Abstract
Knowledge of the microbial consortia participating in the generation of biogas, especially in methane formation, is still limited. To overcome this limitation, the methanogenic archaeal communities in six full-scale biogas plants supplied with different liquid manures and renewable raw materials as substrates were analyzed by a polyphasic approach. Fluorescence in situ hybridization (FISH) was carried out to quantify the methanogenic Archaea in the reactor samples. In addition, quantitative real-time PCR (Q-PCR) was used to support and complete the FISH analysis. Five of the six biogas reactors were dominated by hydrogenotrophic Methanomicrobiales. The average values were between 60 to 63% of archaeal cell counts (FISH) and 61 to 99% of archaeal 16S rRNA gene copies (Q-PCR). Within this order, Methanoculleus was found to be the predominant genus as determined by amplified rRNA gene restriction analysis. The aceticlastic family Methanosaetaceae was determined to be the dominant methanogenic group in only one biogas reactor, with average values for Q-PCR and FISH between 64% and 72%. Additionally, in three biogas reactors hitherto uncharacterized but potentially methanogenic species were detected. They showed closest accordance with nucleotide sequences of the hitherto unclassified CA-11 (85%) and ARC-I (98%) clusters. These results point to hydrogenotrophic methanogenesis as a predominant pathway for methane synthesis in five of the six analyzed biogas plants. In addition, a correlation between the absence of Methanosaetaceae in the biogas reactors and high concentrations of total ammonia (sum of NH3 and NH4+) was observed.
During the last decade the production of biogas from organic materials and residues has increased continuously in order to reduce the greenhouse gas emission resulting from the use of fossil energy sources. The energy-bearing substance of biogas is methane, which is produced as an end product of microbial anaerobic degradation of organic substrates, such as energy crops like maize, grains, grasses, or beets. Research for optimization of biogas production from renewable materials was initially focused on the evaluation of substrate eligibility and on the development and optimization of technical systems. However, biogas formation primarily depends on the structure and activity of the microbial community (28).
The key microorganisms in the biogas formation process are the methane-generating microorganisms (methanogens). The capacity for methanogenesis is limited to members of the domain Archaea and, within this domain, on the phylum Euryarchaeota. With respect to the main metabolic precursors used, methanogens are usually divided into two groups: the aceticlastic methanogens that strictly metabolize acetate and the hydrogenotrophic methanogens that use H2 or formate as an electron donor and CO2 as a carbon source for their metabolism. Besides these major groups, certain methanogens are also able to convert methyl groups, methylamines, or methanol to methane (23, 40). The substrates for the methanogens are provided by several physiological groups of bacteria which degrade organic matter, sometimes in close syntrophic interaction with the methanogens (1).
Several studies on the microbial diversity present in lab-scale biogas reactors supplied with renewable raw material (7, 57) have been recently published. However, analyses under laboratory conditions do not necessarily reflect conditions in full-scale reactors (35). Therefore, further research on the methanogenic community in full-scale biogas reactors is crucial.
Generally, studies regarding the microbial community structure in full-scale biogas reactors have focused on different systems for wastewater treatment or classical biogas plants based on manure digestion (32, 38, 43). In most systems, approximately 70% of the carbon fixed in methane was derived from acetate. Only minor amounts, up to approximately 30%, were deduced from CO2 (1, 42). Together with the presence of huge assemblages of Methanosarcina sp., it was assumed by some authors that aceticlastic methanogenesis was the predominant pathway for methane formation. Moreover, as shown by other studies, the relative contribution of H2/CO2 versus acetate as metabolic precursors for methanogens can be quite different in other anaerobic environments (10, 33, 37). However, the methanogenic microfloras in full-scale biogas reactors supplied with energy crops as a primary or sole substrate have rarely been studied (35, 37, 45).
The aim of this study was to gain insight into the diversity of methane-producing Archaea in six full-scale biogas plants supplied with renewable raw material and different types of liquid manure as substrates. Therefore, a polyphasic approach with three different culture-independent techniques (fluorescence in situ hybridization [FISH], quantitative PCR [Q-PCR], and 16S rRNA gene analysis) to analyze methanogen diversity was carried out to overcome the known limitations of each single approach (15, 46). To analyze potential effects of different process parameters on the methanogenic archaeal community, the reactor performances were correlated with the apparent archaeal diversity.
MATERIALS AND METHODS
Reactor operation and sampling.
