Abstract
One of the oldest unresolved microbiological phenomena is why only a small fraction of the diverse microbiological population grows on artificial media. The “uncultivable” microbial majority arguably represents our planet's largest unexplored pool of biological and chemical novelty. Previously we showed that species from this pool could be grown inside diffusion chambers incubated in situ, likely because diffusion provides microorganisms with their naturally occurring growth factors. Here we utilize this approach and develop a novel high-throughput platform for parallel cultivation and isolation of previously uncultivated microbial species from a variety of environments. We have designed and tested an isolation chip (ichip) composed of several hundred miniature diffusion chambers, each inoculated with a single environmental cell. We show that microbial recovery in the ichip exceeds manyfold that afforded by standard cultivation, and the grown species are of significant phylogenetic novelty. The new method allows access to a large and diverse array of previously inaccessible microorganisms and is well suited for both fundamental and applied research.
It has been known for over a century that the overwhelming majority of microbial species do not grow on synthetic media in vitro and remain unexplored (13, 32, 37, 39, 40, 43). The rRNA and metagenomics approaches demonstrated a spectacular diversity of these uncultivated species (11, 21, 25-27, 30, 36). Accessing this “missing” microbial diversity is of significant interest for both basic and applied sciences and has been recognized as one of the principal challenges for microbiology today (12, 29, 41). In recent years, technical advances in cultivation methodologies have recovered a diverse set of ecologically relevant species (1, 3, 5, 7, 15, 20, 24, 28, 33, 42). However, by and large the gap between microbial diversity in nature and that in culture collections remains unchanged, and most microbial phyla still have no cultivable representatives (25, 29). Earlier, we developed a novel method of in situ cultivation of environmental microorganisms inside diffusion chambers (15). The rationale for such an approach was that diffusion would provide cells inside the chamber with naturally occurring growth components and enable those species that grew in nature at the time of the experiment to also grow inside the diffusion chambers. Expectedly, this method yields a rate of microbial recovery many times larger than those of standard techniques. Even so, this method is laborious and does not allow an efficient, high-throughput isolation of microbial species en masse. This limits the method's applicability, for example, in the drug discovery effort. Here we transform this methodology into a high-throughput technology platform for massively parallel cultivation of “uncultivable” species. Capitalizing on earlier microfluidics methods developed for microbial storage and screening (4, 16), we have designed and tested an isolation chip, or ichip for short, which consists of hundreds of miniature diffusion chambers. If each diffusion minichamber is loaded with a single cell, the resulting culture is monospecific. The ichip thus allows microbial growth and isolation into pure culture in one step. Here we demonstrate that cultivation of environmental microorganisms inside the ichip incubated in situ leads to a significantly increased colony count over that observed on synthetic media. Perhaps even more significantly, species grown in ichips are different from those registered in standard petri dishes and are highly novel.
MATERIALS AND METHODS
Isolation chip (ichip) design and application.
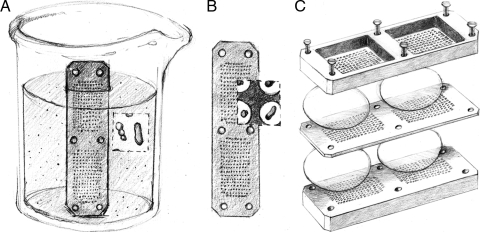
The ichip is an assembly of flat plates containing multiple registered through-holes (Fig. 1C), manufactured by HI-TECH Manufacturing, Schiller Park, IL. The plates are machined from blocks of hydrophobic plastic polyoxymethylene, commonly known under DuPont's brand name Delrin. The central plate (72 by 19 by 1 mm) and the two symmetrical top and bottom plates (72 by 19 by 6.5 mm each), the latter with ridges providing rigidity, have multiple through-holes 1 mm in diameter, arranged in two arrays with 192 through-holes per array. The size of the array is such that it can be completely covered by standard 25- or 47-mm-diameter membranes.
FIG. 1.
