Abstract
Asthma increased dramatically in the last decades of the 20th century and is representative of chronic diseases that have been linked to altered microbial exposure and immune responses. Here we evaluate the effects of environmental exposures typically associated with asthma protection or risk on the microbial community structure of household dust (dogs, cats, and day care). PCR-denaturing gradient gel analysis (PCR-DGGE) demonstrated that the bacterial community structure in house dust is significantly impacted by the presence of dogs or cats in the home (P = 0.0190 and 0.0029, respectively) and by whether or not children attend day care (P = 0.0037). In addition, significant differences in the dust bacterial community were associated with asthma outcomes in young children, including wheezing (P = 0.0103) and specific IgE (P = 0.0184). Our findings suggest that specific bacterial populations within the community are associated with either risk or protection from asthma.
Recent studies have begun to explore the microbial composition of house dust, finding great diversity and a high abundance of Gram-positive organisms (18, 24). Yet this transient community remains relatively unexplored despite increasing evidence of an association between microbial exposure and human health, in particular, the development of chronic diseases such as asthma. Settings with high levels of microbial exposure, such as farms and day care centers, have been associated with protection from asthma (3, 23), while interventions that reduce home microbial exposures have been related to higher rates of allergic sensitization (29). Moreover, variability in home microbial exposures has been linked to differences in immune response and asthma risk in childhood (1, 4). Finally, genes involved in innate immune responses to microbes have been found to interact with microbial exposures, resulting in altered risks for asthma (10). Microbial exposure has primarily been assessed in these studies through measurement of surrogates, such as lipopolysaccharide (LPS, or endotoxin), a component of the outer membrane of Gram-negative bacteria (16), and more recently N-acetyl muramic acid for Gram-positive bacteria (28) and ergosterol for fungi (25). While the use of such surrogates has confirmed a negative relationship between levels of microbial exposure and the development of asthma, exploration is needed beyond a handful of surrogates to more completely represent the complexity of the actual microbial communities present in homes.
A preliminary experiment was performed to characterize the bacterial communities of dust from homes in Tucson, AZ. First, general bacterial community diversity was examined in house dust samples by using a PhyloChip microarray as described by Brodie et al. (5). These samples were obtained from homes of four volunteers in the Tucson area. Array results revealed an average of 295 ± 16 (mean ± standard deviation) unique operational taxonomic units (OTUs) per sample, where a taxon was considered present in the sample when more than 92% of the assigned probe pairs for its corresponding probe set were positive. Of these OTUs, 85% belonged to the same 13 divisions in each dust sample, with the 5 most dominant divisions including the Alphaproteobacteria (17.6 ± 2.6%), Firmicutes (16.3 ± 4.1%), Actinobacteria (13.4 ± 2.2%), Gammaproteobacteria (11.8 ± 1.1%), and Deltaproteobacteria (8.7 ± 0.4%) (see Fig. S1 in the supplemental material). Distributions among the 13 major divisions were fairly consistent but not identical among the four different dust samples. The remaining 15% of OTUs belonged to 33 other bacterial divisions but were present at frequencies of less than 1% each. Thus, the number of OTUs and the phylogenetic distributions of these OTUs among the four households were similar.
A second experiment was performed to see how the structure of house dust communities varies with environmental parameters and asthma outcomes. The dust samples, examined using denaturing gradient gel electrophoresis (DGGE), were obtained from homes of infants enrolled in the Infant Immune Study (IIS), a nonselected birth cohort (12). Dust samples were collected when the child was approximately 3 months old, at which time the home condition was evaluated. Other information available on this cohort includes data on (i) early life exposures potentially related to asthma risk (the presence of a dog or cat in the home and day care attendance in the first months of life) and (ii) asthma-related outcomes of wheezing and aeroallergen sensitization (specific IgE [Sp-IgE]) (7). A subset (n = 44) of the IIS dust samples was used in this analysis. Based upon the premise that health outcomes will be influenced by the most abundant members of the dust bacterial community (in terms of numbers), DGGE of 16S rRNA gene PCR amplicons was selected as the profile technique to compare the microbial community structures of dust samples from the 44 homes. PCR-DGGE profiles evaluate 16S rRNA genes from all bacterial populations whose initial template concentrations represent >1% of the total community DNA (6, 13, 17). PhyloChip analysis was not used for this experiment because the cost would have been prohibitive for the number of households analyzed.