Six full-scale biogas plants (R1 to R6) located in northeastern Germany (Brandenburg, Mecklenburg-Vorpommern, and Sachsen-Anhalt) were chosen for polyphasic analysis of the composition of the methane-producing Archaea in biogas plants supplied with a mixture of different liquid manures and renewable raw materials as substrates (Table 1). All reactors were operated at mesophilic temperatures and under wet fermentation conditions. The substrates of reactors R1 to R3 and of reactors R5 and R6 consisted of mixtures of animal manure and renewable raw materials. Reactor R4 was fed exclusively with renewable raw material.
TABLE 1.
Biogas plants and operating conditionsa
| Biogas plant | Feedstock components (% fresh wt of total feedstock)b | Reactor vol (m3) | Organic loading rate (kgODS m−3 day−1) | Retention time (days) | Total biogas production (mBG3 m−3 day−1)d | CH4 production (mCH43 m−3 day−1)d | Operation temp (°C) | pH | Total ammonia (g liter−1)e | NH3-N (g liter−1) | NH4+-N (g liter−1) | VFA (gHAc eq. liter−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1c | Cattle dung (18%), cattle sewage (41%), maize silage (38%), water (2.5%) | 3,600 | 3.8 | 41 | 2.90 | 1.5 | 36.8 | 7.6 | 1.6 | 0.08 | 1.52 | 1.4 |
| R2 | Pig liquid manure (50%), maize silage (39%), turkey dung (9%) | 1,300 | 2.9 ± 0.2 (2.5) | 30.2 ± 3.1 (35) | 3.6 ± 0.2 (3.4) | 1.9 ± 0.2 (1.8) | 44.6 ± 0.4 (44.0) | 8.2 ± 0.1 (8.3) | 4.3 ± 0.0 (4.3) | 1.15 ± 0.1 (1.23) | 3.11 ± 0.1 (3.03) | 7.6f |
| R3 | Pig liquid manure (57%), maize silage (40%) | 2,700 | 3.9 ± 0.2 (3.9) | 47 ± 1.5 (48) | 2.5 ± 0.1 (2.7) | 1.3 ± 0.1 (1.4) | 38.5 ± 0.3 (38.7) | 7.7 ± 0.1 (7.6) | 2.0 ± 0.5 (1.5) | 0.12 ± 0.04 (0.08) | 1.85 ± 0.41 (1.42) | 2.5 ± 0.5 (2.8) |
| R4c | Maize silage (82%), barley grain (12%), water (6%) | 3,674 | 3.4 | 108 | 2.6 | 1.4 | 44.7 | 7.5 | 2.6 | 0.37 | 2.23 | 2.1 |
| R5 | Cattle liquid manure (64%), maize silage (37%), pig liquid manure (6%), turkey dung (2%) | 2,640 | 4.2 ± 0.7 (4.0) | 36.0 ± 3.2 (34.0) | NA | NA | 41.0 ± 0.1 (41.0) | 7.9 ± 0.1 (7.8) | 4.0 ± 0.0 (4.0) | 0.45 ± 0.04(0.38) | 3.55 ± 0.04(3.62) | 2.4f |
| R6 | Cattle liquid manure (76%), maize silage (13%), grass silage (5%), cattle dung (4%), grains (2%) | 1,950 | 2.5 ± 0.3 (2.4) | 46.9 ± 1.4 (47.0) | 1.3 ± 0.1 (1.3) | 0.7 ± 0.0 (0.7) | 39.9 ± 0.8 (39.3) | 7.9 ± 0.3 (7.4) | 1.7 ± 0.0 (1.7) | 0.20 ± 0.08(0.06) | 1.50 ± 0.08(1.64) | 1.5f |
Data are shown are average values with standard deviations of a time period of 4 weeks before sampling (data of the sampling day are in parentheses). ODS, organic dry substance; BG, biogas; HAc eq., acidic acid equivalent of all organic acids; NA, not analyzed.
For components representing ≥2% of the total.
One-time sampling from biogas plant at the beginning of a measurement period.
Data normalized with 273.15 K and 1,013.25 hPa.
Sum of NH3-N and NH4+-N.
Data from day of sampling.
