Abstract
We investigated the prevalence of sapoviruses (SaVs) in the Tamagawa River in Japan from April 2003 to March 2004 and performed genetic analysis of the SaV genes identified in river water. A total of 60 river water samples were collected from five sites along the river, and 500 ml was concentrated using the cation-coated filter method. By use of a real-time reverse transcription (RT)-PCR assay, 12 (20%) of the 60 samples were positive for SaV. SaV sequences were obtained from 15 (25%) samples, and a total of 30 SaV strains were identified using six RT-PCR assays followed by cloning and sequence analysis. A newly developed nested RT-PCR assay utilizing a broadly reactive forward primer showed the highest detection efficiency and amplified more diverse SaV genomes in the samples. SaV sequences were frequently detected from November to March, whereas none were obtained in April, July, September, or October. No SaV sequences were detected in the upstream portion of the river, whereas the midstream portion showed high positive rates. Based on phylogenetic analysis, SaV strains identified in the river water samples were classified into nine genotypes, namely, GI/1, GI/2, GI/3, GI/5, GI/untyped, GII/1, GII/2, GII/3, and GV/1. To our knowledge, this is the first study describing seasonal and spatial distributions and genetic diversity of SaVs in river water. A combination of real-time RT-PCR assay and newly developed nested RT-PCR assay is useful for identifying and characterizing SaV strains in a water environment.
Sapoviruses (SaVs), formerly called “Sapporo-like viruses,” belong to the family Caliciviridae and cause acute gastroenteritis in humans and swine (5). The prototype strain of SaV, Hu/SV/GI/Sapporo/77/JP (Sapporo virus), was first detected in 1977 in a gastroenteritis outbreak (1). SaVs are nonenveloped viruses possessing a single-stranded, positive-sense RNA genome with two or three open reading frames (2, 8). SaVs show a high level of diversity in their genomes and are currently divided into at least five genetically distinct genogroups, genogroups I to V (GI to GV), of which GI, GII, GIV, and GV strains infect humans and GIII strains infect swine (4, 8). Human SaVs cannot be cultivated in routine cell culture or animal models. However, developments in molecular techniques have facilitated their detection in clinical and environmental samples. Reverse transcription (RT)-PCR is currently the most widely used assay for detection of SaVs in clinical and environmental samples. Moreover, an RT-PCR assay coupled with nucleotide sequencing techniques enables us to obtain valuable information on the SaV strains (9, 10, 11).
SaV-associated gastroenteritis is becoming more prevalent worldwide. Since SaVs have been detected from fecal samples of infected patients (6, 17, 19, 25, 26, 29, 30) and environmental samples such as wastewater, river water, clams, and oysters (3, 9, 11, 16, 18), it is believed that SaVs are transmitted from person to person via fecal-oral routes and through contaminated foods and water. Caliciviruses, namely, SaVs and noroviruses (NoVs), are included in the latest U.S. Environmental Protection Agency's Contaminant Candidate List (CCL 3), a list of emerging contaminants that may pose a public health risk in water environments (27). However, knowledge of the occurrence and fate of SaVs in the environment is limited compared with that of NoVs. Viral contamination of river water is of epidemiological significance because there are many rivers that receive effluents from wastewater treatment plants (WWTPs) upstream and supply water to drinking water treatment plants (DWTPs) downstream.
In this study, we selected the Tamagawa River in Japan as a typical example of such rivers and investigated the prevalence and genetic diversity of SaVs in river water. Furthermore, we comparatively evaluated the detection rates of human SaV genomes in river water samples among different molecular tools, such as the TaqMan-based real-time RT-PCR assay and single-round, nested, and seminested RT-PCR assays, including the newly developed assays utilizing a broadly reactive primer.
MATERIALS AND METHODS
Collection of river water samples.
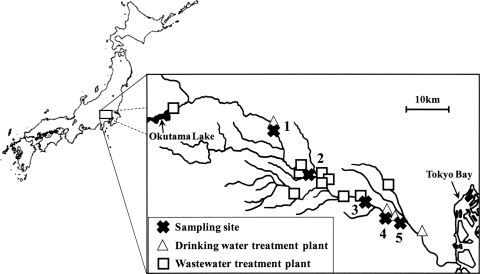
In order to detect human SaV genomes in river water, a total of 60 samples were collected monthly from April 2003 to March 2004 at five sites (sites 1 to 5) along the Tamagawa River in Japan (Fig. 1). All samples were collected on clear days, stored in plastic bottles on ice, and delivered to the laboratory within several hours after collection.
FIG. 1.
Locations of sampling sites, drinking water treatment plants, and wastewater treatment plants along the Tamagawa River.
