Abstract
N-Acylhomoserine lactones (AHLs) are used as quorum-sensing signal molecules by many Gram-negative bacteria. We have reported that Microbacterium testaceum StLB037, which was isolated from the leaf surface of potato, has AHL-degrading activity. In this study, we cloned the aiiM gene from the genomic library of StLB037, which has AHL-degrading activity and shows high homology with the α/β hydrolase fold family from Actinobacteria. Purified AiiM as a maltose binding fusion protein showed high degrading activity of AHLs with both short- and long-chain AHLs with or without substitution at carbon 3. High-performance liquid chromatography analysis revealed that AiiM works as an AHL lactonase that catalyzes AHL ring opening by hydrolyzing lactones. In addition, expression of AiiM in the plant pathogen Pectobacterium carotovorum subsp. carotovorum reduced pectinase activity markedly and attenuated soft rot symptoms on potato slices. In conclusion, this study indicated that AiiM might be effective in quenching quorum sensing of P. carotovorum subsp. carotovorum.
Quorum sensing is a cell-cell communication mechanism that depends on cell population density in bacteria (3, 7). In many Gram-negative bacteria, several kinds of N-acyl-l-homoserine lactones (AHLs) have been identified as signal compounds involved in this mechanism, and these are termed autoinducers (3, 7). AHL-mediated quorum sensing regulates the expression of many genes, including those responsible for bioluminescence, the production of pigments and antibiotics, and other processes (7). Many Gram-negative plant pathogens produce AHLs and regulate their virulence by AHL-mediated quorum sensing (31). For instance, Pectobacterium carotovorum subsp. carotovorum (formerly Erwinia carotovora), which causes soft rot diseases in many plant species, induces the production of various exoenzymes and plant tissue maceration by AHLs (1). Pantoea stewartii and Pantoea ananatis produce AHLs and regulate exopolysaccharide biosynthesis and the infection of plants (15, 32). In general, AHL-negative mutants show defects in pathogenicity, so it is expected that disrupting or manipulating quorum-sensing signals could inhibit the expression of virulence and infection of host cells.
Recently, many AHL-degrading genes have been cloned and characterized from various bacteria. Genes encoding AHL lactonase, which catalyzes AHL ring opening by hydrolyzing lactones, have been cloned from Bacillus sp., Arthrobacter sp., Agrobacterium tumefaciens, and Rhodococcus erythropolis (5, 23, 30, 34). Genes encoding AHL acylase, which hydrolyze the amide bond of AHL, have been cloned from Ralstonia sp., Anabaena sp., Streptomyces sp., Shewanella sp., and Pseudomonas aeruginosa (11, 12, 16, 22, 25). Human and murine paraoxonase degrades AHL by hydrolyzing its lactone ring (21). Novel AHL lactonase genes have been isolated from a metagenomic library which was constructed from environmental soil samples (24, 27). AHL-degrading genes have also been utilized in the biocontrol of plant diseases. Expression of aiiA in transformed P. carotovorum subsp. carotovorum significantly attenuates pathogenicity on some crops (5). Transgenic plants expressing AHL lactonase exhibited significantly enhanced resistance to the infection of P. carotovorum subsp. carotovorum (4).
We have reported the isolation of AHL-degrading Microbacterium testaceum StLB037 from the leaf surface of potato (Solanum tuberosum) (17). In coinfections, we found that StLB037 interrupted quorum-sensing-dependent bacterial infection by the plant pathogen P. carotovorum subsp. carotovorum. In this study, we report the cloning and characterization of a novel AHL lactonase gene (aiiM) from the chromosome of StLB037. In addition, we evaluated the potential use of heterologous aiiM gene expression in quenching quorum sensing in the plant pathogen P. carotovorum subsp. carotovorum.
MATERIALS AND METHODS
Bacterial strains, plasmids, compounds, and growth conditions.
Selected bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani (LB) medium (26). All other bacteria were grown at 30°C in tryptic soy broth (TSB; Nippon Becton Dickinson, Tokyo, Japan). Solid bacterial medium was made by the addition of agar at a final concentration of 1.5%. Antibiotics were added as required at final concentrations of 100 μg/ml ampicillin, 10 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 10 μg/ml gentamicin. The AHLs used in this study, N-hexanoyl-l-homoserine lactone (C6-HSL), N-octanoyl-l-homoserine lactone (C8-HSL), N-decanoyl-l-homoserine lactone (C10-HSL), N-dodecanoyl-l-homoserine lactone (C12-HSL), N-(3-oxohexanoyl)-l-homoserine lactone (3OC6-HSL), N-(3-oxooctanoyl)-l-homoserine lactone (3OC8-HSL), N-(3-oxodecanoyl)-l-homoserine lactone (3OC10-HSL), and N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL), were synthesized by a previously described method (2).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Nippon Gene |
| C. violaceum | ||
| CV026 | ATCC 31532 derivative; cviI::Tn5xylE; Kmr Smr | 13 |
| VIR07 | ATCC 12472 derivative; cviI::Kmr; Apr | 14 |
| M. testaceum | ||
| StLB037 | AHL-degrading strain isolated from potato leaf surface | 17 |
| R. erythropolis | ||
| NBRC 100887 | Crude-oil-degrading marine bacterium | 8 |
| R. opacus | ||
| B-4 | Benzene-tolerant strain | 18 |
| P. carotovorum subsp. carotovorum | ||
| NBRC 3830 | Wild type | NBRCa |
| Plasmids | ||
| pUC118 | Cloning vector; Apr | Takara Bio |
| pST37-1 | 4.4-kb Sau3AI fragment from StLB037 genomic DNA in pUC118 | This study |
| pUC118-aiiM | pUC118 containing aiiM gene | This study |
| pSTV28 | Cloning vector; Cmr | Takara Bio |
| pHV200 | 8.8-kb SalI fragment with V. fischeri ES184 lux regulon | 6 |
| pLux28 | 8.8-kb SalI fragment from pHV200 in pSTV28 | This study |
| pGEM-T easy | Cloning vector; Apr | Promega |
| pMAL-c2X | Cloning vector to make MBP fusions; Apr | New England Biolabs |
| pMAL-aiiM37 | pMAL-c2X containing aiiM gene from StLB037 | This study |
| pMAL-aiiMRe | pMAL-c2X containing aiiM gene homolog from NBRC 100887 | This study |
| pMAL-aiiMRo | pMAL-c2X containing aiiM gene homolog from B-4 | This study |
| pJN105Z | Broad-host-range vector pJN105 containing lacZα from pUC118 cloning vector | 16 |
| pJN105Z-aiiM | pJN105Z containing aiiM gene | This study |
National Institute of Technology and Evaluation Biological Resource Center, Japan.