The biogas plants were sampled once in a time period from August 2006 to July 2007. At the date of sampling all biogas plants had been operated for at least 1 year. From each of the six biogas plants, four samples of 5 liters each were taken from the stirred reactor contents in time intervals of 15 min. From each individual sample 500 ml was pooled and stored at room temperature for a maximum of 12 h until further processing. Corresponding operating parameters are summarized in Table 1. Total ammonia nitrogen is defined as the sum of NH3-N and NH4+-N. The NH3-N (ammonia nitrogen) concentration was calculated with following formula after Anthonisen et al. (4) and Gallert and Winter (22): (total ammonia-N × 10pH)/(Kb/Kw + 10pH), where N (nitrogen) concentration is in g liter−1, Kb/Kw is e(6344/273+T), and T is the temperature in °C.
All organic acids including volatile fatty acids (VFA) were calculated as acetic acid equivalents (HAc eq.).
FISH.
A total of 25 ml of pooled reactor sample was mixed with 1 volume of 96% ethanol and kept at −20°C for a maximum of 12 h. The fixation of the samples with 3.7% formaldehyde was carried out according to a slightly modified protocol published by Daims et al. (19). To disperse the extracellular polymeric substances (EPS) which promote the adhesion of cells to plant particles and the formation of cell aggregates, an enzyme mixture of α-glucosidase, β-galactosidase, and lipase (Sigma-Aldrich Chemie GmbH, Munich, Germany) was used according to a modified protocol developed by Böckelmann et al. (8). A total of 100 μl of each formaldehyde-fixed sample was washed twice with 1× phosphate-buffered saline ([PBS] 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 5.7); the pellet was mixed with 2 U of α-glucosidase, 2 U of β-galactosidase, 10 U of lipase, and 2 mM MgCl2. PBS (1×; pH 5.7) was added to a final volume of 500 μl. After incubation for 1 h (at 30°C and 1,000 × g), a two-step ultrasonic treatment (Sonoplus GW2070; Bandelin, Berlin, Germany) for 30 s each at pulse level 1 with 50% power (approximately 35 W) was carried out to support the dispersion of EPS. For each reactor sample, a dilution series (100-, 500-, and 1,000-fold) was performed.
For identification of total archaeal and bacterial populations the ARC915 (58) and the EUB223 (2) probes, respectively, were used in experiments performed as triplicates. Specific probes for Methanomicrobiales (MG1200) (50), Methanobacteriales (mix of MB310 and MB1174) (50), Methanosarcinaceae (Ms821) (50), and Methanosaetaceae (Mx825) (18, 50) were applied only for biogas plants R1, R4, R5, and R6 in duplicates. The oligonucleotide probes were used under optimal stringency conditions as described in the probeBase data bank (41). All probes were Cy3 labeled and purchased from metabion (Martinsried, Germany). 4′,6′-Diamidino-2-phenylindole (DAPI) was used for total cell detection.
As positive controls for the specific probes the following reference cultures were used: Methanoculleus marisnigri (DSM1498), Methanospirillum hungatei (Mh1), Methanosaeta concilii (DSM6752), Methanobacterium formicicum (DSM1525), Methanobrevibacter arboriphilus (DSM1125), and Methanothermobacter thermautotrophicus (DSM1053).
Prior to hybridization each of the 10 wells of a Teflon-coated slide was treated with 0.1% gelatin and 0.01% CrK(SO4)2. Hybridizations and washing procedures were performed according to a modified protocol of Daims et al. (19). The hybridization buffer as described by Daims et al. (19) was supplemented with 10× Denhardts reagent (20) to speed up the hybridization and to prevent nonspecific binding of the probes. Hybridization was performed in a buffer-saturated humidity chamber at 46°C for 2 h in a hybridization oven (HL-2000 HybriLinker; UVP Laboratory Products, Cambridge, United Kingdom).
Prior to microscopic analysis, 5 μl of the antifading reagent Citifluor AF1 and 0.1 μl of DAPI (70 μg ml−1) were added to each sample. Fluorescence was detected by a Nikon Optiphot-2 microscope (Nikon, Düsseldorf, Germany) fitted for epifluorescence microscopy with a 100-W mercury high-pressure bulb (HBO 103W/2) and filter sets for DAPI (DAPI AMCA) and Cy3 (HQ:Cy3). Digital images of the samples were taken with a Nikon Digital Sight DS-2Mv (Nikon, Düsseldorf, Germany) and the software NIS-Elements, version 2.2. For determination of total cell counts, approximately 1,000 DAPI-stained cells from independent, randomly chosen microscopic fields were counted at a magnification of ×630. In the case of aggregate-forming Methanosaeta spp. and Methanosarcina spp., the area of cell aggregates was determined using the software NIS-Elements, version 2.2. Based on aggregate area, the corresponding cell numbers were estimated as follows: members of the genus Methanosarcina have an average diameter of 2 μm (23), which corresponds to an area of 3.1415 μm2. A Methanosaeta sp. cell is 1.05 μm wide and 4.5 μm long on average (23), resulting in a two-dimensional image with an average area of 4,725 μm2.
