presented are the averages and standard deviations of results from four independent experiments.
DISCUSSION
Cell surface display in LAB has been studied using various anchor proteins. The LysM domain was a new type of anchoring device and offered an alternative approach for cell surface attachment in LAB (5, 26). AcmA, containing three typical LysM repeats, can bind noncovalently to the peptidoglycan of Gram-positive bacteria, allowing the display of functional proteins or enzymes on the surfaces of genetically unmodified Gram-positive bacteria (1, 2, 18, 25, 28).
In the present study, the C terminus of Lyb5, containing three LysM motifs, has been proven to have the ability to target different heterologous proteins (GFP and β-Gal) onto the surfaces of LAB cells (Fig. 2 and 7), raising the possibility of cell surface display of other proteins (antigens or functional enzymes) by using Ly5C as the anchoring motif. To our knowledge, this is the first report describing the use of the C terminus of an endolysin from a bacteriophage as an anchor for surface display. This strategy has practical applications for the development of oral vaccines, biocatalysts, whole-cell absorbents, and so on. Moreover, cell-free supernatants containing GFP-Ly5C or β-Gal-Ly5C could be directly mixed with LAB cells, and heterologous proteins were targeted to the non-genetically modified bacteria. Therefore, the protein-displaying cells could be probiotic wild-type strains which lack an efficient genetic manipulation system, and the concentration of protein displayed on the cells can be controlled by mixing different amounts of purified protein with the cells. As shown in Fig. 2 and 7, the GFP or β-Gal fused to Ly5C at its N or C terminus did not influence the cell wall binding capacity of Ly5C and also maintained its activity, suggesting that Ly5C was compatible with the heterologous protein to be fused. Finally, Ly5C had the ability to direct the GFP or β-Gal to the surfaces of a broad range of LAB cells (Fig. 3 and 7), providing the advantage of being able to use various probiotic LAB strains isolated from different environments as the host strains to display functional proteins.
For the development of a cell surface display system, it is desirable to obtain anchor proteins that are endowed with a high capacity for cell surface binding. Previous research has demonstrated that TCA treatment is an effective strategy to solve the problem of inefficient binding of the cA domain to the cell wall (4, 23). As shown in Fig. 2, more GFP-Ly5C bound to TCA-pretreated cells than to nonpretreated cells and the binding took place on the entirety of the exposed surfaces of L. lactis and Lb. casei cells, while GFP-Ly5C was located mainly at separate sites (e.g., at the midcell and each pole) on nonpretreated cells. However, TCA may be less desirable for use in large-scale production and causes bacterial cell death. As shown in our study, the NaCl concentration and pH had a significant impact on binding (Fig. 5). The highest capacity for GFP-Ly5C binding to the TCA-pretreated LAB cells was obtained under optimum conditions (at pH 11 in the presence of 0.5 M NaCl). Surprisingly, an 8-fold increase in fluorescence intensity of the nonpretreated cells was also achieved under these conditions, and the fluorescence of nonpretreated cells reached the same level as that of TCA-pretreated cells (Fig. 4B). Therefore, we offer an efficient approach to improve the binding capacity of Ly5C for living cells, thus making the surface display system more advantageous for vaccine development purposes.
In conclusion, Ly5C was firstly described as an anchor domain to display heterologous proteins on the cell surfaces of LAB. A versatile, inexpensive, and effective surface display system based on Ly5C was constructed and used to functionally display β-Gal on the cell surfaces of LAB. Additionally, the binding capacity of the heterologous protein on the cells was significantly enhanced by pretreating cells with TCA or changing the NaCl concentration and pH. Altogether, the novel surface display system based on Ly5C should be useful in enzyme immobilization, the delivery of vaccines, the development of whole-cell absorbents, and so on.
Acknowledgments
We are grateful to Jan-Willem de Gier for kindly providing the plasmid pWaldo-Control. We also thank M. van de Guchte, S. Hazebrouck, and N. Galleron for their generous gifts of Lb. delbrueckii subsp. bulgaricus ATCC 11842, Lb. casei BL23, and L. lactis NZ9000, respectively.