Cloning of an AHL-degrading gene from StLB037.
A standard protocol for genetic manipulation was used as described previously (26). An AHL-responsive plasmid, designated pLux28, was constructed by the following method. An 8.8-kb SalI fragment of pHV200 was cloned into the SalI site of cloning vector pSTV28. For the construction of a genomic library, chromosomal DNA of StLB037 was partially digested with Sau3AI, and the fragments were inserted into the BamHI site of cloning vector pUC118 dephosphorylated by bacterial alkaline phosphatase (Takara Bio, Shiga, Japan). Both the genomic library of StLB037 and the pLux28 plasmid were transformed into E. coli DH5α. The transformants were grown on LB agar plates containing ampicillin and chloramphenicol. The formed colonies were picked and inoculated into 200 μl of fresh LB medium containing ampicillin, chloramphenicol, and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in a 96-well plate. After incubation at 30°C for 20 h with gentle shaking, the cell cultures in each well were evaluated for luminescence activities, using a Luminescenser JNR-II (Atto, Tokyo, Japan). A positive clone, which contains AHL-degrading plasmid, shows a very low level of luminescence activity. The positive clone was sequenced by a BigDye Terminator, version 3.1, and ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Tokyo, Japan).
Cloning of the aiiM gene.
The aiiM coding region of the StLB037 genome was amplified with GoTaq DNA polymerase (Promega, Tokyo, Japan) and the primers 5′-AGATCTAGGGTGGTCGACATCGAGGCATCC-3′ and 5′-AAGCTTTCGACGACGACATCCAGCTCCACG-3′ containing BglII and HindIII restriction sites, respectively (underlined). PCR was performed with the following cycling parameters: 94°C for 30 s, 60°C for 30 s, and 74°C for 1 min for 27 cycles. The PCR products were cloned into a pGEM-T easy cloning vector (Promega). The aiiM coding region was cut out by BglII and HindIII digestion and inserted into the BamHI-HindIII site of the broad-host-range vector pJN105Z for construction of pJN105Z-aiiM and into the BamHI-HindIII site of pUC118 for construction of pUC118-aiiM.
Purification of AiiM as a maltose binding protein (MBP) fusion.
The aiiM coding regions on the genome of M. testaceum StLB037, R. erythropolis NBRC 100887, and Rhodococcus opacus B-4, were amplified with GoTaq DNA polymerase and the following primer sets: 5′-AGATCTATGATCCTCGCCCACGACGTGTCG-3′ and 5′-AAGCTTTCGACGACGACATCCAGCTCCACG-3′ for StLB037; 5′-AGATCTATGACTTTGTCGCACGACATCTCC-3′ and 5′-AAGCTTAACCATCTAGCCTGCAGCACTCAC-3′ for NBRC 100887; and 5′-AGATCTATGGACTTGCCTCACGACATCGCC-3′ and 5′-AAGCTTTCAGCGCGGGGAGTCGCTGGAACT-3′ for B-4 (restriction sites are underlined). PCR was performed with the following cycling parameters: 94°C for 30 s, 60°C for 30 s, and 74°C for 1 min for 27 cycles. The PCR products were cloned into the pGEM-T easy cloning vector. The aiiM coding region was cut out by BglII and HindIII digestion and inserted into the BamHI-HindIII site of pMAL-c2X (New England Biolabs, Tokyo, Japan) for construction of pMAL-aiiM37 (aiiM from StLB037), pMAL-aiiMRe (aiiM homolog from NBRC 100887), and pMAL-aiiMRo (aiiM homolog from B-4).