Total cell counts, total bacteria, and total archaea per ml of reactor contents were calculated by following formula (49): Awell/Acount × Xm × v, where Awell is the effective area of well surface (35.26 mm2), Acount is the surface area of a microscopic field (1.44 mm2), Xm is the average cell number per microscopic field, and v is the dilution factor (1 ml−1). The sum of the average cell counts determined by specific FISH probes served as a basis for the calculation of percent distribution of the four main methanogenic groups.
DNA extraction and purification.
From the pooled reactor samples (total volume, 2 liters) four parallel subsamples of 40 ml each were taken and processed separately. The genomic DNA was extracted and purified as described previously (45). This included sample purification by centrifugal steps; enzymatic cell lysis with lysozyme, proteinase K, and sodium dodecyl sulfate (SDS); purification steps with cetyl-trimethylammonium-bromide (CTAB) and chloroform-isoamylalcohol (24:1); and a subsequent isopropyl alcohol precipitation.
For the biogas plants R1 and R6, an additional purification step was necessary to remove PCR inhibitors. A purification technique based on low-melting point (LMP) agarose (AppliChem GmbH, Darmstadt, Germany) was carried out according to a modification of the protocol of Moreira (44).
Microscopic verification of cell lysis.
Before and after the cell lysis procedure, microscopic verification of complete cell lysis was conducted: the samples were examined by phase-contrast light microscopy (Nikon Optiphot-2 with a 40× objective; Nikon, Düsseldorf, Germany) and UV fluorescence microscopy (Nikon Optiphot-2 with a 40× objective and filter block DM430; Nikon, Düsseldorf, Germany). Fluorescence microscopy was used to detect hydrogenotrophic methanogens by their coenzyme F420 autofluorescence (56). In all samples only very few unlysed cells were visible by phase-contrast microscopy, which indicated a sufficient treatment for cell lysis. Accordingly, the samples showed none of the F420 fluorescence signals that are characteristic for many methanogens, indicating lysis of the majority of methanogenic cells.
Q-PCR.
For Q-PCR a 5′ nuclease assay (TaqMan assay) was applied and performed on an ABI 7300 System (Applied Biosystems, Darmstadt, Germany). Primer sets and TaqMan probes, including their PCR conditions, for the domains Archaea and Bacteria, the orders Methanomicrobiales and Methanobacteriales, and the families Methanosarcinaceae and Methanosaetaceae as published by Yu et al. (64, 65) were applied as described by Nettmann et al. (45). Standard amplification curves were constructed according to Nettmann et al. (45). The Q-PCR results were analyzed with the 7300 Real-Time PCR System Sequence Detection Software, version 1.3 (Applied Biosystems, Darmstadt, Germany). The 16S rRNA gene copies in the samples were calculated as described previously (45).
Construction and analysis of 16S rRNA gene clone libraries.
Construction of 16S rRNA gene clone libraries and the subsequent library screening by amplified rRNA gene restriction analysis (ARDRA) was carried out as described previously (45). The following 16S rRNA gene primers were used: Arch16S-Forw2 (5′-YGAYTAAGCCATGCRAGT-3′) modified after (24) and Univ16S-Rev5 (5′-TGCTCCCCCGCCAATTCCT-3′), modified according to Fernandez et al. (21). Subsequently, the amplified DNA fragments (860 bp) were digested with the restriction enzymes BsuRI and Hin6I (Fermentas, St. Leon-Rot, Germany). Clones with similar ARDRA patterns were taken as operational taxonomic units (OTUs). For DNA sequencing (MWG Biotech AG, Martinsried, Germany) and subsequent phylogenetic classification, one clone representative of a group of clones with identical ARDRA patterns was chosen.
Phylogenetic analysis.
All obtained 16S rRNA gene sequences were checked for chimeric artifacts by the Chimera Check software tool (14). Alignments of 16S rRNA gene sequences and reference sequences from NCBI GenBank were performed with the software package Mega, version 4.0 (61), and ClustalW, version 1.6 (62), using a neighbor-joining algorithm (51) with the Kimura-2 parameter (34) as a distance correction model, applying standard settings. For direct comparison of determined 16S rRNA gene nucleotide sequences (approximately 860 bp) with individual reference sequences, the uncorrected p-distances (Mega, version 4.0, software package) were calculated as indicators of nucleotide sequence dissimilarities.