This research was supported by grants from the National Natural Science Foundation of China (NSFC; grant no. 30870023) and from the 863 Hi-Tech Research and Development Program of China (grant no. 2006AA10Z321 and 2008AA10Z335).
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Audouy, S. A., M. L. van Roosmalen, J. Neef, R. Kanninga, E. Post, M. van Deemter, H. Metselaar, S. van Selm, G. T. Robillard, K. J. Leenhouts, and P. W. Hermans. 2006. Lactococcus lactis GEM particles displaying pneumococcal antigens induce local and systemic immune responses following intranasal immunization. Vaccine 24:5434-5441. [DOI] [PubMed] [Google Scholar]
- 2.Audouy, S. A., S. van Selm, M. L. van Roosmalen, E. Post, R. Kanninga, J. Neef, S. Estevão, E. E. Nieuwenhuis, P. V. Adrian, K. Leenhouts, and P. W. Hermans. 2007. Development of lactococcal GEM-based pneumococcal vaccines. Vaccine 25:2497-2506. [DOI] [PubMed] [Google Scholar]
- 3.Avall-Jääskeläinen, S., K. Kylä-Nikkilä, M. Kahala, T. Miikkulainen-Lahti, and A. Palva. 2002. Surface display of foreign epitopes on the Lactobacillus brevis S-layer. Appl. Environ. Microbiol. 68:5943-5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosma, T., R. Kanninga, J. Neef, S. A. Audouy, M. L. van Roosmalen, A. Steen, G. Buist, J. Kok, O. P. Kuipers, G. Robillard, and K. Leenhouts. 2006. Novel surface display system for proteins on non-genetically modified gram-positive bacteria. Appl. Environ. Microbiol. 72:880-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buist, G., A. Steen, J. Kok, and O. P. Kuipers. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838-847. [DOI] [PubMed] [Google Scholar]
- 6.Buist, G., J. Kok, K. J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, S. A., T. Hain, U. Technow, A. Darji, P. Pashalidis, S. W. Joseph, and T. Chakraborty. 2003. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J. Bacteriol. 185:6801-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J. C. Piard. 2001. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donovan, D. M. 2007. Bacteriophage and peptidoglycan degrading enzymes with antimicrobial applications. Recent Pat. Biotechnol. 1:113-122. [DOI] [PubMed] [Google Scholar]
- 10.Hazebrouck, S., L. Pothelune, V. Azevedo, G. Corthier, J. M. Wal, and P. Langella. 2007. Efficient production and secretion of bovine beta-lactoglobulin by Lactobacillus casei. Microb. Cell Fact. 6:6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S. Y., J. H. Choi, and Z. Xu. 2003. Microbial cell-surface display. Trends Biotechnol. 21:45-52. [DOI] [PubMed] [Google Scholar]
- 12.Liu, X., L. A. Lagenaur, P. P. Lee, and Q. Xu. 2008. Engineering of a human vaginal Lactobacillus strain for surface expression of two-domain CD4 molecules. Appl. Environ. Microbiol. 74:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 14.Lu, W., W. Kong, Z. Sun, J. Kong, and M. Ji. 2008. Cloning and expression of the beta-galactosidase gene of Paenibacillus sp. K1 in E. coli. J. Shandong Univ. (Nat. Sci.) 43:69-73. [Google Scholar]
- 15.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narita, J., K. Okano, T. Kitao, S. Ishida, T. Sewaki, M. H. Sung, H. Fukuda, and A. Kondo. 2006. Display of α-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein, and production of lactic acid from starch. Appl. Environ. Microbiol. 72:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okano, K., Q. Zhang, S. Kimura, J. Narita, T. Tanaka, H. Fukuda, and A. Kondo. 2008. System using tandem repeats of the cA peptidoglycan-binding domain from Lactococcus lactis for display of both N- and C-terminal fusions on cell surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 74:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raha, A. R., N. R. Varma, K. Yusoff, E. Ross, and H. L. Foo. 2005. Cell surface display system for Lactococcus lactis: a novel development for oral vaccine. Appl. Microbiol. Biotechnol. 68:75-81. [DOI] [PubMed] [Google Scholar]