For expression and purification of the MBP-AiiM fusion, a full-grown culture of E. coli DH5α harboring an AiiM-expressing plasmid was inoculated into 300 ml of fresh LB medium containing ampicillin and incubated for 2 h at 37°C with shaking. Expression of the recombinant MBP-AiiM fusion was induced upon addition of 0.1 mM IPTG after 2 h, and expression was continued for an additional 8 h at 37°C. After incubation, cells were harvested by centrifugation and resuspended with column buffer (20 mM Tris-HCl buffer and 200 mM NaCl, pH 7.4). Lysozyme from egg white (Wako, Osaka, Japan) was added to the suspension at a final concentration of 250 μg/ml. After incubation for 4 min at 37°C, the suspension was sonicated, centrifuged at 10,000 × g for 5 min to remove the cell debris, and filtered. Protein purification was performed by ÄKTAprime systems (GE Healthcare, Tokyo, Japan). The filtered sample was loaded on an MBPTrap affinity chromatography column (GE Healthcare) equilibrated with column buffer and eluted with 10 mM maltose after unbound bacterial protein was washed out. For a negative control, we also purified an MBP-LacZα fusion from E. coli DH5α harboring pMAL-c2X by the same method used for MBP-AiiM. Expression and purification of recombinant MBP-AiiM and MBP-LacZα were checked by SDS-PAGE analysis.
Detection of the AHL-degrading activity of AiiM.
For in vivo assays, E. coli DH5α harboring the constructed plasmids was inoculated into 4 ml of LB medium containing ampicillin and 0.1 mM IPTG and incubated for 18 h at 37°C. The culture was diluted 1:100 in 4 ml of fresh LB medium containing ampicillin and 10 μM C10-HSL. After incubation at 37°C for 2 h, cells were removed by centrifugation. The culture supernatants were collected and used for the following AHL bioassays. For in vitro assays, the purified protein solutions were mixed with equal volumes of 200 μM C10-HSL and incubated at 37°C for 2 h. Residual C10-HSL was detected using the AHL biosensor Chromobacterium violaceum VIR07, which responds to long-chain AHLs by producing the purple pigment violacein (14). An overnight culture of VIR07 was mixed with 25 ml of LB agar medium and poured in the plates. Paper disks, 8 mm in diameter (Advantec, Tokyo, Japan), were placed on an agar plate, and the AHL samples were applied. Assay plates were incubated overnight at 30°C, and the appearance of pigment was determined.
HPLC analysis.
High-performance liquid chromatography (HPLC) analysis was performed according to a previously described method, with slight modification (28). To analyze AHL degradation products, 50 μl of the purified protein solution was mixed with 49 μl of the column buffer and 1 μl of 200 mM C10-HSL stock solution (in methanol). After incubation at 37°C for 15 min, reactions were stopped with an equal volume of acetonitrile, and the mixture was vortexed and centrifuged to pellet the precipitated protein. The hydrolyzed C10-HSL, as a comparable control, was made by incubating C10-HSL in 10 mM NaOH at room temperature for 30 min. Samples (20 μl) were chromatographed on an HPLC system (Jasco, Tokyo, Japan) with a UV/visible light (VIS) detector set at 205 nm by use of a Crestpak C18T-5 reverse-phase column (Jasco). Samples were eluted isocratically with water-acetonitrile-acetic acid (32:68:0.2 [vol/vol/vol]) at 1 ml/min. For kinetic assays, 1 μl of the purified protein solution was mixed with 89 μl of the column buffer and 10 μl of 20 mM AHL solution (in methanol). After incubation for 1 min, reactions were stopped, and samples were chromatographed by HPLC. The amount of AHL was estimated by comparing the reduction in peak areas for a given retention time with an AHL solution of known concentration.
Extracellular pectolytic enzyme assay and pathogenicity test.
P. carotovorum subsp. carotovorum NBRC 3830 was used for the following experiments as a plant-pathogenic bacterium (17). The broad-host-range plasmids pJN105Z and pJN105Z-aiiM were transformed into NBRC 3830 by electroporation. The activity of extracellular pectinase was determined by a previously described method, with slight modification (1). Briefly, NBRC 3830 harboring pJN105Z or pJN105Z-aiiM was grown for 18 h in M63 minimal medium (26) with 0.2% glycerol, 0.1% peptone, and 0.4% polygalacturonic acid (PGA). Paper disks, 8 mm in diameter, were placed on the pectinase assay plates (1% PGA, 1% yeast extract, 0.38 μM CaCl2, 100 mM Tris-HCl, pH 8.5, and 0.8% agar), and 50 μl of culture supernatants was applied. After incubation for 18 h at 28°C, 1 N HCl was poured on the assay plates for the development of clear zones around the paper disks. For pathogenicity tests, NBRC 3830 harboring pJN105Z or pJN105Z-aiiM was grown in 4 ml of TSB medium containing gentamicin for 15 h at 30°C. The potato slices were placed in petri dishes with wet filter paper to keep them moist. The full-grown culture (5 μl) of NBRC 3830 was inoculated on the surfaces of the potato slice and incubated for 24 h at 30°C.
Nucleotide sequence accession number.
The nucleotide sequences of aiiM from M. testaceum StLB037 have been deposited in the DDBJ/EMBL/GenBank databases under accession number AB513359.
RESULTS AND DISCUSSION
Identification of the M. testaceum gene involved in AHL degradation.