Statistical analyses of 16S rRNA gene clone libraries.
Rarefaction analyses to obtain richness curves were performed with the software Analytical Rarefaction, version 3.1 (http://www.uga.edu/∼strata/software/Software.html). The interpolating rarefaction method evaluates how the number of OTUs in a sample changes with the number of individuals (29) and reflects the OTU richness of a clone library. Additionally, the extrapolating Chao I index (11) was calculated with the software EstimateS, version 8.0.0 (R. K. Colwell, University of Connecticut, Storrs [http://viceroy.eeb.uconn.edu/EstimateS]). This nonmetric estimate index considers the OTUs represented by one or two clones in the sample. The 95% confidence intervals represent the significance of measuring points.
The following formula described by Good (26) was used to calculate the coverage of the clone libraries: 1 − (n/N), where n is the number of OTUs represented by one clone, and N is the total number of clones.
In order to quantify the diversity of archaea, the Shannon index (H) was calculated (55). This index gives the proportional abundance of species and reacts sensitively to rare species. Evenness (E) was computed to describe the uniformity of the distribution of the individuals over the number of OTUs (47). These analyses were performed using the software package PAST, version 1.75b (P. D. Ryan, University of Oslo, Oslo, Norway [http://folk.uio.no/ohammer/past]).
Comparison of archaeal communities in the biogas plants.
Similarities between archaeal communities and archaeal OTU abundances in the biogas plants were calculated by the Chao-Jaccard (CJ) similarity index (12). This index is slightly sensitive to the sample size because it accounts for the unseen shared species. The calculation of the CJ index is based upon the diversity of methanogens as determined by the abundance of OTUs and ARDRA patterns. All CJ similarity values were computed with the software EstimateS, version 8.0.0 (see above). The distances for multidimensional scaling (MDS), calculated by the software program PERMAP (R. B. Heady and J. L. Lucas, University of Louisiana, Lafayette [http://www.ucs.louisiana.edu/∼rbh8900/]), were used to provide a two-dimensional illustration of the diversity similarities between the samples. To analyze potential effects of main substrates on methanogenic biocoenosis, these parameters were correlated with the determined archaeal diversities.
Nucleotide sequence accession numbers.
All nucleotide sequences obtained in this study have been deposited in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession numbers EU447678, EU636905, FJ222201 and FJ222202, FJ222204 and FJ222205, FJ222208 to FJ222225, FJ222228 and FJ222229, FJ222231 to FJ222236, and FJ356063 to FJ222266.
RESULTS
Reactor performance.
All biogas plants were sampled during a stable operating phase, where the biogas and methane production rates were maintained at constant rates. All biogas plants were operated in a mesophilic temperature range between 36.8 (R1) and 44.6°C (R2). The pH values of all six biogas reactors varied from pH 7.4 (R6) to 8.2 (R2) and were thus in the lower alkaline range (Table 1). Biogas production rates of the biogas plants ranged between 1.3 (R6) and 3.6 m3 m−3 day−1 (R2) (Table 1). Methane yields amounted to between 51 and 52% (vol/vol) of total biogas yield. These yields were comparable to production rates and methane yields, respectively, of other agricultural biogas plants (9).
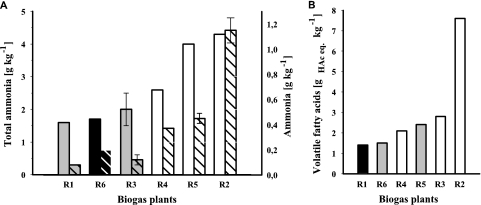
The VFA concentrations in biogas plants R2 (7.6 g liter−1), R3 (2.8 g liter−1), and R5 (2.4 g liter−1), supplied with pig liquid manure (R2 and R3) and cattle liquid manure (R5), were higher than in R1 (1.4 g liter−1) and R6 (1.5 g liter−1), fed with cattle liquid manure, and in R4 (2.1 g liter−1), fed with maize silage (Table 1). The total ammonia concentration (sum of NH3 and NH4+-nitrogen) in reactors R2, R4, and R5 ranged between 2.6 and 4.3 g liter−1 and was higher than the concentrations in reactors R1, R3, and R6, with total ammonia concentrations between 1.6 and 2.0 g liter−1 (Table 1). The calculated NH3-N concentrations ranged from 0.01 g liter−1 (R4) to 1.15 g liter−1 (R2) (Table 1).