The Tamagawa River has a total length of 138 km and a catchment area of 1,240 km2, with a small WWTP and DWTP in the upstream area, nine WWTPs in the midstream area, and three DWTPs in the downstream area (Fig. 1). Chlorination has been adopted for disinfecting microorganisms at all of these WWTPs. The effluents from these WWTPs account for nearly half of the river water in the midstream and downstream areas. These WWTPs serve 7%, 94%, and 100% of the populations in the upstream, midstream, and downstream areas, respectively. Septic tank systems are also installed in the upstream and midstream areas.
Concentration of river water samples.
A virus concentration method named the “cation-coated filter method,” developed for concentrating viruses in freshwater (13, 15), was used to concentrate viruses in the river water samples, as previously described (13). Briefly, 5 ml of 250 mM AlCl3 was passed through an electronegative filter (0.45-μm pore size and 90-mm diameter; Millipore, Tokyo, Japan) to form a cation (Al3+)-coated filter, and then 500 ml of the sample was passed through the filter. Aluminum ions were removed by passing 200 ml of 0.5 mM H2SO4 (pH 3.0) through the filter, and viruses were eluted with 10 ml of 1.0 mM NaOH (pH 10.8). The filtrate was recovered in a tube containing 50 μl of 100 mM H2SO4 (pH 1.0) and 100 μl of 100× Tris-EDTA buffer (pH 8.0) for neutralization, followed by further centrifugal concentration using a Centriprep YM-50 (Millipore) to obtain a final volume of 700 μl.
RNA extraction and RT reaction.
Viral RNA was extracted from 140 μl of concentrated sample using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) to obtain a final volume of 60 μl, according to the manufacturer's protocol.
The RT reaction was performed using the High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Briefly, 10 μl of extracted RNA was added to 10 μl of RT mixture containing 2 μl of 10× reverse transcription buffer, 0.8 μl of 25× deoxynucleoside triphosphates (dNTPs), 2 μl of 10× random hexamers, 50 units of MultiScribe reverse transcriptase, and 20 units of RNase inhibitor. The RT reaction mixture was incubated at 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min to inactivate the enzyme.
Real-time PCR.
TaqMan-based real-time PCR for human SaVs was performed by using an ABI PRISM 7500 sequence detection system (SDS) (Applied Biosystems) as described previously (22), with a minor modification. Briefly, 5 μl of cDNA was mixed with 20 μl of a reaction buffer containing 12.5 μl of 2× QuantiTect Probe PCR master mix (Qiagen), 400 nM each primer (SaV124F, SaV1F, SaV5F, and SaV1245R) (Table 1), and 5 pmol of TaqMan minor groove-binding (MGB) probes (SaV124TP and SaV5TP) (Table 1). The single-round PCR was performed under the following conditions: initial denaturation at 95°C for 15 min to activate DNA polymerase, followed by 50 cycles of amplification with denaturation at 94°C for 15 s, and annealing and extension at 62°C for 1 min. Amplification data were collected and analyzed with SDS software version 1.3 (Applied Biosystems). The threshold value was adjusted to 0.01, as recommended by Qiagen, and the threshold cycle (CT) was determined. Two PCR tubes were used for each sample, and the sample was considered positive only when the fluorescence signal was obtained from both tubes and the average CT value was not more than 40.
TABLE 1.
Primers and probes used in this study
| PCR type | Primer or probe | Name | Sequence (5′-3′)a | Polarityb | Location (nt) | Reference |
|---|---|---|---|---|---|---|
| Real time | Primer | SaV124F | GAYCASGCTCTCGCYACCTAC | + | 5078-5098c | 22 |
| SaV1F | TTGGCCCTCGCCACCTAC | + | 700-717d | 22 | ||
| SaV5F | TTTGAACAAGCTGTGGCATGCTAC | + | 5112-5135e | 22 | ||
| SaV1245R | CCCTCCATYTCAAACACTA | − | 5181-5163c | 22 | ||
| TaqMan MGB probe | SaV124TP | FAM-CCRCCTATRAACCA-MGB-NFQ | − | 5118-5105c | 22 | |
| SaV5TP | FAM-TGCCACCAATGTACCA-MGB-NFQ | − | 5157-5142e | 22 | ||
| Single round | Primer | SV-F11 | GCYTGGTTYATAGGTGGTAC | + | 5098-5117f | 23 |
| SV-R1 | CWGGTGAMACMCCATTKTCCAT | − | 5878-5857f | 23 | ||
| Nested/seminested | Primer | SV-F13 | GAYYWGGCYCTCGCYACCTAC | + | 5074-5094f | 24 |
| SV-F14 | GAACAAGCTGTGGCATGCTAC | + | 5074-5094f | 24 | ||
| SV-F22 | SMWAWTAGTGTTTGARATG | + | 5154-5172f | 24 | ||
| 1245Rfwd | TAGTGTTTGARATGGAGGG | + | 5163-5181c | This study | ||
| SV-R13 | GGTGANAYNCCATTKTCCAT | − | 5876-5861f | 24 | ||
| SV-R14 | GGTGAGMMYCCATTCTCCAT | − | 5876-5861f | 24 | ||
| SV-R2 | GWGGGRTCAACMCCWGGTGG | − | 5591-5572f | 24 |
Mixed bases in degenerate primers are as follows: K, G or T; M, A or C; N, A, C, G, or T; R, A or G; S, G or C; W, A or T; Y, C or T.