Isolation chip, or ichip, for high-throughput microbial cultivation in situ. (A) Dipping a plate with multiple through-holes into a suspension of mixed environmental cells leads to capturing (on average) a single cell (B). (C) Ichip assembly: membranes cover arrays of through-holes from each side; upper and bottom plates with matching holes press the membranes against the central (loaded) plate. Screws provide sufficient pressure to seal the content of individual through-holes, each becoming a miniature diffusion chamber containing (on average) a single cell. (Artwork by Stacie Bumgarner, Whitehead Institute for Biomedical Sciences, Cambridge, MA.)
The preparation of an ichip for microbial incubation starts with sterilizing its plastic components in ethanol, followed by drying in a laminar flow hood and rinsing in particle-free DNA-grade water (Fisher Scientific, Hampton, NH). The central plate is then dipped into a suspension of cells targeted for cultivation (Fig. 1A). A 50-ml Falcon tube (Fisher Scientific, Hampton, NH) is well suited to house the cell suspension. As a result of dipping, each through-hole captures a volume of suspension containing a certain number of cells. The cell number depends on the degree of dilution and can be on average one cell per through-hole (Fig. 1B). When the cells are suspended in a liquid agar-based medium, cells get immobilized inside small agar plugs that are formed once the agar solidifies. The cells thus become individually “trapped” in their respective through-holes and separated from each other. The next step in the assembly is application of membranes to each array of through-holes from both sides of the central plate (Fig. 1C); four membranes are required for the assembly. In a typical experiment, we used 0.03-μm-pore-size, 47-mm polycarbonate membranes (Osmonics Inc., Westborough, MA). These prevent cell migration in and out of the agar plugs. Lastly, the top and bottom plates are applied and aligned and screws are tightened to provide pressure. The pressure seals, without adhesive, the contents of the individual through-holes and agar plugs within and transforms the assembly into a combination of 384 miniature diffusion chambers, containing on average one cell per through-hole. Subsequent in situ incubation in the cell's original environmental habitat provides the immobilized cells with their naturally occurring nutrients and growth factors. After incubation, ichips are washed vigorously in particle-free DNA-grade water (Fisher Scientific, Hampton, NH) and disassembled. The central plate can then be examined under compound or high-power dissecting microscope for colony count. Agar plugs are extracted with unwound and sterile no. 1 gauge paper clips for further analyses.
Verification of the ichip's seal.
In the ichip tested, the contents of individual through-holes were separated from the environment by membranes. It was important to show that microorganisms from the environment could not invade agar in through-holes through spaces between membranes and the plastic parts. The latter were pressed against each other by means of screws, and we verified how well the pressure applied by tightening these screws sealed the inner space of the through-holes. Triplicate ichips were loaded with sterile 1% agar (BD, Franklin Lakes, NJ), assembled, submerged into 40 ml of Escherichia coli K-12 culture growing in 2.5% (wt/vol) Luria-Bertrani broth (LB) (BD, Franklin Lakes, NJ) in 50-ml Falcon tubes, and incubated for 24 h. After incubation, the ichips were removed and disassembled, and the contents of the through-holes were examined for growth under a compound microscope (Zeiss Axioskop 50 compound microscope) equipped for differential interference contrast (DIC) and fluorescence (Carl Zeiss, Jena, Germany) at ×100 magnification. In parallel, triplicate ichips were loaded with E. coli K-12 cells mixed with 1% warm LB agar, assembled, and incubated for 24 h in sterile LB. The external medium was then examined for growth.
Sources of environmental cells and their enumeration.
Seawater samples were obtained from the flowthrough seawater system at the Marine Science Center of Northeastern University, Nahant, MA (42°26′N, 70°56′W). Soil samples were collected from a (fresh) waterlogged wetland area on the grounds of the center, a few hundred feet away from the ocean. Soil samples were mixed with DNA-grade water (Fisher, Hampton, NH), and cells were dislodged by sonication using two 10-s-long pulses at amplitude setting 40 (Sonics Vibra-Cell VC130; 3-mm stepped microtip; Sonics & Materials, Inc., Newtown, CT). Particles were allowed to settle for 60 s before aliquots of supernatant were used for counting and as inocula in microbial growth experiments. Seawater samples were used without sonication. Environmental cells were enumerated with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Aldrich, St. Louis, MO) (23).
Microbial growth experiments.