Methanogenic archaeal communities in agricultural biogas reactors.
For the detection and quantification of main methanogenic groups in six agricultural biogas plants, two different techniques, microscopic analysis by FISH and 16S rRNA gene quantification by Q-PCR analysis, were applied.
The concentrations of bacteria determined by FISH (EUB338 probe) for the six biogas plants ranged between 9.6 × 107 ± 7.0 × 107 (R2) and 1.0 × 109 ± 0.2 × 109 (R4) cells per ml of reactor content (Table 2), which corresponds roughly to 51% ± 4% (R2) and 65% ± 12% (R1) of total cell counts determined by DAPI staining. The low percentage of bacteria detected by FISH probe EUB338 might be caused by the high background fluorescence in the samples, which interferes with probe signal. Archaeal abundances ranged between 1.7 × 107 ± 1.4 × 107 (R2) and 1.3 × 108 ± 0.9 × 108 (R6) cells per ml of reactor content (Table 2). The percentage of archaea determined by FISH ranged between 3% ± 0.4% (R6) and 7% ± 3.9% (R1) of total cell counts.
TABLE 2.
Cell and 16S rRNA gene copy concentrations in the six biogas reactors determined by FISH and Q-PCR
| Biogas plant | Total cell count (ml−1)a | Bacterial abundance determined by: |
Archaeal abundance determined by: |
||
|---|---|---|---|---|---|
| FISH (cells ml−1) | Q-PCR (16S rRNA gene copies ml−1) | FISH (cells ml−1) | Q-PCR (16S rRNA gene copies ml−1) | ||
| R1 | (8.5 ± 5.6) × 108 | (6.2 ± 2.7) × 108 | (2.5 ± 0.4) × 1011 | (6.5 ± 5.8) × 107 | (2.0 ± 0.3) × 1010 |
| R2 | (3.1 ± 2.5) × 108 | (9.6 ± 7.0) × 107 | (9.3 ± 2.8) × 1011 | (1.7 ± 1.4) × 107 | (3.1 ± 0.2) × 1010 |
| R3 | (7.6 ± 0.0) × 108 | (4.6 ± 2.3) × 108 | (9.6 ± 3.7) × 1010 | (3.0 ± 1.2) × 107 | (5.0 ± 0.8) × 109 |
| R4 | (1.9 ± 5.4) × 108 | (1.0 × 0.2) × 109 | (1.2 ± 0.6) × 1011 | (7.9 ± 2.9) × 107 | (3.0 ± 0.9) × 109 |
| R5 | (1.1 ± 1.0) × 108 | (5.6 ± 4.1) × 108 | (4.1 ± 0.9) × 1011 | (7.8 ± 6.0) × 107 | (2.9 ± 0.9) × 1010 |
| R6 | (1.9 ± 8.7) × 108 | (9.8 ± 5.3) × 108 | (2.5 ± 0.2) × 1011 | (1.3 ± 0.9) × 108 | (2.1 ± 0.7) × 1010 |
Detected by DAPI staining.
The detected numbers of bacterial and archaeal 16S rRNA gene copies, respectively, were higher than the counted bacterial and archaeal cell densities, respectively, as determined by FISH (Table 2). Because of different 16S rRNA gene copy numbers in microbial genomes, the results of Q-PCR show only relative abundances of the microorganisms.
Table 3 shows the percent distribution of the main methanogenic groups based on results of ARDRA, Q-PCR, and FISH analyses of the six biogas reactors. The hydrogenotrophic Methanomicrobiales represented the dominant order in reactors R1, R4, and R5, determined as 60 to 64% of archaeal cell counts (FISH) and 61 to 97% of archaeal 16S rRNA gene copies (Q-PCR). In addition, the Q-PCR analyses showed a dominance of Methanomicrobiales in R2 with 99% and in R3 with 85% of archaeal 16S rRNA gene copies (Table 3).
TABLE 3.