+, sense; −, antisense.
Corresponding nucleotide position of Mc10 virus (accession no. AY237420).
Corresponding nucleotide position of Parkville virus (accession no. U73124).
Corresponding nucleotide position of NK24 virus (accession no. AY646856).
Corresponding nucleotide position of Manchester virus (accession no. X86560).
PCR amplification of human SaV genomes. (i) Conventional single-round PCR assay.
The single-round PCR was performed in 50 μl of reaction volume containing 5 μl of cDNA, 2.5 U of Ex Taq DNA polymerase (TaKaRa Bio Co., Shiga, Japan), and 20 pmol of each primer. For single-round PCR, we used SV-F11 and SV-R1 primers (Table 2; RT-PCR number 1), which amplify approximately 780 bp (23).
TABLE 2.
RT-PCR assays to amplify partial capsid sequences of human sapoviruses
| RT-PCR no. | RT-PCR type | First PCR |
Second PCR |
Reference | ||||
|---|---|---|---|---|---|---|---|---|
| Forward primer | Reverse primer | Product size (bp)a | Forward primer | Reverse primer | Product size (bp)a | |||
| 1 | Single-round | SV-F11 | SV-R1 | 780 | 23 | |||
| 2 | Nested | SV-F13 SV-F14 | SV-R13 SV-R14 | 800 | SV-F22 | SV-R2 | 440 | 24 |
| 3 | Nested | SaV124F SaV1F SaV5F | SV-R13 SV-R14 | 800 | 1245Rfwd | SV-R2 | 430 | This study |
| 4 | Seminested | SaV124F SaV1F SaV5F | SV-R13 SV-R14 | 800 | 1245Rfwd | SV-R13 SV-R14 | 710 | This study |
| 5 | Seminested | SaV124F SaV1F SaV5F | SV-R13 SV-R14 | 800 | SaV124F SaV1F SaV5F | SV-R2 | 520 | This study |
| 6 | Seminested | SaV124F SaV1F SaV5F | SV-R2 | 520 | 1245Rfwd | SV-R2 | 430 | This study |
Approximate product size.
(ii) Conventional nested PCR assay.
Nested PCR was performed as described previously (24), with a minor modification (Table 2; RT-PCR number 2). Briefly, the first PCR was performed in 50 μl of reaction volume containing 5 μl of cDNA, 2.5 U of Ex Taq DNA polymerase, and 20 pmol of SV-F13, SV-F14, SV-R13, and SV-R14 primers (Table 1). These primers generated approximately an 800-bp product. PCR amplification was performed under the following conditions: initial denaturation at 94°C for 2 min, followed by 35 cycles of amplification with denaturation at 94°C for 30 s, primer annealing at 48°C for 30 s, extension reaction at 72°C for 2 min, and then a final extension at 72°C for 10 min. The second PCR was performed in 50 μl of reaction volume containing 2 μl of the first PCR product, 2.5 U of Ex Taq DNA polymerase, and 20 pmol of SV-F22 and SV-R2 primers (Table 1). PCR amplification was performed under the following conditions: initial denaturation at 94°C for 2 min, followed by 35 cycles of amplification with denaturation at 94°C for 30 s, primer annealing at 48°C for 30 s, extension reaction at 72°C for 2 min, and then a final extension at 72°C for 10 min. These primers generated approximately a 420-bp product.
(iii) Newly designed nested and seminested PCR assay.
Nested and seminested RT-PCR assays in combination with 1245Rfwd (newly designed forward primer to react with diverse SaV strains) or SaV-124F/1F/5F for the forward primer(s) and SV-R13/R14 or SV-R2 for the reverse primer(s) (Table 1) were performed. The primer combination is described in Table 2 (RT-PCR numbers 3, 4, 5, and 6).