Environmental cells were brought to a concentration of 103 cells/ml in diluted (0.1% [wt/vol]), warm (45°C) LB agar. Note that the ichip method does not depend on using agar, which can be replaced by low-melting-point agarose, alginate, gellan gum, and other gelling agents. For the seawater treatment, the above medium was supplemented with 4% (wt/vol) sea salts (Sigma Aldrich, St. Louis, MO). For both seawater and soil treatments, triplicate diffusion chambers received 3 ml of cell-agar mix and were established as described previously (15). Triplicate ichips were prepared from the same cell-agar mixes as described above. The volume of an agar plug forming in the through-hole upon agar solidification, with the volume of the two menisci factored in, is approximately 1.25 μl. Therefore, each ichip received approximately 500 μl of cell-agar mix and thus 500 cells. In order for standard cultivation conditions, referred to hereinafter as petri dish cultivation, to approximate the species diversity inoculated into ichips, triplicate petri dishes also received 500 μl of the cell-agar mixes (with no additional agar present, so that the cell/volume ratio stayed the same in all experiments). These were established in 24-well culture plates (Corning Costar, Corning, NY), which we used as analogues of small conventional petri dishes.
Cells were allowed to grow for 2 weeks. For incubation, diffusion chambers and ichips were returned to the environments that served as the sources of cells; they were either suspended in seawater on water tables of the flowthrough seawater system or buried in waterlogged soil. Petri dishes were incubated in the lab at room temperature.
After incubation, ichips were disassembled and 45 to 47 random cores were individually removed from each chip, flattened by coverslips on standard microscope slides, and examined for growth using a Zeiss Axioskop 50 compound microscope at ×1,000 magnification equipped for DIC. Triplicate, 5- to 10-μl samples of agar material from diffusion chambers and petri dishes were counted similarly. Microbial recovery was calculated as the percentage of cells forming microcolonies.
Microorganisms grown in ichips and petri dishes were identified using 16S rRNA gene sequences. Agar material was removed, separately from each ichip, using sterile paper clips (see above), disrupted in sterile deionized water, and homogenized by vortexing and passaging it through 25-gauge PrecisionGlide needles (Fisher Scientific, Hampton, NH). Genomic DNA was extracted using the DNeasy tissue kit (Qiagen, Valencia, CA) following the manufacturer's protocol for Gram-positive microorganisms. Fragments of 16S rRNA gene were amplified by seminested PCR, separately for each ichip, using Platinum PCR SuperMix (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The first PCR employed eubacterial universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) (17). The second PCR used eubacterial universal primers 27F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 518R (5′-ATT ACC GCG GCT GCT GG-3′) (18). The PCR products were combined, purified with a MinElute Kit (Qiagen, Valencia, CA), cloned using a TA cloning kit with TOP10 cells (Invitrogen, Carlsbad, CA), and commercially sequenced at Agencourt Bioscience Corporation (Beverly, MA). Agar material from the three petri dishes per treatment was processed in an identical manner.
Sequences were edited using the 4peaks software package (A. Griekspoor and T. Groothuis, http://www.mekentosj.com) and clustered into operational taxonomic units (OTUs) based on 99, 97, 95, and 90% sequence similarity cutoff values. This was achieved by first making all possible pairwise sequence alignments using ClustalW at default settings (34) and calculating percent sequence similarities, followed by clustering of the sequences into OTUs employing the mean unweighted-pair group method using average linkages as implemented in the OC clustering program (http://www.compbio.dundee.ac.uk/Software/OC/oc.html). From each OTU, the sequence least different from the other members of the cluster was compared to the NCBI database using the BLAST search function. The top hits were used to establish identity and relatedness of the OTUs.
RESULTS
To verify the completeness of the ichip's seal, we loaded the central plates of the ichips with sterile agar, assembled the units, and incubated them in growing E. coli culture. After incubation, we observed no growth inside the ichips. In the second set of controlling experiments, we loaded the central plates with agar containing E. coli cells and incubated the assembled units in sterile medium. After incubation, no growth was observed outside ichips. This indicated that the seal provided by tightening screws was sufficient to prevent cells from migrating in and out of ichips, and we proceeded with the microbial growth experiments.