- 19.Ribeiro, L. A., V. Azevedo, Y. Le Loir, S. C. Oliveira, Y. Dieye, J. C. Piard, A. Gruss, and P. Langella. 2002. Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 68:910-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruhland, G. J., M. Hellwig, G. Wanner, and F. Fiedler. 1993. Cell-surface location of Listeria-specific protein p60—detection of Listeria cells by indirect immunofluorescence. J. Gen. Microbiol. 139:609-616. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 22.Steen, A., G. Buist, G. J. Horsburgh, G. Venema, O. P. Kuipers, S. J. Foster, and J. Kok. 2005. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 272:2854-2868. [DOI] [PubMed] [Google Scholar]
- 23.Steen, A., G. Buist, K. J. Leenhouts, M. El Khattabi, F. Grijpstra, A. L. Zomer, G. Venema, O. P. Kuipers, and J. Kok. 2003. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278:23874-23881. [DOI] [PubMed] [Google Scholar]
- 24.Steidler, L., J. Viaene, W. Fiers, and E. Remaut. 1998. Functional display of a heterologous protein on the surface of Lactococcus lactis by means of the cell wall anchor of Staphylococcus aureus protein A. Appl. Environ. Microbiol. 64:342-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarahomjoo, S., Y. Katakura, E. Satoh, and S. Shioya. 2008. Bidirectional cell-surface anchoring function of C-terminal repeat region of peptidoglycan hydrolase of Lactococcus lactis IL1403. J. Biosci. Bioeng. 105:116-121. [DOI] [PubMed] [Google Scholar]
- 26.Turner, M. S., L. M. Hafner, T. Walsh, and P. M. Giffard. 2004. Identification and characterization of the novel LysM domain-containing surface protein Sep from Lactobacillus fermentum BR11 and its use as a peptide fusion partner in Lactobacillus and Lactococcus. Appl. Environ. Microbiol. 70:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Guchte, M., S. Penaud, C. Grimaldi, V. Barbe, K. Bryson, P. Nicolas, C. Robert, S. Oztas, S. Mangenot, A. Couloux, V. Loux, R. Dervyn, R. Bossy, A. Bolotin, J. M. Batto, T. Walunas, J. F. Gibrat, P. Bessières, J. Weissenbach, S. D. Ehrlich, and E. Maguin. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U. S. A. 103:9274-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Roosmalen, M. L., R. Kanninga, M. El Khattabi, J. Neef, S. Audouy, T. Bosma, A. Kuipers, E. Post, A. Steen, J. Kok, G. Buist, O. P. Kuipers, G. Robillard, and K. Leenhouts. 2006. Mucosal vaccine delivery of antigens tightly bound to an adjuvant particle made from food-grade bacteria. Methods 38:144-149. [DOI] [PubMed] [Google Scholar]
- 29.Waldo, G. S., B. M. Standish, J. Berendzen, and T. C. Terwilliger. 1999. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17:691-695. [DOI] [PubMed] [Google Scholar]
- 30.Wang, S., J. Kong, and X. Zhang. 2008. Identification and characterization of the two-component cell lysis cassette encoded by temperate bacteriophage phiPYB5 of Lactobacillus fermentum. J. Appl. Microbiol. 105:1939-1944. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, H., Y. Miyake, M. Hisaoka, S. Kurosawa, and J. Sekiguchi. 2008. The major and minor wall teichoic acids prevent the sidewall localization of vegetative dl-endopeptidase LytF in Bacillus subtilis. Mol. Microbiol. 70:297-310. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, X., J. Kong, and Y. Qu. 2006. Isolation and characterization of a Lactobacillus fermentum temperate bacteriophage from Chinese yogurt. J. Appl. Microbiol. 101:857-863. [DOI] [PubMed] [Google Scholar]
- 33.Ziedaite, G., R. Daugelavicius, J. K. Bamford, and D. H. Bamford. 2005. The holin protein of bacteriophage PRD1 forms a pore for small-molecule and endolysin translocation. J. Bacteriol. 187:5397-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]