The first PCR was performed in 50 μl of reaction volume containing 5 μl of cDNA, 2.5 U of Ex Taq DNA polymerase, and 20 pmol of each primer. PCR amplification was performed under the following conditions: initial denaturation at 94°C for 2 min, followed by 40 cycles of amplification with denaturation at 94°C for 30 s, primer annealing at 50°C for 30 s, extension reaction at 72°C for 2 min, and then a final extension at 72°C for 10 min. The second PCR was performed in 50 μl of reaction volume containing 2 μl of the first PCR product, 2.5 U of Ex Taq DNA polymerase, and 20 pmol of each primer. PCR amplification was performed under the following conditions: initial denaturation at 94°C for 2 min, followed by 45 cycles of amplification with denaturation at 94°C for 30 s, primer annealing at 50°C for 30 s, extension reaction at 72°C for 1 min, and then a final extension at 72°C for 10 min.
Cloning and sequencing.
The PCR products were separated by electrophoresis on a 2% agarose gel and visualized under a UV lamp after ethidium bromide staining. PCR products of expected size were excised from the gel and purified using the QIAquick gel extraction kit (Qiagen). The purified products were cloned into a pCR 2.1 vector (Invitrogen, Carlsbad, CA), and the constructs were then transformed into Escherichia coli One-Shot IVN αF′ competent cells (Invitrogen). The transformants were incubated at 37°C on a Luria broth agar plate containing 50 μg/ml of ampicillin and 50 μg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal). Four to eight colonies were picked up, and insertion sizes were checked by direct colony PCR amplifications with an M13 forward (5′-GTAAAACGACGGCCAG-3′) and reverse (5′-CAGGAAACAGCTATGAC-3′) primer set. The PCR products were purified with a PCR purification kit (Qiagen), and both strands were sequenced with the BigDye cycle sequencing kit, version 3.1, and the 3130 genetic analyzer (Applied Biosystems). Nucleotide sequences were assembled using the program Sequencher version 4.2.2 (Gene Codes Corporation, Ann Arbor, MI) and aligned with Clustal W version 1.83 (http://clustalw.ddbj.nig.ac.jp/top-j.html). The distances were calculated using Kimura's two-parameter method (20), and phylogenetic dendrograms from a bootstrap analysis with 1,000 replicates were generated by the neighbor-joining method.
Equivalent original river water volumes tested for human SaV sequences.
Out of 700 μl of the concentrated sample, 140 μl was applied to RNA extraction, and 2.5 μl out of 60 μl of the resulting extracted RNA was used for detection of SaV sequences using the respective RT-PCR assays, followed by sequence analysis. Therefore, the volumes tested for SaV sequences by respective RT-PCR assays were equivalent to 1/120 of 500 ml of original river water sample, or approximately 4.2 ml.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in GenBank under accession numbers AB504233 to AB504269.
RESULTS
Detection of human SaVs by real-time RT-PCR assay.
Results for detection of human SaV genomes in river water samples during a 1-year investigation, from April 2003 to March 2004, are summarized in Table 3. By use of a real-time RT-PCR assay, 12 (20%) of the 60 samples were positive for human SaV genomes (Table 3), demonstrating that the river water samples contained a quite high percentage of human SaV genomes.
TABLE 3.
Detection of sapoviruses in the Tamagawa River
| Yr and month of isolation | Site 1 |
Site 2 |
Site 3 |
Site 4 |
Site 5 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reala | RT- PCRb | Sequence | Real | RT- PCR | Sequence | Real | RT- PCR | Sequence | Real | RT- PCR | Sequence | Real | RT- PCR | Sequence | |
| 2003 | |||||||||||||||
| Apr | —c | — | — | — | — | — | — | — | — | — | |||||
| May | — | — | — | — | — | — | 39.0 | 2 | GI/2 | — | — | ||||
| Jun | — | — | — | — | 37.8 | 1 | GI/3 | — | — | — | — | ||||
| Jul | — | — | — | — | — | — | — | — | — | — | |||||
| Aug | — | — | — | — | — | 2 | GI/1 | — | — | — | — | ||||
| Sep | — | — | — | — | — | — | — | — | — | — | |||||
| Oct | — | — | — | — | — | — | — | — | — | — | |||||
| Nov | — | — | 36.8 | 3 | GII/3 | 37.5 | 3 | GII/2 | 37.1 | 3 | GV/1 | — | — | ||
| Dec | — | — | 37.0 | — | — | 3 | GII/2 | — | — | — | — | ||||
| 2004 | |||||||||||||||
| Jan | — | — | 39.3 | 3 | GI/3 | 39.1 | 3 | GI/3, GII/1 | 37.1 | 3, 6 | GI/3, GV/1 | 37.1 | 6 | GI/3 | |
| Feb | — | — | — | 3, 4 | GI/3 | — | — | 39.8 | — | — | — | ||||
| Mar | — | — | — | — | — | 2 | GI/5 | 39.2 | 6 | GI/untyped | — | 3 | GII/2 | ||
| % positive | 0% | 0% | 25% | 25% | 25% | 50% | 42% | 33% | 8% | 17% | |||||
Detection of human SaVs by RT-PCR assays.