DNA extraction was performed on the IIS dust samples by direct lysis using the Fast DNA spin kit for soil (MP Biomedicals, Solon, OH) as described previously (9). The amount of dust used for extraction ranged from 21 to 35 mg. For each set of extractions a DNA blank (no dust) was included to monitor potential contamination during the extraction process. The extracted DNA was labeled with the PicoGreen double-stranded DNA (dsDNA) quantitation reagent (Molecular Probes, Eugene, OR) and quantified using the Turner Biosystems TBS 380 fluorometer. DNA yields ranged from 0.2 to 41.2 ng DNA/mg of dust.
For DGGE analysis, the community DNA obtained from the dust samples was PCR amplified (25 cycles) using the V7/V8 variable region of the 16S rRNA gene and primers 1070f and 1392R with a 40-bp GC clamp (11), following the protocol of Colores et al. (8). Unacetylated bovine serum albumin (Sigma, St. Louis, MO) was added to the PCR mixture at a concentration of 0.4 g liter−1 to relieve PCR inhibition. Amplicons were separated on DGGE gels by using the Bio-Rad Laboratories system (D-Code; Hercules, CA) with a 6% acrylamide gel and a 45% to 65% urea-formamide denaturing gradient. Each DGGE gel run in this study examined either an environmental exposure or an asthma-related outcome variable, as detailed in Tables 1 and 2. For example, children reported to have actively wheezed on at least two out of three questionnaires obtained at 1, 2, and 3 years were compared with children for whom all three questionnaires were completed but for whom no wheezing was reported. Fourteen samples were analyzed on each gel, including seven different households from the negative group and seven different households from the positive group. Two lanes of the gel were also loaded with negative controls. DGGE gels were run for 15.5 h at a constant voltage of 50 V, stained with 3× SYBR green I (Molecular Probes, Eugene, OR) for 40 min, and then visualized.
TABLE 1.
Gel comparison groups for early life environmental exposures
| Variable | Assessment age | Negative gel group (not exposed) | Positive gel group (exposed) |
|---|---|---|---|
| Indoor dogs | 2 wks | No pets (dogs or cats) plus no evidence of pets in home | 2 or more indoor dogs with evidence of dogs in home and no cats |
| Indoor cats | 2 wks | No pets (dogs or cats) plus no evidence of pets in home | Any cat plus evidence of cat in home and no dogs |
| Day care attendance | 1, 2, or 3 mos | No day care by 3 mos | Day care outside home by 3 mos |
| Home condition | 3 mos | Better home condition | Worse home condition |
TABLE 2.
Gel comparison groups for asthma-related outcomes
| Variable | Assessment ages (yrs) | Negative gel group | Positive gel group |
|---|---|---|---|
| Wheeze | 1, 2, and 3 | Questionnaires completed at all three ages but no wheeze | Wheeze reported at two or more ages |
| Total IgE | 1, 2, and 3 | Lowest quartile, all ages | Highest quartile, all ages |
| Sp-IgE | 1, 2, and 3 | No detectable IgE to the specific allergens tested | Detectable specific IgE |
Among the dust samples analyzed, the average number of bands in each DGGE profile (lane) analyzed was 20 ± 5 (range, 10 to 34), where each band is theoretically equivalent to one unique bacterial OTU (17). This can be compared to the 295 unique OTUs identified by PhyloChip analysis, revealing that DGGE detects approximately 7% of the OTUs detected by the microarray.
The DGGE community banding patterns were evaluated using Quantity One 4.5.2 software. Briefly, each possible vertical band location on the gel was assigned a number, in numerical order starting at the top of the gel. Banding patterns were analyzed using canonical correspondence analysis (CCA), a form of correspondence analysis widely used in community ecology (14, 19, 26). CCA finds axes of variation in OTU composition (in our case, profiles or banding patterns) that are maximally related to explanatory variables (e.g., presence of dogs). CCA eigenvalues represent the strength of the relationship between OTUs (DGGE profile bands) and one explanatory variable (exposure or outcome) and are tested against the null model of no relationship by using a permutation test (27). CCA also allows simultaneous visualization of explanatory variables, samples, and OTUs in a few dimensions by using a triplot. In our case, each analysis had only one explanatory variable (exposure or outcome) and hence there was only one canonical axis (20). Binary variables in CCA triplots are typically represented by centroids, i.e., points that reflect the average location of the class (e.g., exposure or outcome versus control). The second and higher axes represent residual axes, i.e., variations in profile OTU patterns that are unrelated to variations linked to the explanatory variable. According to the centroid rule (15), the proximity of an OTU band number to a centroid or a sample is directly related to the occurrence of that OTU in that centroid or sample.