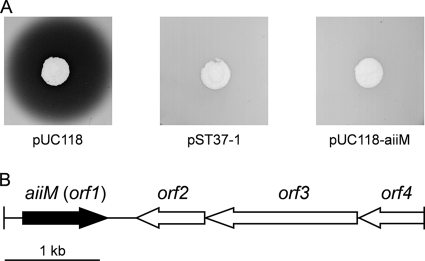
For cloning the AHL-degrading gene, we used the E. coli DH5α/pLux28 reporter system. The reporter plasmid pLux28, constructed in this study, carries an 8.8-kb region of the lux operon of Vibrio fischeri strain ESl14, which contains the AHL synthase gene (luxI), AHL receptor gene (luxR), and bioluminescence gene cluster (luxCDABE). E. coli DH5α harboring pLux28 produces AHL and expresses luminescence under AHL-mediated regulation. Furthermore, a pUC118-based genomic library of StLB037 was prepared. The prepared genomic library was transformed into E. coli DH5α harboring pLux28. The formed colonies were grown in fresh LB medium, and their luminescence activities were measured. When approximately 3,000 transformants were screened, five clones expressed luminescence to a very low degree. To elucidate whether the reductions in luminescence activities resulted from degrading AHL, E. coli DH5α cells harboring the positive clones were inoculated into LB medium containing 10 μM C10-HSL. After incubation for 2 h, the residual AHL in the culture supernatant was detected by C. violaceum strain VIR07, which produced the purple pigment violacein in response to the presence of AHL. E. coli DH5α harboring one of these clones, designated pST37-1, showed obvious AHL-degrading activity, with disappearance of the purple pigment in comparison to the control (Fig. 1A).
FIG. 1.
(A) AHL-degrading activity of E. coli DH5α harboring pUC118, pST37-1, and pUC118-aiiM. A subculture of E. coli DH5α was mixed with 10 μM C10-HSL and incubated at 37°C for 2 h. The residual AHL was detected by C. violaceum VIR07, which produced the purple pigment violacein in response to the presence of AHL. (B) Arrangement of predicted ORFs on the original genomic clone pST37-1. The scale represents a 1-kb length of nucleotides.
The sequence of the genomic DNA fragment (4,398 bp), which was cloned into pST37-1, contained one incomplete open reading frame (ORF) and three complete ORFs (Fig. 1B). The incomplete ORF (orf4) showed similarities to a putative ABC transporter permease protein. The third complete ORF (orf3) is predicted to encode a putative sensor protein of 530 amino acids. The second complete ORF (orf2) is predicted to encode a hypothetical protein of 238 amino acids. The first complete ORF (orf1) is predicted to encode a protein of 295 amino acids related to members of the α/β hydrolase fold family and is most closely related to the α/β hydrolase fold family protein of Kineococcus radiotolerans SRS30216 (accession number A6WBL1). To test if the orf1 encodes an AHL-degrading enzyme, the complete ORF of orf1 was amplified by PCR and subcloned into the pUC118 vector. E. coli DH5α harboring the Orf1-expressing plasmid as well as cells harboring pST37-1 showed AHL-degrading activity (Fig. 1A). These results demonstrated that the orf1 gene product has AHL-degrading activity. Therefore, orf1 has been named aiiM (auto-inducer inactivation gene from M. testaceum) since it is the first AHL-degrading gene identified in genus Microbacterium.
Characterization of the AHL-degrading activity of AiiM.
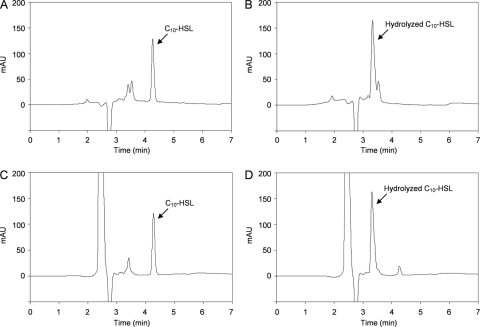
We purified AiiM as an MBP fusion for an in vitro AHL-degrading assay. The MBP-AiiM fusion protein was overproduced in E. coli DH5α harboring pMAL-aiiM37. MBP-AiiM was purified by maltose affinity chromatography, and purified proteins were analyzed by 10% SDS-PAGE. As expected, the results from SDS-PAGE analysis revealed the overexpression of products that were approximately 74 kDa in size (Fig. 2A). When the AHL-degrading activity of the purified proteins was examined, purified MBP-AiiM completely degraded 100 μM C10-HSL within 2 h while MBP-LacZα did not (Fig. 2B). In our previous study, the putative AHL lactonase activities were detected from StLB037 (17). Therefore, to determine whether AiiM works as an AHL lactonase, C10-HSL degraded by AiiM was analyzed by HPLC. Fractionation of the C10-HSL standard revealed one major HPLC peak, with a retention time of about 4.2 min (Fig. 3A). To prepare the lactone ring-opened C10-HSL, C10-HSL was hydrolyzed by 10 mM NaOH. Fractionation of the hydrolyzed C10-HSL revealed one major HPLC peak with a retention time of about 3.3 min (Fig. 3B). To examine the enzymatic property of AiiM, solutions of MBP-LacZα and MBP-AiiM were mixed with C10-HSL and incubated at 37°C for 15 min. Fractionation of MBP-LacZα-treated C10-HSL revealed one major HPLC peak, which corresponded to that of the C10-HSL standard (Fig. 3C). This result indicated that the MBP domain did not have AHL-degrading activity. In contrast, fractionation of MBP-AiiM-treated C10-HSL revealed two HPLC peaks, which corresponded to those of the C10-HSL standard and the lactone ring-opened C10-HSL (Fig. 3D). MBP-LacZα and MBP-AiiM solutions, which were not mixed with C10-HSL, displayed no distinct peaks (data not shown). These results indicated that AiiM works as an AHL lactonase that catalyzes AHL ring opening by hydrolyzing lactones.