To identify SaV strains in the river water samples, we also performed conventional RT-PCR assays. Human SaV sequences were obtained from only one (2%) and three (5%) of the 60 samples using single-round and nested RT-PCR assays (numbers 1 and 2 in Table 2) as shown in Table 3. Only one (2%) sample was positive for SaV when genogroup-specific PCR using SV-F13 and SV-F14 forward primers and SV-G1-R, SV-G2-R, SV-G4-R, and SV-G5-R reverse primers was performed (reference 24 and data not shown). When the single-round RT-PCR targeting the polymerase gene was examined (28), there was no amplification of the SaV sequence, although there were multiple nonspecific bands (data not shown). These results suggest that the SaV detection rate of any published RT-PCR assay followed by sequencing is lower than that of the real-time RT-PCR assay. To overcome this problem, we designed a forward primer (1245Rfwd) based on highly conserved sequences within the polymerase-capsid junction region. We also utilized forward primers used for real-time RT-PCR (SaV-124F/1F/5F) and reverse primers used for nested RT-PCR (SV-R2 and SV-R13/R14) and performed nested and seminested RT-PCR assays (numbers 3, 4, 5, and 6 in Table 2). The detection rates of human SaV genomes with these four RT-PCR assays were 15% (9 of 60), 2% (1 of 60), 0% (0 of 60), and 5% (3 of 60), respectively. Although the detection rate varied depending on the primer sets, the RT-PCR, especially number 3, showed a much higher detection rate than previously published assays. These results demonstrated that the newly developed primer is valuable for the efficient detection of human SaV genomes.
Seasonal and spatial distributions of human SaVs in the Tamagawa River.
Human SaV sequences were frequently detected from November to March, whereas none were obtained in April, July, September, or October (Table 3).
No SaV sequence was detected in the upstream area (site 1), whereas a high positive ratio was observed in the midstream area (sites 2, 3, and 4) (Table 3). A minimum CT value of 36.8 was obtained from the sample collected at site 2 in November 2003, indicating that this sample contained the highest concentration of human SaV genomes.
Genetic diversity of human SaVs in the Tamagawa River.
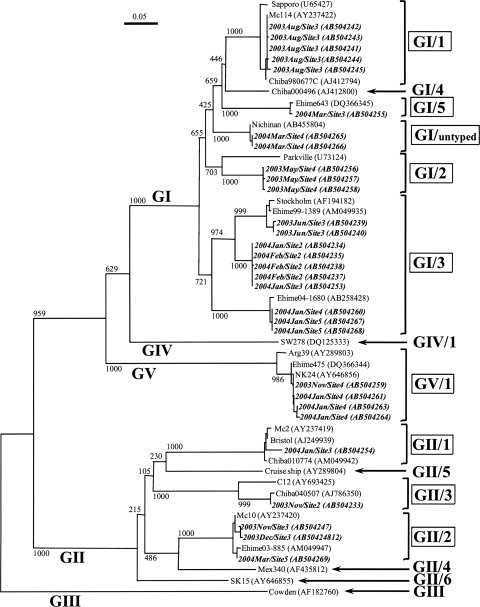
To investigate the genetic diversity of human SaV strains in the Tamagawa River, the nucleotide sequences of the cloned PCR products (approximately 400 bp) were aligned and a phylogenetic dendrogram was constructed based on the partial capsid gene sequences (Fig. 2). SaV sequences were obtained from 15 (25%) samples: four samples by conventional single-round or nested RT-PCR (RT-PCR numbers 1 and 2 in Table 3) and 11 samples by nested or seminested RT-PCR developed in this study (RT-PCR numbers 3, 4, and 6 in Table 3). A total of 30 SaV strains were identified, and these were classified into three human SaV genogroups; GI, GII, and GV sequences were detected in 10 (17%), 5 (8%), and 2 (3%) samples, respectively. Five different GI genotypes were identified in the water samples: GI/1, GI/2, GI/3, GI/5, and GI/untyped. GI/3 strains were most frequently detected (six samples), whereas the other four GI genotypes were detected in only one sample each. GII strains were subdivided into three different genotypes: GII/1, GII/2, and GII/3. GII/2 strains were detected in three samples, whereas GII/1 and GII/3 genotypes were detected in only one sample each. GV/1 strains were detected in two samples. These results demonstrated the presence of highly diverse human SaV strains in river water.