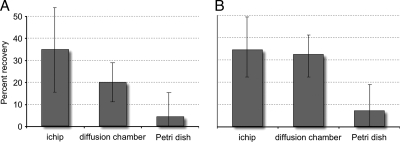
In this study, we used cells from two environments, seawater and soil; DAPI counts showed extant abundances of 3.6 × 106 cells/ml and 3.4 × 109 cells/g, respectively. The fractions of these cells forming colonies in the ichips, diffusion chambers, and petri dishes are given in Fig. 2. The colony counts were higher in the ichips than in diffusion chambers or petri dishes regardless of the environment studied. In ichips, growing cells constituted over 40% (seawater) and 50% (soil) of the number of cells inoculated. These counts were not statistically different from those obtained from the diffusion chambers (Student's t test, P > 0.05). Colony counts in petri dishes were approximately 5-fold lower, with statistically significant difference from either the ichip- or diffusion chamber-derived recoveries (Student's t test, P < 0.02).
FIG. 2.
Microbial recovery in ichip, diffusion chamber, and standard petri dish as percentage of inoculated cells forming colonies. (A) Seawater data. (B) Soil data.
To compare diversities of microorganisms grown in the ichips and petri dishes, we constructed and compared libraries of PCR-amplified 16S rRNA gene fragments from the ichip- and petri dish-grown material. Four libraries were established, comprising the following numbers of clones: 525 (soil, ichips), 314 (soil, petri dishes), 635 (seawater, ichips), and 265 (seawater, petri dishes). These numbers do not include sequences from 173 clones that were either too short or chimeric. Sequences longer than 420 nucleotides (nt) were considered adequate for the comparisons among the libraries. These were grouped into OTUs at various level of sequence similarity, as shown in Table 1. The ichip-reared microorganisms were dominated by Deltaproteobacteria, Firmicutes, and Gammaproteobacteria in soil and Firmicutes, Deltaproteobacteria, and Spirochaetes in seawater treatments (Table 2). In contrast, the petri dish-reared microorganisms were mostly representatives of Betaproteobacteria and Gammaproteobacteria in soil and Betaproteobacteria, Firmicutes, and Gammaproteobacteria in seawater treatments. The reasons why representatives of other phyla (e.g., Acidobacteria) were absent from either treatment are unclear. Also, Actinobacteria were not detected in the ichip-grown material. While most Actinobacteria require relatively long incubation times, often in excess of 2 weeks, they were observed in conventional petri dishes. Perhaps growth of actinobacteria is slowed down in situ versus in vitro, necessitating in situ incubations longer than 3 weeks.
TABLE 1.
Sequence similarities under growth conditions
| Condition | OTU at indicated % similarity levela |
|||
|---|---|---|---|---|
| 99 | 97 | 95 | 90 | |
| Ichip, seawater | 102 | 54 | 43 | 19 |
| Ichip, soil | 133 | 81 | 60 | 26 |
| Petri dish, seawater | 69 | 53 | 39 | 20 |
| Petri dish, soil | 68 | 49 | 37 | 17 |
Number of OTUs registered in ichips and petri dishes inoculated with seawater and soil microorganisms, at different percentages of rRNA gene sequence similarity to the cluster criterion.
TABLE 2.
Occurrences of phyla and other taxa under growth conditions
| Taxon | No. of occurrences with cultivation methoda: |
|||
|---|---|---|---|---|
| Ichip |
Petri dish |
|||
| Seawater | Soil | Seawater | Soil | |
| Alphaproteobacteria | 1 | 1 | 2 | 6 |
| Betaproteobacteria | 3 | 0 | 33 | 33 |
| Deltaproteobacteria | 16 | 54 | 0 | 0 |
| Epsilonbacteria | 2 | 1 | 0 | 0 |
| Gammaproteobacteria | 9 | 12 | 8 | 17 |
| Actinobacteria | 0 | 0 | 4 | 4 |
| Bacteroidetes | 0 | 1 | 6 | 3 |
| Firmicutes | 18 | 14 | 15 | 6 |
| Planctomycetes | 0 | 0 | 0 | 1 |
| Verrucomicrobia | 0 | 1 | 0 | 0 |
The numbers of occurrences of bacterial phyla and other taxa were registered in ichips and petri dishes inoculated with seawater and soil microorganisms.