FIG. 2.
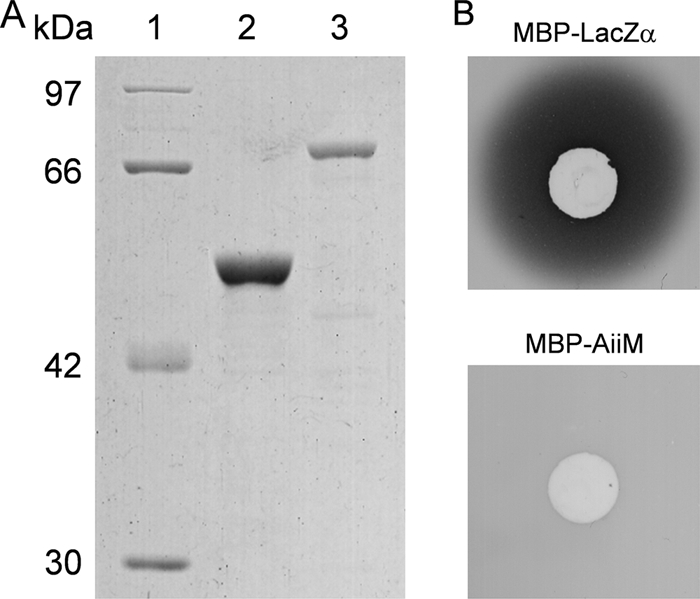
(A) Purification of MBP-LacZα and MBP-AiiM fusion proteins. Lane 1, protein molecular mass marker (Fermentas, Hanover, MD); lane 2, purified MBP-LacZα; and lane 3, purified MBP-AiiM from M. testaceum StLB037. Samples were analyzed by SDS-PAGE in a 10% polyacrylamide gel. (B) AHL-degrading activity of MBP-AiiM. The solutions of purified MBP-LacZα and MBP-AiiM were mixed with equal volumes of 1 mM C10-HSL solutions and incubated at 37°C for 15 min. The residual AHL was detected by C. violaceum VIR07.
FIG. 3.
HPLC profiles of C10-HSL (A), C10-HSL hydrolyzed by 10 mM NaOH (B), C10-HSL treated with MBP-LacZα (C), and C10-HSL treated with MBP-AiiM (D). The peaks corresponding to C10-HSL (a retention time of about 4.2 min) and hydrolyzed C10-HSL (3.3 min) are indicated by arrows. AU, arbitrary units.
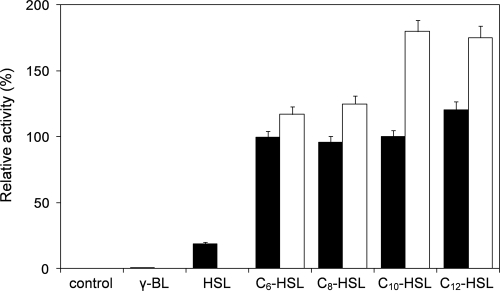
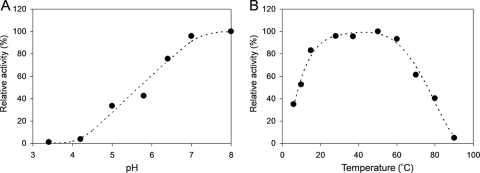
To assess the AHL substrate range of AiiM, its degrading activities against various AHLs were determined using HPLC. MBP-AiiM and AHLs were mixed and incubated for 1 min at 37°C and pH 7.4 and chromatographed on an HPLC. MBP-AiiM exhibited high relative activities toward all of the tested AHLs (Fig. 4). Although MBP-AiiM worked slightly better than C6-, C8-, and C10-HSLs against C12-HSL, differences in acyl chain length and substitution did not significantly affect enzyme activity. In a previous study, AiiA from Bacillus sp. 240B1 worked better against AHLs without a 3-oxo substitution than did the substituted derivatives (33). In contrast, MBP-AiiM worked better against 3-oxo-substituted AHLs than against unsubstituted AHLs. AiiM also showed slight degrading activity toward l-homoserine lactone (approximately 20% activity of that for C10-HSL) but could not degrade γ-butyrolactone (Fig. 4). The optimal pH for the AHL-degrading activity of MBP-AiiM was examined using C10-HSL as a substrate at 37°C. AHL-degrading activity was enhanced with increasing pH and reached a maximum at pH 8, but no or little activity was detected when the pH was adjusted to 4.2 or below (Fig. 5A). Strain StLB037 could grow at temperatures ranging from 10 to 60°C, reaching its optimum temperature at 37°C. Correspondingly, the optimal temperature for the AHL-degrading activity of MBP-AiiM was examined using C10-HSL as a substrate at pH 7.4. MBP-AiiM displayed over 80% of its maximum activity at 15 to 60°C, but the relative activity was greatly reduced at temperatures over 70°C (Fig. 5B).
FIG. 4.