FIG. 2.
SaV strain phylogenetic tree based on partial capsid gene sequences (approximately 400 nucleotides). The numbers on each branch indicate the bootstrap values for the genotype, and bootstrap values of 950 or higher were considered statistically significant for the grouping. The scale represents nucleotide substitutions per site. Strains shown in italic bold are SaVs detected in this study, representing the year and month of sampling, sampling site, and GenBank accession number. GI/1, GI/2, GI/3, GI/5, GI/untyped, GII/1, GII/2, GII/3, and GV/1 (boxed) are genotypes identified in this study.
DISCUSSION
Although SaVs have been detected among gastroenteritis patients, the frequency of their occurrence in the environment remains unknown. In the present study, we investigated their prevalence, seasonality, and genetic diversity in river water in Japan.
We investigated the presence of human SaV genomes in the samples by using the TaqMan-based real-time RT-PCR assay targeting the polymerase-capsid junction region (22), which is widely used for the rapid and sensitive detection of human SaVs in clinical specimens (7, 10, 12, 17, 19, 25, 26, 29, 30). When the real-time RT-PCR assay was used, 12 (20%) of the 60 samples were positive for human SaV genomes (Table 3). Our previous studies using TaqMan-based real-time PCR assays (13, 14, 21) also demonstrated that the Tamagawa River water was contaminated with several other human enteric viruses, namely, NoVs, enteroviruses, torque teno viruses, and adenoviruses. The positive ratio of SaVs in river water was lower than that of NoVs and adenoviruses but higher than that of enteroviruses and torque teno viruses, suggesting a relatively high prevalence of SaVs in the samples. However, the detection rate of SaV sequences obtained using several published RT-PCR assays (23, 24, 28) was lower than that obtained using the real-time RT-PCR assay. Therefore, we designed a broadly reactive forward primer (Table 1; 1245Rfwd), performed nested and seminested RT-PCR assays using the newly designed primer (Table 2; RT-PCR numbers 3, 4, 5, and 6), and successfully amplified SaV genomes with an increased detection rate. SaV sequences were identified by RT-PCR with the novel primer in 10 (83%) of 12 samples positive for the real-time RT-PCR assay (Table 3). We also identified SaV sequences in five samples negative for the real-time RT-PCR assay (Table 3). The newly developed nested RT-PCR assay (Table 2; RT-PCR number 3) was capable of amplifying SaV genomes in nine samples, including three samples negative for the real-time RT-PCR assay. The nested RT-PCR assay, number 3 shown in Table 3, had the highest amplification efficiency among the RT-PCR assays tested. Although some previous studies have attempted to identify SaV sequences in environmental samples, such as wastewater, river water, and clams, by conventional RT-PCR (number 1 or 2 in Table 2) (9, 11, 18), the sensitivity is low and the detection from environmental samples might be underestimated in these studies.
Human SaV sequences were frequently detected in winter, from November to March, whereas none were obtained in April, July, September, or October (Table 3). The number of gastroenteritis patients with SaV infection increases in winter in Japan (12, 23), and a recent clinical study of SaVs indicated that there are asymptomatic people with high viral loads (30) and that SaV excretion continues even after recovery from gastroenteritis symptoms (19). Moreover, our previous study demonstrated that a higher SaV concentration was observed in wastewater during the winter (16), suggesting that larger amounts of SaVs were discharged into rivers from WWTP in winter. Monitoring of river water could be one of the suitable approaches to understand the actual prevalence of viruses in the river catchment area, because most urban rivers receive effluents from multiple WWTPs that contain viruses shed from all patients in the catchment area. In this study, we collected the water samples at five sampling sites along the Tamagawa River to identify the differences in occurrence of human SaVs among sampling sites. No SaV sequence was detected upstream (site 1), whereas sampling sites 2, 3, and 4 showed high positive rates. This is probably because midstream and downstream areas of the Tamagawa River are highly contaminated with effluents from several upstream WWTPs. In this study, we identified SaV genomes in river water but were unable to confirm whether they were derived from viable SaV particles. However, the results obtained may provide information to understand the occurrence of SaVs in river water, and it should be noted that there is a potential risk of infection when river water is used for drinking and recreational purposes.