The above differences are highlighted by the small size of overlap between the lists obtained from the ichips incubated in seawater and soil, which shared only 6 species out of the total 129 species detected in the ichips (defined as OTUs composed of 16S rRNA gene sequences sharing over 97% identity [31]). These species were related, as determined by the percent 16S rRNA gene similarity, to Desulfovibrio brasiliensis (88%), three Desulfovibrio spp. (98, 96, and 93%), Eubacterium oxidoreducens (93%), and a Spirochaeta sp. (97%). This overlap was larger with petri dishes: while 68 species out of 85 total registered in petri dishes were unique to their respective habitats, 17 were shared between them. Of the latter, three were registered multiple times, all of them previously described: a Ralstonia sp. (100% 16S rRNA gene sequence similarity to one of our OTUs), an Acidovorax sp. (99%), and Cloacibacterium normanense (100%). However, when species grown in ichips are compared with those cultivated in petri dishes, virtually no overlap is observed regardless of the environment tested. Only one species, in the seawater treatment, was shared between ichip- and petri dish-derived species lists; this exhibited 100% 16S rRNA gene similarity to Vibrio sp. strain ATCC EU655333. Even at a level above species, e.g., comparing lists of OTUs clustered at 90% of the 16S rRNA gene sequence similarity, overlap between the ichip- and petri dish-derived lists is minimal. Collectively, this indicates that the small overlap between ichip- and petri dish-reared species is not due to undersampling of respective diversities but is more likely due to differences in performance of the incubation devices themselves.
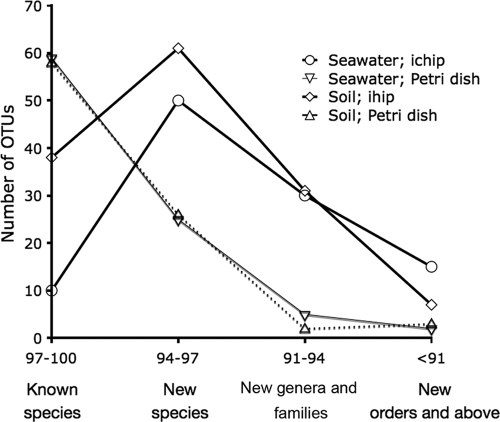
Ichip- and petri dish-derived strains (defined as OTUs composed of 16S rRNA gene sequences sharing over 99% identity) were different in the degrees of their phylogenetic novelty (Fig. 3). In petri dishes, the most frequently observed class of strains shares 97 to 100% identity with previously cultivated species. In contrast, there were few such strains registered in ichips from either treatment. Instead, the most frequently observed class of strains detected in ichips exhibit 94 to 97% identity with known species.
FIG. 3.
Novelty of seawater and soil microbial strains grown in ichips and petri dish. The equation of sequence novelty, in percent divergence from the known species, and taxonomic rank of novelty (genus level, family level, etc.) is very approximate.
DISCUSSION
The number of microbial species in nature is unknown but is likely large, with thousands of species present in a single gram of soil or aquatic sediment (10, 11, 26). However, most of this diversity is inaccessible for either basic or applied research. The cultivable microbial species are widely considered overmined for secondary metabolites (2, 38), and the probability of discovery of a novel bioactive compound is low. For example, discovery of a novel antibiotic from these (cultivable microorganisms) is a very unlikely event, with a probability of 10−7 per isolate (2). With such a discovery being seemingly impractical, microbial exploration has refocused on metagenomics and high-throughput screening of synthetic compound libraries (9, 22). These are promising approaches, but their application is not without difficulties. In environmental microbiology research, even the largest metagenomics studies are capable of sampling only a fraction of genes from the most abundant taxa (11); in applied microbiology, either approach has yet to produce a pipeline of novel drug candidates. The easiest way to study and exploit a novel species is through having it available in culture (19). The fact that the overwhelming part of microbial diversity remains uncultivated and unexplored presents exciting opportunities for basic and applied discoveries.