Substrate specificity of purified MBP-AiiM. One microliter of purified MBP-AiiM was mixed with 89 μl of the column buffer (pH 7.4) and 10 μl of 20 mM AHL solution, γ-butyrolactone (γ-BL), or l-homoserine lactone (HSL). After incubation for 1 min, the residual substrate was quantified by HPLC. The activity toward C10-HSL was defined as 100%. The C10-HSL-degrading activity without MBP-AiiM is represented as the control. Filled and open bars represent unsubstituted and 3-oxo-substituted AHLs, respectively. The results were reproduced at least three times, and error bars indicate standard deviations.
FIG. 5.
(A) Optimal pH of AHL-degrading activity of purified MBP-AiiM. One microliter of purified MBP-AiiM was mixed with 89 μl of the buffer ranging from pH 3 to pH 8 and 10 μl of 20 mM AHL solution. (B) Optimal temperature of AHL-degrading activity of purified MBP-AiiM. One microliter of purified MBP-AiiM was mixed with 89 μl of the buffer ranging from pH 3.4 to pH 8 and 10 μl of 20 mM AHL solution. After incubation for 1 min, the residual substrate was quantified by HPLC. The maximum activity was defined as 100%.
AiiM is a new member of the AHL lactonase family.
The deduced amino acid sequence of AiiM was used to perform a BLAST search of the DDBJ/EMBL/GenBank databases. A phylogenetic tree was constructed by the neighbor-joining method with the ClustalW program (29). AiiM had less than 15% identity to each of the known AHL lactonases, which were AiiA (9.6% identity) from Bacillus sp. 240B1 (4), AttM (9.9% identity) from A. tumefaciens C58 (34), AhlD (11.7% identity) from Arthrobacter sp. IBN110 (23), QsdA (12.5% identity) from R. erythropolis W2 (30), and QlcA (14.9% identity), BpiB01 (12.9% identity), BpiB04 (13.3% identity), and BpiB07 (12.4% identity) from the soil metagenome (24, 27). A conserved HXHXDH sequence is a zinc-binding motif that is found in the metallo-β-lactamase superfamily and is common to many AHL lactonase enzymes (5). This motif was found in AiiA, AttM, AhlD, and QlcA but not in Bpi01, Bpi04, Bpi07, and AiiM. The QsdA protein belongs to the PTE family, another zinc-dependent metalloprotein family which is unrelated to the metallo-β-lactamase superfamily, and the two conserved zinc binding domains are found in the amino acid sequence of QsdA (30). However, these conserved domains of QsdA were not found in AiiM (Table 2). Therefore, these results demonstrated that AiiM is a novel AHL lactonase.
TABLE 2.
A comparison of the known AHL lactonases
| Name | Strain or source | Protein family | Zinc-binding motif | Reference or source |
|---|---|---|---|---|
| AiiM | M. testeceum StLB037 | α/β Hydrolase fold family | NFa | This study |
| AiiA | Bacillus sp. 240B1 | Metallo-β-lactamase superfamily | HXHXDH | 5 |
| AttM | A. tumefaciens C58 | Metallo-β-lactamase superfamily | HXHXDH | 34 |
| AhlD | Arthrobacter sp. IBN110 | Metallo-β-lactamase superfamily | HXHXDH | 23 |
| QsdA | R. erythropolis W2 | PTE superfamily | PTE domain | 30 |
| BpiB01 | Soil metagenome | Hypothetical protein family | NF | 27 |
| BpiB04 | Soil metagenome | Glycosyl hydrolase family | NF | 27 |
| BpiB07 | Soil metagenome | Dienelactone hydrolase family | NF | 27 |
| QlcA | Soil metagenome | Metallo-β-lactamase superfamily | HXHXDH | 24 |
NF, not found.
The α/β hydrolase fold is one of the most versatile and widespread protein architectures, and the α/β hydrolase fold family includes functionally diverse enzymes such as esterases, proteases, lipases, dehalogenases, haloperoxidases, lyases, and epoxide hydrolases. Although the enzymes belonging to this family do not share any significant overall sequence similarity, the common shared structure, the α/β hydrolase fold, provides them a stable scaffold for the catalytic residues (10, 19, 20). A BLAST search revealed that AiiM showed similarity to predicted α/β hydrolase fold family proteins from K. radiotolerans SRS30216 (26.2% identity), R. erythropolis PR4 (21.3% identity), and R. opacus B-4 (20.2% identity). Both of these bacteria and M. testaceum belong to the class Actinobacteria. According to the AiiM sequence from the Blocks database, an α/β hydrolase fold signature was identified in the two conserved domains (positions 35 to 50 and 84 to 97) (9). The two domains were highly conserved among AiiM and α/β hydrolase fold family proteins from Actinobacteria (data not shown). To our knowledge, there is no report of an AHL lactonase that belongs to the α/β hydrolase fold family. To confirm whether an aiiM-homologous gene from other actinobacteria has AHL-degrading activity, aiiM-homologous genes from R. erythropolis PR4 (also designated NBRC 100887) and R. opacus B-4, which showed high similarity to AiiM from StLB037, were amplified by PCR and expressed as MBP fusion proteins. However, MBP-AiiM from Rhodococcus species did not have any AHL-degrading activity (data not shown). These results suggested that the functional aiiM might not be present in the two strains.
AiiM quenched the virulence in P. carotovorum subsp. carotovorum.