Percentage of Methanomicrobiales, Methanobacteriales, Methanosarcinaceae, Methanosaetaceae, and unclassified Euryarchaeota calculated on basis of the total archaeal community as determined by ARDRA, Q-PCR, and FISH
| Reactor and analysis method | Distribution of the indicated methanogenic group (%)a |
||||
|---|---|---|---|---|---|
| MM | MB | Ms | Mx | NCE | |
| R1 | |||||
| ARDRA | 75 | 4 | ND | 21 | ND |
| Q-PCR | 61 | 2 | <1 | 36 | ND |
| FISH | 68 | 11 | ND | 21 | ND |
| R2 | |||||
| ARDRA | 99 | 1 | ND | ND | ND |
| Q-PCR | 99 | <1 | ND | ND | ND |
| FISH | NA | NA | NA | NA | NA |
| R3 | |||||
| ARDRA | 98 | ND | 1 | 1 | ND |
| Q-PCR | 85 | 3 | ND | 12 | ND |
| FISH | NA | NA | NA | NA | NA |
| R4 | |||||
| ARDRA | 96 | 1 | ND | ND | 3 |
| Q-PCR | 83 | 16 | <1 | ND | ND |
| FISH | 64 | 36 | ND | ND | ND |
| R5 | |||||
| ARDRA | 93 | 1 | 4 | ND | 2 |
| Q-PCR | 97 | 2 | <1 | ND | ND |
| FISH | 60 | 10 | 30 | ND | ND |
| R6 | |||||
| ARDRA | 49 | 5 | ND | 40 | 6 |
| Q-PCR | 24 | 3 | <1 | 72 | ND |
| FISH | 30 | 6 | ND | 64 | ND |
MM, Methanomicrobiales; MB, Methanobacteriales; Ms, Methanosarcinaceae; Mx, Methanosaetaceae; NCE, not classified Euryarchaeota; ND, not detected; NA, not analyzed.
The aceticlastic Methanosaetaceae were detected in reactors R1 and R6, with 21% and 64% of archaeal cell counts (FISH), respectively. Q-PCR analysis resulted in 36% (R1) and 72% (R6) Methanosaetaceae of archaeal 16S rRNA gene copies, respectively. In reactor R3 Methanosaetaceae were assigned to 12% of archaeal 16S rRNA gene copies by Q-PCR analysis. However, this archaeal group was not detected in reactors R2, R4, and R5 (Table 3).
In reactors R1, R4, R5, and R6 Methanosarcinaceae were detected with less than 1% of archaeal 16S rRNA gene copies by Q-PCR. With FISH analysis this family was detected with 30% of archaeal cell counts only in reactor R5 (Table 3).
Methanobacteriales seemed to be underrepresented by the Q-PCR analysis in contrast to FISH analysis: the Q-PCR values of reactors R1 to R6 ranged between <1 and 16% of archaeal 16S rRNA gene copies, whereas the FISH values in R1, R4, R5, and R6 ranged between 8 and 36% of archaeal cell counts (Table 3).
For the differentiated capture of methanogenic diversity, one 16S rRNA gene clone library with approximately 100 clones was constructed for each biogas reactor (Table 4). In total, 643 clones of six clone libraries were analyzed by ARDRA; 29 clones considered to be possible chimeras were excluded from subsequent analysis. By 16S rRNA gene sequencing, 35 OTUs representative of 488 clones were found to be of archaeal origin. The remaining 126 clones were of bacterial origin.
TABLE 4.
Statistical analyses of 16S rRNA gene clone libraries
| Reactor | Total no. of clones | No. of archaeal clones | No. of archaeal OTUs | Shannon index | Evenness | Chao I indexa | Coverage (%)b |
|---|---|---|---|---|---|---|---|
| R1 | 102 | 28 | 7 | 1.60 | 0.71 | 7 (1-15) | 75 |
| R2 | 108 | 99 | 11 | 0.96 | 0.24 | 39 (18-116) | 92 |
| R3 | 103 | 88 | 10 | 1.26 | 0.35 | 25 (13-78) | 93 |
| R4 | 100 | 80 | 9 | 1.43 | 0.47 | 10 (9-18) | 96 |
| R5 | 113 | 105 | 11 | 1.04 | 0.29 | 32 (16-97) | 90 |
| R6 | 117 | 88 | 13 | 1.99 | 0.56 | 16 (13-35) | 94 |
Numbers in parentheses represent the 95% confidence intervals.
Based on the formula of Good (26).
The sample sizes of reactors R2, R4, R5, and R6 were sufficient for analysis of the diversity of the major methanogenic groups, as shown by a rarefaction analysis of the six clone libraries (data not shown). In the case of R1 and R3 the rarefaction curves showed no asymptotic progression. Hence, the Chao I index was additionally calculated to estimate a sufficient sample size (Table 4). The Chao I values of R1, R4, and R6 showed only small differences from the actual OTU number of detected archaea, indicating that the sample size of the clone libraries was sufficient and confirming the results from the rarefaction analysis. In contrast, the results of the Chao I analyses of R2, R3, and R5 revealed higher OTU estimation values than actual OTU numbers. However, the lower 95% confidence intervals were near the actual sample size (Table 4), indicating that the majority of methanogens in the samples were detected. The coverage values (75 to 96%) of all clone libraries supported this assumption (Table 4).