The renewed interest in microbial cultivation (1, 5, 7, 8, 14, 15, 33, 42) has led to the development of several innovative approaches to bring new species into culture. Most of these approaches share one basic strategy: to mimic the environment of target organisms. An ultimate move in this direction is to replace in vitro growth with in vivo cultivation in natural habitats (1, 5, 7, 15). However, these approaches also share an important drawback: presently, they do not provide a steady flow of new microbial cultures because they are either laborious, technologically complex, or limited in application to specific environments. Here we have capitalized on the success of the principle of in situ growth of “missing” microbial species and developed on its basis a novel, high-throughput technology platform (ichip) for a massively parallel microbial isolation. The essential elements of the new method are the use of naturally occurring compounds to meet the nutritional requirements of target microorganisms and the cultivation of these microorganisms as (predominantly) single colonies in isolated microchambers. The first allows for growth of novel species, and the second provides for their convenient isolation into pure culture. We show the advantage of this method in two ways.
Comparing to traditional technologies, the number of cells forming colonies in ichips is substantially higher than that in standard petri dishes (Fig. 2). Expectedly, the ichip performance in this regard is at least as good as that of the diffusion chamber we developed earlier (15) (Fig. 2). Though counts in petri dishes were much lower than those in ichips, they were uncharacteristically high for conventional cultivation. This elevated recovery in petri dishes is likely to be only apparent. Colony counts include microcolonies visible only under a compound microscope. We have shown previously (15) that a significant number of environmental cells form such microcolonies in vitro, but, unlike the diffusion chamber-reared microcolonies, the former do not regrow on subculturing. Though counted here, the petri dish-grown microcolonies are likely unimportant for cultivation efforts, while those from diffusion chambers, and by extension from ichips, can be propagated, domesticated, and scaled up.
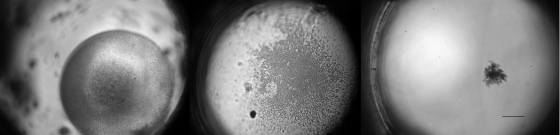
We also note the ease with which colonies grown in ichips can be visualized under a microscope (Fig. 4). The principal finding of this study is that organisms growing in ichips are more likely to be novel than those grown by standard approaches (Fig. 3). In fact, the level of novelty of the majority of organisms in the ichip-reared material is so high that known species appear to be almost “discriminated” against by this approach. There is essentially no overlap between species isolated by the ichip and those isolated by standard petri dish. Interestingly, the same is observed even when we consider more inclusive OTUs formed by sequences with >90% rRNA gene sequence identity. This means that ichip- and petri dish-based methods recover not only entirely different microbial strains and species but also different genera and possibly different families. Even microbial phyla recovered are different between the two cultivation approaches: out of 10 phyla detected, 5 were unique to one or another technique (Table 2). We recognize that the number of 16S rRNA gene sequences was larger in the case of ichip-derived material, and this affects the number of OTUs registered (Table 1). However, presence-absence analysis, which is less sensitive to the inequality in sample size, still shows considerable differences between the two approaches, especially in the case of recovery of Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria, as well as Actinobacteria and Bacteroidetes.
FIG. 4.
Examples of microcolonies grown in ichips as seen under a compound microscope equipped for differential interference contrast at ×100. The diameter of the through-holes is 1 mm.
The noted lack of overlap between pools of species, genera, etc., obtained by the two cultivation approaches is also unlikely to be due to trivial undersampling, because we did observe a measurable overlap between species diversities within treatment. For example, petri dishes inoculated with seawater and soil environments shared 20% of grown species (likely due to the close proximity of the sampling points). Notably, at the level of microbial phyla, differences within treatment—but not between the treatments—essentially disappear (Table 2).
Comparing the ichip to the original diffusion chamber (3, 15), we emphasize the ease of ichip operation: its assembly and disassembly take under 5 min; scoring growth is straightforward because the amount of agar in each through-hole is minimal, so small that it does not interfere with visual inspection (but a quality dissecting microscope is required); and removal of growth is quite simple. Also, the array could be reconfigured to match the size of a standard microtiter plate, enabling high-throughput applications, further facilitated by the use of, e.g., pin replicators to transfer growth into a microtiter plate in a single step or by using robotic colony pickers. Based on our current results, we would expect the rate of microbial recovery within ichip treatment to be high—with up to 50% of the loaded cells forming colonies (Fig. 2)—and dozens of novel (Fig. 3) and pure isolates to be produced in each ichip growth experiment.