The plant pathogen P. carotovorum subsp. carotovorum, which causes soft rot diseases on many plant species, produces 3OC6-HSL and induces production of various exoenzymes and plant tissue maceration by AHL-mediated quorum sensing (1). In a previous study, we investigated the effects of the AHL-degrading activity of StLB037 on the development of plant disease by P. carotovorum subsp. carotovorum. The maceration of the potato tuber by P. carotovorum subsp. carotovorum NBRC 3830 was inhibited by coinoculation of StLB037 (17). In this study, we evaluated the potential use of heterologous expression of the aiiM gene for interfering with quorum sensing in NBRC 3830. The AiiM-expressing plasmid, pJN105Z-aiiM, and the control plasmid, pJN105Z, were transformed into NBRC 3830 by electroporation. We tested the AHL production of NBRC 3830 harboring pJN105Z or pJN105Z-aiiM by C. violaceum CV026, which responds to short-chain AHL by producing the purple pigment violacein (13). AHL production was detected in NBRC 3830 harboring pJN105Z but not in NBRC 3830 harboring pJN105Z-aiiM (data not shown). Expression of aiiM in NBRC 3830 did not affect growth (data not shown). Pectinase, which is an enzyme that breaks down pectin, is one of the major virulence factors and is regulated by AHL-mediated quorum sensing in P. carotovorum subsp. carotovorum (1). The pectinase activity of NBRC 3830 harboring pJN105Z-aiiM was drastically decreased compared with that of NBRC 3830 harboring pJN105Z (Fig. 6A). The pathogenicity of NBRC 3830 was determined on potato slices. Although NBRC 3830 harboring pJN105Z caused severe tissue maceration, NBRC 3830 harboring pJN105Z-aiiM showed attenuated soft rot symptoms on potato slices (Fig. 6B). These results indicated that the expression of AiiM in NBRC 3830 contributed to the self-degradation of AHLs and the interruption of expression of pectinase and pathogenicity, which were regulated by AHL-mediated quorum sensing.
FIG. 6.
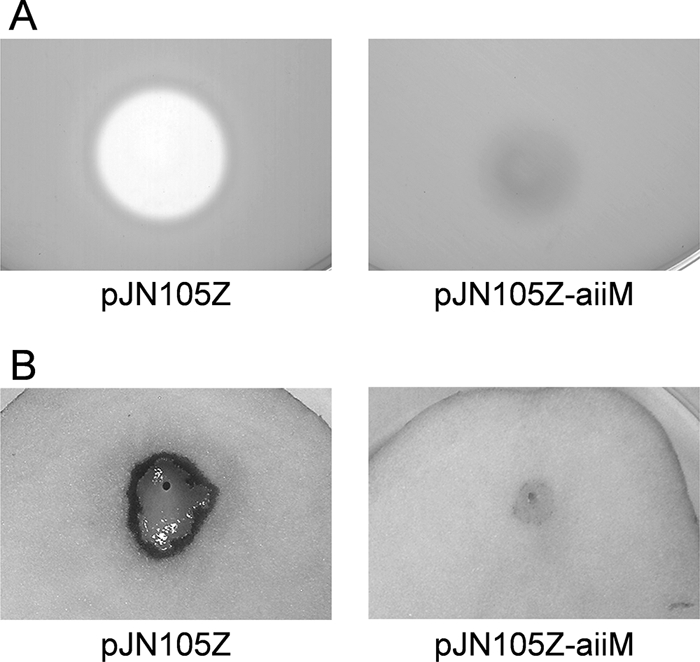
Characterization of P. carotovorum subsp. carotovorum NBRC 3830 harboring pJN105Z and pJN105Z-aiiM. (A) Extracellular pectinase activity in the culture supernatants of NBRC 3830. For agar plate assays of pectinase, 50 μl of culture supernatant was applied to the paper disks. Clear zones around the paper disks indicate pectinase activities. (B) Plant tissue maceration activities in NBRC 3830. The full-grown culture (5 μl) of NBRC 3830 was inoculated on the potato slices. The inoculated potato slices were incubated for 24 h at 30°C.
In summary, our work is the first report that AiiM, which belongs to the α/β hydrolase fold family, has AHL lactonase activity. In our previous study, AHL-degrading M. testaceum strains could be isolated from potato leaves, which were collected from various areas (17). Therefore, the aiiM gene homolog might be widespread among the leaf-associated M. testaceum. In addition, M. testaceum is an endophytic bacterium which resides within plant hosts without causing disease symptoms (35). The AHL-degrading activity of AiiM might perform useful functions, such as antipathogenic activity, which protect plants from pathogens in the leaves.
Acknowledgments
We are grateful to Junichi Kato (Hiroshima University, Japan) for providing R. opacus B-4.
This work was supported in part by a research grant (2009 to 2011) of the Institute for Fermentation, Osaka, Japan.