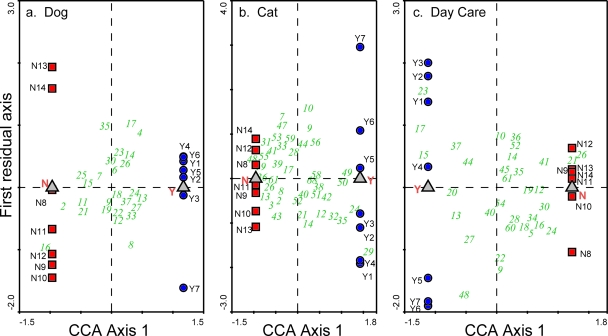
DGGE CCA of four environmental exposure variables (dog, cat, day care attendance, home condition) showed a significant difference in the dust bacterial community structures for three of the variables which have been linked to risk for asthma (21, 22). Specifically, community structure was different in homes that had dogs versus no dogs (P = 0.0190) and cats versus no cats (P = 0.0029) and where children attended or did not attend day care (P = 0.0037) (Fig. 1). There was no significant difference in the dust bacterial community structure by home condition (P = 0.1209).
FIG. 1.
CCA triplot analysis of DGGE gel OTU banding patterns, comparing exposure and control microbial communities in household dust samples as a function of a single environmental variable. (a) Dogs (P = 0.0190); (b) cats (P = 0.0029); (c) day care attendance (P = 0.0037). Dust samples from the same households were used as the control (N) group for the dog and cat exposure analyses, and six out of seven of these dust samples were used as the control group for the day care analysis. No household dust samples were in common between the dog and cat or the day care and cat exposure (Y) groups. One household was in common between the dog and day care exposure (Y) groups. The red squares in each triplot represent the seven control (N) samples (e.g., no dog), and the blue circles represent the seven exposure (Y) samples (e.g., dog). The gray triangles in each triplot represent the centroids, which are the average location of the exposure or control sample. A permutation test was used to evaluate the null model of no relationship between the exposure and controls (P ≤ 0.05). The vertical distribution of sample points above and below the centroids (the residual axis) represents the variation in profile OTU banding patterns that is unrelated to variation between the exposure and control averages (centroids). The green numbers in each panel represent the OTUs from the DGGE gel profiles that have >20% of the maximum abundance. Each number represents a unique vertical OTU location on the DGGE gel. In numerical order, beginning at the top of the gel, a number was assigned to each possible vertical location for a band (OTU) among the 14 lanes analyzed. Thus, the location of these numbers can be used to distinguish the relative frequency of occurrence of an OTU in control and exposure samples. From the CCA plot then, the horizontal distribution of each green number (OTU) provides information about whether it is found more often in the exposure (located closer to the exposure centroid) or control (located closer to the control centroid) samples or is shared equally between them.
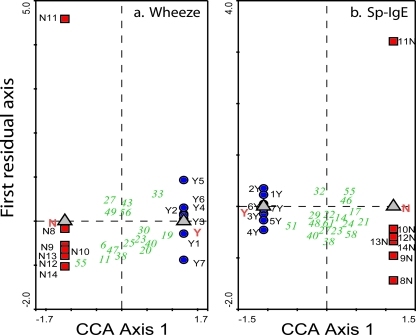
Analysis of asthma-related outcomes showed that there was also a significant difference in the dust bacterial community structure in homes where children exhibited wheeze versus no wheeze (P = 0.0103) and where children were Sp-IgE positive versus Sp-IgE negative (P = 0.0184) (Fig. 2). There was no significant difference in the dust bacterial community structure in homes where children had high total IgE versus low total IgE (P = 0.1998).
FIG. 2.
CCA triplot analysis of DGGE gel OTU banding patterns, comparing outcome and control microbial communities in household dust samples as a function of a single outcome variable. (a) Wheeze (P = 0.0103); (b) Sp-IgE (P = 0.0184). In these analyses, one of the seven household dust samples was the same for the asthma and Sp-IgE control (N) groups. Two out of seven households were the same for the wheeze and specific IgE outcome (Y) analyses. For a detailed explanation of the symbols on the triplot, see the legend for Fig. 1.