The ichip-grown isolates are amenable to a number of analyses, including genomic analyses, but detailed experimentation and exploration of biotechnological potential will require their in vitro domestication. In previous research, we explored ways to domesticate diffusion chamber-grown isolates. We discovered that their multiple transfers through the chamber typically led to the ability of the cells to sustainably grow in standard petri dishes (Fig. 2 in reference 20), with 26% of chamber-reared colonies domesticating after the first round and up to 40% after two rounds. We confirmed this observation in a separate study of “uncultivables” from a fresh pond environment (3), noting that that domesticated pool included representatives of rarely cultivated classes and phyla, such as Deltaproteobacteria, Verrucomicrobia, Spirochaetes, and Acidobacteria. In a follow-up study of Actinobacteria, we again observed a substantial number of strains appearing on petri dishes after one to several rounds of cultivation in situ (A. Bollmann, A. Adams, K. Lewis, and S. S. Epstein, unpublished results). The nature of the domestication process is unknown and discussed in detail elsewhere (6). The practical implication is that in situ incubation facilitates the appearance of cells that have fewer growth restrictions and are capable of growing in vitro, which provides convenient access to a substantial pool of novel microbial diversity. We expect that ichip-grown isolates can be domesticated similarly because the ichip and diffusion chamber (15) share the same basic principle: in situ growth on natural sources supplied by diffusion. If so, we can estimate the overall microbial recovery as the percentage of cells inoculated into the ichip that would form colonies on synthetic media as a result of a two-step process, cultivation in an ichip followed by subculturing on standard media. This estimate is the product of recovery in the ichip itself (up to 50%; Fig. 2) and success of domestication (26% after a single ichip incubation), or about 10 to 15%. This is at least an order of magnitude above the level of a typical recovery by standard cultivation alone and affords access to significantly novel species that otherwise do not seem to grow in vitro. Note that, with this recovery rate, chances of mixed cultures become low. In the event of two cells loaded into one diffusion minichamber of the ichip, and since each has a probability of being domesticated of 10 to 15%, only about 1 to 2% of minichambers with two cells will produce mixed cultures in vitro. Considering that (i) a single researcher can conservatively establish 10 to 20 ichips in a day, (ii) incubation of ichips requires no labor, (iii) disassembly of 10 to 20 ichips and subculturing material grown therein on standard media can be done in one day, and (iv) mixed cultures are unlikely to represent a significant problem, our method will likely result in producing domesticated and pure cultures of novel microbial species at a rate of >100/researcher/day. Consequently, the limiting step in obtaining microbial novelty no longer seems to be in the cultivation step but rather in the downstream analyses, such as, e.g., microbial identification via 16S rRNA gene sequencing.
In conclusion, the ichip represents a practical device for massively parallel in situ cultivation of environmental microorganisms. The grown isolates exhibit substantial phylogenetic novelty, and their list overlaps little with that of the collection obtained by standard techniques. Based on our earlier reports, around a quarter of these isolates should grow in standard petri dishes after a single in situ incubation; more can be domesticated by serial passaging of the ichip-grown material through several generations of ichips. The application of ichips will contribute to the resolution of the “great plate count anomaly” and may form a basis for drug discovery from the previously uncultivated microbial majority.
Acknowledgments
We are grateful to several individuals who contributed significantly to this research. Ron Ortenberg (Northeastern University, Boston, MA) first brought our attention to OpenArray plates by Biotrove, Inc. (Woburn, MA), as a device to sample precise amounts of liquids. Alex Schering, Ron Ortenberg, and Tammy Hartke (Northeastern University) performed preliminary cultivation experiments that led to the development of the ichip. Alex Volchek and Simon Sorsher (Hi-Tech Manufacturing, LLC, Shiller Park, IL) manufactured ichips and provided practical advice. Danbi Choi and Jennifer Trost (Northeastern University) assisted with several trials of the ichip. Lucy Ling (Novobiotic Pharmaceuticals, LLC, Cambridge, MA) helped with microphotography.
This work was supported in part by NSF grants OCE-0221267, MCB-0348341, and DEB-0816840 to S.S.E., DOE grant DE-FG02-04ER63782 to K.L. and S.S.E., and DOE grants DE-FG02-07ER64507 and DE-FG02-04ER63782 to S.S.E. and K.L.