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Chatterjee, A., Y. Cui, Y. Liu, C. K. Dumenyo, and A. K. Chatterjee. 1995. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl. Environ. Microbiol. 61:1959-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chhabra, S. R., C. Harty, D. S. Hooi, M. Daykin, P. Williams, G. Telford, D. I. Pritchard, and B. W. Bycroft. 2003. Synthetic analogues of the bacterial signal (quorum sensing) molecule N-(3-oxododecanoyl)-l-homoserine lactone as immune modulators. J. Med. Chem. 46:97-104. [DOI] [PubMed] [Google Scholar]
- 3.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong, Y. H., L. H. Wang, J. L. Xu, H. B. Zhang, X. F. Zhang, and L. H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 5.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. U. S. A. 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg, E. P. 1997. Quorum sensing in gram-negative bacteria. ASM News 63:371-377. [Google Scholar]
- 8.Harayama, S., K. Venkateswaren, H. Toki, S. Komukai, M. Goto, H. Tanaka, and M. Ishihara. 1996. Degradation of crude oil by marine bacteria. J. Mar. Biotechnol. 3:239-243. [Google Scholar]
- 9.Henikoff, J. G., E. A. Greene, S. Pietrokovski, and S. Henikoff. 2000. Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Res. 28:228-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmquist, M. 2000. Alpha/beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 1:209-235. [DOI] [PubMed] [Google Scholar]
- 11.Huang, J. J., J. I. Han, L. H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, Y. H., J. L. Xu, J. Hu, L. H. Wang, S. L. Ong, J. R. Leadbetter, and L. H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 13.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycrof, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 14.Morohoshi, T., M. Kato, K. Fukamachi, N. Kato, and T. Ikeda, T. 2008. N-Acylhomoserine lactone regulates violacein production in Chromobacterium violaceum type strain ATCC 12472. FEMS Microbiol. Lett. 279:124-130. [DOI] [PubMed] [Google Scholar]
- 15.Morohoshi, T., Y. Nakamura, G. Yamazaki, A. Ishida, N. Kato, and T. Ikeda. 2007. The plant pathogen Pantoea ananatis produces N-acylhomoserine lactone and causes center rot disease of onion by quorum sensing. J. Bacteriol. 189:8333-8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morohoshi, T., S. Nakazawa, A. Ebata, N. Kato, and T. Ikeda. 2008. Identification and characterization of N-acylhomoserine lactone-acylase from the fish intestinal Shewanella sp. strain MIB015. Biosci. Biotechnol. Biochem. 72:1887-1893. [DOI] [PubMed] [Google Scholar]
- 17.Morohoshi, T., N. Someya, and T. Ikeda. 2009. Novel N-acylhomoserine lactone-degrading bacteria isolated from the leaf surface of Solanum tuberosum and their quorum-quenching properties. Biosci. Biotechnol. Biochem. 73:2124-2127. [DOI] [PubMed] [Google Scholar]
- 18.Na, K. S., A. Kuroda, N. Takiguchi, T. Ikeda, H. Ohtake, and J. Kato. 2005. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains. J. Biosci. Bioeng. 99:378-382. [DOI] [PubMed] [Google Scholar]
- 19.Nardini, M., and B. W. Dijkstra. 1999. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9:732-737. [DOI] [PubMed] [Google Scholar]
- 20.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, J. Schrag, J. L. Sussman, K. H. G. Verschueren, and A. Goldman. 1992. The α/β hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 21.Ozer, E. A., A. Pezzulo, D. M. Shih, C. Chun, C. Furlong, A. J. Lusis, E. P. Greenberg, and J. Zabner. 2005. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol. Lett. 253:29-37. [DOI] [PubMed] [Google Scholar]
- 22.Park, S. Y., H. O. Kang, H. S. Jang, J. K. Lee, B. Y. Koo, and D. Y. Yum. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, S. Y., S. J. Lee, T. K. Oh, J. W. Oh, B. T. Koo, D. Y. Yum, and J. K. Lee. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 24.Riaz, K., C. Elmerich, D. Moreira, A. Raffoux, Y. Dessaux, and D. Faure. 2008. A metagenomic analysis of soil bacteria extends the diversity of quorum-quenching lactonases. Environ. Microbiol. 10:560-570. [DOI] [PubMed] [Google Scholar]
- 25.Romero, M., S. P. Diggle, S. Heeb, M. Cámara, and A. Otero. 2008. Quorum quenching activity in Anabaena sp. PCC 7120: identification of AiiC, a novel AHL-acylase. FEMS Microbiol. Lett. 280:73-80. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schipper, C., C. Hornung, P. Bijtenhoorn, M. Quitschau, S. Grond, and W. R. Streit. 2009. Metagenome-derived clones encoding two novel lactonase family proteins involved in biofilm inhibition in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75:224-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teiber, J. F., S. Horke, D. C. Haines, P. K. Chowdhary, J. Xiao, G. L. Kramer, R. W. Haley, and D. I. Draganov. 2008. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-l-homoserine lactone. Infect. Immun. 76:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uroz, S., P. M. Oger, E. Chapelle, M. T. Adeline, D. Faure, and Y. Dessaux. 2008. A Rhodococcus qsdA-encoded enzyme defines a novel class of large-spectrum quorum-quenching lactonases. Appl. Environ. Microbiol. 74:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Bodman, S. B., W. D. Bauer, and E. L. Coplin. 2003. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 41:455-482. [DOI] [PubMed] [Google Scholar]
- 32.von Bodman, S. B., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, L. H., L. X. Weng, Y. H. Dong, and L. H. Zhang. 2004. Specificity and enzyme kinetics of the quorum-quenching N-acyl homoserine lactone lactonase (AHL-lactonase). J. Biol. Chem. 279:13645-13651. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinniel, D. K., P. Lambrecht, N. B. Harris, Z. Feng, D. Kuczmarski, P. Higley, C. A. Ishimaru, A. Arunakumari, R. G. Barletta, and A. K. Vidaver. 2002. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 68:2198-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]