These results provide the first evidence that the dominant bacterial populations in household dust are significantly influenced by environmental variables such as domestic animals and day care attendance. Further, the dominant bacterial populations are significantly correlated to asthma-related outcomes, supporting the hypothesis that the types of microorganisms present in homes in early life may play key roles in the development of childhood asthma. This work shows that we can begin to define the relationship between childhood development of chronic immune system-related disease and the bacterial community found within the child's immediate environment. The success of the DGGE CCA suggests that this process can focus initially on the most abundant bacterial populations in the dust samples, a small subset of the entire dust community (as demonstrated by the number of OTUs represented in the DGGE profiles, compared to the PhyloChip results). CCA can potentially help refine this subset even further. The location of any particular bacterial OTU (see green numbers in the triplots in Fig. 1 and 2) in relation to the exposure/outcome versus the control centroids on a CCA triplot provides information about whether it is associated primarily with one centroid or the other or is shared between them. For example, in Fig. 1C the OTUs labeled 15, 17, and 23 are associated with homes where children attended day care. Specifically, each of these three OTUs was found in six (OTU 15), five (OTU 17), and four (OTU 23) of the seven day care home samples tested, while they were found in none of the homes without children who attended day care. Similarly, the OTUs labeled 21 and 26 were each found in four of the seven homes in which no children attended day care and in none of the day care homes. These data suggest that the OTUs associated with a common environmental exposure or asthma-related outcome variable can be identified and the biological mechanisms through which they influence the development of the disease further explored.
In the last century, human disease in the industrialized world has largely shifted from acute infectious illnesses to chronic conditions such as Crohn's disease, multiple sclerosis, and asthma, which stem from abnormal immunologic responses (2). The increase in these conditions coincides with dramatic alterations in human microbial exposure that have occurred following improved sanitation, reduced rural living, and widespread use of antibiotics and antimicrobials. These parallels suggest that unidentified differences in exposure to microbial communities in the industrialized world may have fundamentally changed human immune responses, thereby enhancing susceptibility to autoimmune and allergic diseases. The research presented here provides a foundation for the directed discovery and exploration of specific bacteria that may stimulate or have prophylactic effects on the lifetime development of asthma and other chronic immune diseases.
Supplementary Material
Acknowledgments
This study was supported with funding to R.M.M. from National Science Foundation grant MCB-0604300. A.L.W., M.J.H., and D.A.S. were supported with funding from the National Institute of Allergy and Infectious Diseases (AI61811 and AI 42268).
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abraham, J. H., P. W. Finn, D. K. Milton, L. M. Ryan, D. L. Perkins, and D. R. Gold. 2005. Infant home endotoxin is associated with reduced allergen-stimulated lymphocyte proliferation and IL-13 production in childhood. J. Allergy Clin. Immunol. 116:431-437. [DOI] [PubMed] [Google Scholar]
- 2.Bach, J. F. 2002. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347:911-920. [DOI] [PubMed] [Google Scholar]
- 3.Ball, T. M., J. A. Castro-Rodriguez, K. A. Griffith, C. J. Holberg, F. D. Martinez, and A. L. Wright. 2000. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N. Engl. J. Med. 343:538-543. [DOI] [PubMed] [Google Scholar]
- 4.Braun-Fahrlander, C., J. Riedler, U. Herz, W. Eder, M. Waser, L. Grize, S. Maisch, D. Carr, F. Gerlach, A. Bufe, R. P. Lauener, R. Schierl, H. Renz, D. Nowak, and E. von Mutius. 2002. Environmental exposure to endotoxin and its relation to asthma in school-age children. N. Engl. J. Med. 347:869-877. [DOI] [PubMed] [Google Scholar]
- 5.Brodie, E. L., T. Z. DeSantis, J. P. Parker, I. X. Zubietta, Y. M. Piceno, and G. L. Andersen. 2007. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 104:299-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casamayor, E. O., H. Schafer, L. Baneras, C. Pedros-Alio, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro-Rodriguez, J. A., C. J. Holberg, A. L. Wright, and F. D. Martinez. 2000. A clinical index to define risk of asthma in young children with recurrent wheezing. Am. J. Respir. Crit. Care Med. 162:1403-1406. [DOI] [PubMed] [Google Scholar]
- 8.Colores, G. M., R. E. Macur, D. M. Ward, and W. P. Inskeep. 2000. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 66:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drees, K. P., J. W. Neilson, J. L. Betancourt, J. Quade, D. A. Henderson, B. M. Pryor, and R. M. Maier. 2006. Bacterial community structure in the hyperarid core of the Atacama Desert, Chile. Appl. Environ. Microbiol. 72:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eder, W., W. Klimecki, L. Yu, E. von Mutius, J. Riedler, C. Braun-Fahrlander, D. Nowak, O. Holst, and F. D. Martinez. 2006. Association between exposure to farming, allergies and genetic variation in CARD4/NOD1. Allergy 61:1117-1124. [DOI] [PubMed] [Google Scholar]
- 11.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra, S., I. C. Lohman, M. Halonen, F. D. Martinez, and A. L. Wright. 2004. Reduced interferon gamma production and soluble CD14 levels in early life predict recurrent wheezing by 1 year of age. Am. J. Respir. Crit. Care Med. 169:70-77. [DOI] [PubMed] [Google Scholar]
- 13.Konopka, A., T. Bercot, and C. Nakatsu. 1999. Bacterioplankton community diversity in a series of thermally stratified lakes. Microb. Ecol. 38:126-135. [DOI] [PubMed] [Google Scholar]
- 14.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English ed. Elsevier, Amsterdam, Netherlands.