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Aoi, Y., T. Kinoshita, T. Hata, H. Ohta, H. Obokata, and S. Tsuneda. 2009. Hollow fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol. 75:3826-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltz, R. H. 2005. Antibiotics from Actinomycetes: will a renaissance follow the decline and fall? SIM News 55:186-196. [Google Scholar]
- 3.Bollmann, A., K. Lewis, and S. S. Epstein. 2007. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 73:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenan, C. J. H., T. Morrison, K. Stone, T. Heitner, A. Katz, T. Kanigan, R. Hess, S.-J. Kwon, H.-C. Jung, and J.-G. Pan. 2002. A massively parallel microfluidics platform for storage and ultra high throughput screening. Proc. Soc. Photo. Opt. Instrum. Eng. 4626:560-569. [Google Scholar]
- 5.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein, S. S. 2009. General model of microbial uncultivability, p. 131-159. In S. S. Epstein (ed.), Uncultivated microorganisms. Springer, Heidelberg, Germany.
- 7.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari, B. C., and M. R. Gillings. 2009. Cultivation of fastidious bacteria by viability staining and micromanipulation in a soil substrate membrane system. Appl. Environ. Microbiol. 75:3352-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong, S.-H., J. Bunge, S.-O. Jeon, and S. Epstein. 2006. Predicting microbial species richness. Proc. Natl. Acad. Sci. U. S. A. 103:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber, J. A., D. B. Mark Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 12.Hurst, C. J. 2005. Divining the future of microbiology. ASM News 71:262-263. [Google Scholar]
- 13.Jannasch, H. W., and G. E. Jones. 1959. Bacterial populations in seawater as determined by different methods of enumeration. Limnol. Oceanogr. 4:128-139. [Google Scholar]
- 14.Joseph, S. J., P. Hugenholtz, P. Sangwan, C. A. Osborne, and P. H. Janssen. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 16.Kanigan, T., C. Brenan, S. Lafontaine, L. Soswoski, P. Madden, and I. Hunter. 2000. Living chips for drug discovery. Proc. Soc. Photo. Opt. Instrum. Eng. 3926:172-180. [Google Scholar]
- 17.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 18.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols, D. 2007. Cultivation gives context to the microbial ecologist. FEMS Microbiol. Ecol. 60:351-357. [DOI] [PubMed] [Google Scholar]
- 20.Nichols, D., K. Lewis, J. Orjala, S. Mo, R. Ortenberg, P. O'Connor, C. Zhao, P. Vouros, T. Kaeberlein, and S. S. Epstein. 2008. Short peptide induces an “uncultivable” microorganism to grow in vitro. Appl. Environ. Microbiol. 74:4889-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 22.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 23.Porter, K. G., and Y. C. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 24.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 25.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 26.Roesch, L. F., R. R. Fulthorpe, A. Riva, G. Casella, A. K. Hadwin, A. D. Kent, S. H. Daroub, F. A. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, and K. Remington. 2007. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5(3):e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sangwan, P., S. Kovac, K. E. Davis, M. Sait, and P. H. Janssen. 2005. Detection and cultivation of soil Verrucomicrobia. Appl. Environ. Microbiol. 71:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sogin, M. L., H. G. Morrison, J. A. Huber, D. Mark Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteria. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 32.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 33.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, and W. Nelson. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 37.Waksman, S. A., and M. Hotchkiss. 1937. Viability of bacteria in sea water. J. Bacteriol. 33:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 39.Winslow, C.-E. A., and G. E. Willcomb. 1905. Tests of a method for the direct microscopic enumeration of bacteria. J. Infect. Dis. Suppl. 1:273-283. [Google Scholar]
- 40.Winterberg, H. 1898. Zur Methodik der Bakterienzählung. Zentralbl. Hyg. 29:75-93. [Google Scholar]
- 41.Young, P. 1997. Major microbial diversity initiative recommended. ASM News 63:417-421. [Google Scholar]
- 42.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. U. S. A. 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.ZoBell, C. E. 1946. Marine microbiology: a monograph on hydrobacteriology. Chronica Botanica Co., Waltham, MA.