The genetic analysis of SaVs in environmental samples, such as wastewater, river water, clams, and oysters, has been reported previously (3, 9, 11, 18). The present study provides additional evidence for the genetic diversity of human SaVs in river water. SaV strains detected in river water were genetically diverse, and a total of nine different human genotypes were identified in the water samples: GI/1, GI/2, GI/3, GI/5, GI/untyped, GII/1, GII/2, GII/3, and GV/1 (Fig. 2). Two strains classified as GI/untyped were genetically close (99.7% identity) to Nichinan strains identified from gastroenteritis patients in Japan in 2005 (GenBank accession numbers AB455803 and AB455804) (19). Moreover, the newly developed nested RT-PCR assay (Table 2; RT-PCR number 3) successfully amplified the GI, GII, and GV genes, whereas the other RT-PCR assays could detect only GI strains. This result demonstrated that the newly designed primer was capable of reacting with a broad spectrum of SaV strains and consequently enhanced detection efficiency when it was applied to river water samples.
In conclusion, this study provides novel evidence of the prevalence, seasonality, and genetic diversity of human SaV strains in river water. Furthermore, we developed a nested RT-PCR assay utilizing a newly designed primer that can efficiently amplify diverse human SaV genomes in river water samples. A combination of a real-time RT-PCR assay and a newly developed nested RT-PCR assay is useful to identify and characterize human SaV strains in water environments. Further studies of the occurrence of SaVs in water samples are needed for a better understanding of SaV epidemics, temporal and spatial distribution, environmental stability, and potential health risks.
Acknowledgments
This study was supported, in part, by a grant from the Ministry of Health, Labour, and Welfare of Japan.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Chiba, S., Y. Sakuma, R. Kogasaka, M. Akihara, K. Horino, T. Nakao, and S. Fukui. 1979. An outbreak of gastroenteritis associated with calicivirus in an infant home. J. Med. Virol. 4:249-254. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, I. N., and P. R. Lambden. 2000. Organization and expression of calicivirus genes. J. Infect. Dis. 181:S309-S316. [DOI] [PubMed] [Google Scholar]
- 3.Costantini, V., T. Loisy, L. Joens, F. S. Le Guyader, and L. J. Saif. 2006. Human and animal enteric caliciviruses in oysters from different coastal regions of the United States. Appl. Environ. Microbiol. 72:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farkas, T., W. M. Zhong, Y. Jing, P. W. Huang, S. M. Espinosa, N. Martinez, A. L. Morrow, G. M. Ruiz-Palacios, L. K. Pickering, and X. Jiang. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149:1309-1323. [DOI] [PubMed] [Google Scholar]
- 5.Green, K. Y. 2007. Caliciviridae: the noroviruses, p. 949-979. In D. Knipe and P. Howley (ed.), Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 6.Hansman, G. S., L. T. Doan, T. A. Kguyen, S. Okitsu, K. Katayama, S. Ogawa, K. Natori, N. Takeda, Y. Kato, O. Nishio, M. Noda, and H. Ushijima. 2004. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch. Virol. 149:1673-1688. [DOI] [PubMed] [Google Scholar]
- 7.Hansman, G. S., S. Ishida, S. Yoshizumi, M. Miyoshi, T. Ikeda, T. Oka, and N. Takeda. 2007. Recombinant sapovirus gastroenteritis, Japan. Emerg. Infect. Dis. 13:786-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansman, G. S., T. Oka, K. Katayama, and N. Takeda. 2007. Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 17:133-141. [DOI] [PubMed] [Google Scholar]
- 9.Hansman, G. S., T. Oka, R. Okamoto, T. Nishida, S. Toda, M. Noda, D. Sano, Y. Ueki, T. Imai, T. Omura, O. Nishio, H. Kimura, and N. Takeda. 2007. Human sapovirus in clams, Japan. Emerg. Infect. Dis. 13:620-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansman, G. S., H. Saito, C. Shibata, S. Ishizuka, M. Oseto, T. Oka, and N. Takeda. 2007. Outbreak of gastroenteritis due to sapovirus. J. Clin. Microbiol. 45:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansman, G. S., D. Sano, Y. Ueki, T. Imai, T. Oka, K. Katayama, N. Takeda, and T. Omura. 2007. Sapovirus in water, Japan. Emerg. Infect. Dis. 13:133-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada, S., M. Okada, S. Yahiro, K. Nishimura, S. Matsuo, J. Miyasaka, R. Nakashima, Y. Shimada, T. Ueno, S. Ikezawa, K. Shinozaki, K. Katayama, T. Wakita, N. Takeda, and T. Oka. 2009. Surveillance of pathogens in outpatients with gastroenteritis and characterization of sapovirus strains between 2002 and 2007 in Kumamoto Prefecture, Japan. J. Med. Virol. 81:1117-1127. [DOI] [PubMed] [Google Scholar]