- 15.Lepš, J., and P. Šmilauer. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, United Kingdom.
- 16.Litonjua, A. A., D. K. Milton, J. C. Celedon, L. Ryan, S. T. Weiss, and D. R. Gold. 2002. A longitudinal analysis of wheezing in young children: the independent effects of early life exposure to house dust endotoxin, allergens, and pets. J. Allergy Clin. Immunol. 110:736-742. [DOI] [PubMed] [Google Scholar]
- 17.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pakarinen, J., A. Hyvarinen, M. Salkinoja-Salonen, S. Laitinen, A. Nevalainen, M. J. Makela, T. Haahtela, and L. von Hertzen. 2008. Predominance of Gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environ. Microbiol. 10:3317-3325. [DOI] [PubMed] [Google Scholar]
- 19.Palmer, M. W. 1993. Putting things in even better order: the advantages of canonical correspondence analysis. Ecology 74:2215-2230. [Google Scholar]
- 20.Palmer, M. W., D. J. McGlinn, L. Westerberg, and P. Milberg. 2008. Indices for detecting differences in species composition: some simplifications of RDA and CCA. Ecology 89:1769-1771. [DOI] [PubMed] [Google Scholar]
- 21.Perzanowski, M. S., E. Ronmark, T. A. Platts-Mills, and B. Lundback. 2002. Effect of cat and dog ownership on sensitization and development of asthma among preteenage children. Am. J. Respir. Crit. Care Med. 166:696-702. [DOI] [PubMed] [Google Scholar]
- 22.Remes, S. T., J. A. Castro-Rodriguez, C. J. Holberg, F. D. Martinez, and A. L. Wright. 2001. Dog exposure in infancy decreases the subsequent risk of frequent wheeze but not of atopy. J. Allergy Clin. Immunol. 108:509-515. [DOI] [PubMed] [Google Scholar]
- 23.Riedler, J., C. Braun-Fahrlander, W. Eder, M. Schreuer, M. Waser, S. Maisch, D. Carr, R. Schierl, D. Nowak, and E. von Mutius. 2001. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet 358:1129-1133. [DOI] [PubMed] [Google Scholar]
- 24.Rintala, H., M. Pitkaranta, M. Toivola, L. Paulin, and A. Nevalainen. 2008. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sebastian, A., and L. Larsson. 2003. Characterization of the microbial community in indoor environments: a chemical-analytical approach. Appl. Environ. Microbiol. 69:3103-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terbraak, C. J. F. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167-1179. [Google Scholar]
- 27.Terbraak, C. J. F., and J. Wiertz. 1994. On the statistical analysis of vegetation change: a wetland affected by water extraction and soil acidification. J. Vegetation Sci. 5:361-372. [Google Scholar]
- 28.van Strien, R. T., R. Engel, O. Holst, A. Bufe, W. Eder, M. Waser, C. Braun-Fahrlander, J. Riedler, D. Nowak, and E. von Mutius. 2004. Microbial exposure of rural school children, as assessed by levels of N-acetyl-muramic acid in mattress dust, and its association with respiratory health. J. Allergy Clin. Immunol. 113:860-867. [DOI] [PubMed] [Google Scholar]
- 29.Woodcock, A., L. A. Lowe, C. S. Murray, B. M. Simpson, S. D. Pipis, P. Kissen, A. Simpson, and A. Custovic. 2004. Early life environmental control: effect on symptoms, sensitization, and lung function at age 3 years. Am. J. Respir. Crit. Care Med. 170:433-439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.