- 13.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2005. Application of cation-coated filter method to detection of noroviruses, enteroviruses, adenoviruses, and Torque Teno viruses in the Tamagawa River in Japan. Appl. Environ. Microbiol. 71:2403-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haramoto, E., H. Katayama, K. Oguma, and S. Ohgaki. 2007. Quantitative analysis of human enteric adenoviruses in aquatic environments. J. Appl. Microbiol. 103:2153-2159. [DOI] [PubMed] [Google Scholar]
- 15.Haramoto, E., H. Katayama, and S. Ohgaki. 2004. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 70:2154-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haramoto, E., H. Katayama, C. Phanuwan, and S. Ohgaki. 2008. Quantitative detection of sapoviruses in wastewater and river water in Japan. Lett. Appl. Microbiol. 46:408-413. [DOI] [PubMed] [Google Scholar]
- 17.Ishida, S., S. Yoshizumi, M. Miyoshi, T. Ikeda, T. Okui, K. Katayama, N. Takeda, and T. Oka. 2008. Characterization of sapoviruses detected in Hokkaido, Japan. Jpn. J. Infect. Dis. 61:504-506. [PubMed] [Google Scholar]
- 18.Iwai, M., S. Hasegawa, M. Obara, K. Nakamura, E. Horimoto, T. Takizawa, T. Kurata, S. Sogen, and K. Shiraki. 2009. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008). Appl. Environ. Microbiol. 75:1264-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwakiri, A., H. Ganmyo, S. Yamamoto, K. Otao, M. Mikasa, S. Kizoe, K. Katayama, T. Wakita, N. Takeda, and T. Oka. 2009. Quantitative analysis of fecal sapovirus shedding: identification of nucleotide substitutions in the capsid protein during prolonged excretion. Arch. Virol. 154:689-693. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Kitajima, M., E. Haramoto, C. Phanuwan, H. Katayama, and S. Ohgaki. 2009. Detection of genogroup IV norovirus in wastewater and river water in Japan. Lett. Appl. Microbiol. 49:655-658. [DOI] [PubMed] [Google Scholar]
- 22.Oka, T., K. Katayama, G. S. Hansman, T. Kageyama, S. Ogawa, F.-T. Wu, P. A. White, and N. Takeda. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 78:1347-1353. [DOI] [PubMed] [Google Scholar]
- 23.Okada, M., K. Shinozaki, T. Ogawa, and I. Kaiho. 2002. Molecular epidemiology and phylogenetic analysis of Sapporo-like viruses. Arch. Virol. 147:1445-1451. [DOI] [PubMed] [Google Scholar]
- 24.Okada, M. Y. Yamashita, M. Oseto, and K. Shinozaki. 2006. The detection of human sapoviruses with universal and genogroup-specific primers. Arch. Virol. 151:2503-2509. [DOI] [PubMed] [Google Scholar]
- 25.Ootsuka, Y., Y. Yamashita, T. Ichikawa, R. Kondo, M. Oseto, K. Katayama, N. Takeda, and T. Oka. 2009. Molecular characterization of sapoviruses detected in sporadic gastroenteritis cases in 2007 in Ehime prefecture, Japan. Jpn. J. Infect. Dis. 62:246-248. [PubMed] [Google Scholar]
- 26.Pang, X. L., B. E. Lee, G. J. Tyrrell, and J. K. Preiksaitis. 2009. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004-2007. J. Infect. Dis. 199:547-551. [DOI] [PubMed] [Google Scholar]
- 27.U.S. Environmental Protection Agency. 2009. Drinking water contaminant candidate list 3—final. Fed. Regist. 74:51850-51862. [Google Scholar]
- 28.Vinje, J., H. Deijl, R. van der Heide, D. Lewis, K.-O. Hedlund, L. Svensson, and M. P. G. Koopmans. 2000. Molecular detection of Sapporo-like viruses. J. Clin. Microbiol. 38:530-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, F. T., T. Oka, N. Takeda, K. Katayama, G. S. Hansman, C. H. Muo, S. Y. Liang, C. H. Hung, D. Dah-Shyong Jiang, J. Hsin Chang, J. Y. Yang, H. S. Wu, and C. F. Yang. 2009. Acute gastroenteritis caused by GI/2 sapovirus, Taiwan, 2007. Emerg. Infect. Dis. 14:1169-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida, T., S. Kasuo, Y. Azegami, Y. Uchiyama, K. Satsumabayashi, T. Shiraishi, K. Katayama, T. Wakita, N. Takeda, and T. Oka. 2009. Characterization of sapoviruses detected in gastroenteritis outbreaks and identification of asymptomatic adults with high viral load. J. Clin. Virol. 45:67-71. [DOI] [PubMed] [Google Scholar]