Biogas reactors R1 and R6 showed the highest methanogenic diversity, which was determined by Shannon index values (H) of 1.60 and 1.99, respectively. The lowest diversity was found in R2 (H of 0.96). The OTUs determined by ARDRA of all biogas plants were assigned to the orders Methanomicrobiales, Methanobacteriales, and Methanosarcinales belonging to the phylum Euryarchaeota. The calculation of uncorrected p-distances (see the data at http://www.atb-potsdam.de/veroeffent/AEM_Nettmann_et_al_2010_Supplement.pdf) between nucleotide sequences determined in this study and NCBI GenBank reference sequences resulted in similarity values between 83 and 100%. In accordance with FISH and Q-PCR analyses, the majority of all detected archaeal clones in five of six biogas plants was allocated to the hydrogenotrophic order Methanomicrobiales (Table 3) and within this order to the genera Methanoculleus (97 to 100% nucleotide similarity) and Methanospirillum (95% nucleotide similarity) (see the data at the URL mentioned above). This was supported by the low evenness values (0.24 to 0.56) of the analyzed reactors, except for reactor R1 (E of 0.71), which indicated the existence of some dominant OTUs (Table 4).
In R1 (21% of archaeal clones), R3 (1% of archaeal clones), and R6 (4% of archaeal clones), the family Methanosaetaceae were detected (Table 3), which is known to be the only methanogenic group exclusively utilizing acetate for methanogenesis. The uncorrected p-distance analysis of detected OTUs and reference sequences resulted in a nucleotide similarity of 97% with known Methanosaeta species (see data at the URL mentioned above).
In addition, in reactors R3 (1% of archaeal clones) and R5 (4% of archaeal clones), OTUs were detected that were assigned to the genus Methanosarcina (see the data at the URL mentioned above). This genus utilizes acetate as well as methyl compounds and CO2 with H2 or formate as electron donors.
With 1 to 5% of archaeal clones (Table 3), the hydrogenotrophic order Methanobacteriales, represented by the genera Methanobacterium (91 to 94% nucleotide similarity) and Methanobrevibacter (94 to 95% nucleotide similarity) (see the data at the URL mentioned above), was detected in all digesters, except in R3.
Detection of hitherto uncultivated potential methanogens.
In reactors R4 and R5 OTUs were detected by ARDRA which could not be assigned to known species. The calculation of the uncorrected p-distances between the 16S rRNA gene sequence of these OTUs and known reference sequences showed only 83% similarity to DNA sequences of the CA-11 cluster (see the data at the URL mentioned above). Members of this cluster were first found in a fluidized-bed reactor fed by wine distillation waste (25).
Furthermore, the 16S rRNA gene sequence of one OTU detected in reactor R6 showed 98% nucleotide sequence similarity to a hitherto unclassified member of the ARC-I cluster (see the data at the URL mentioned above). Members of the ARC-I cluster were first found in an anaerobic slugged digester by Chouari et al. (13).
Similarities in the methanogenic archaeal community structure between the analyzed biogas plants.
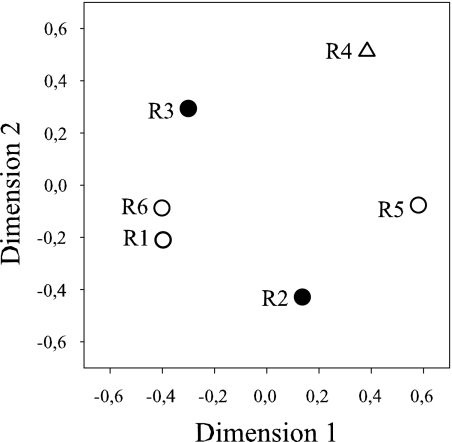
The pairwise diversity similarities based on the OTU abundance of archaeal origin in the 16S rRNA gene clone libraries were calculated by the CJ index (data not shown). The results of the CJ calculation were illustrated by multidimensional scaling (Fig. 1). The highest similarity of methanogenic diversity was found in the 16S rRNA gene clone libraries of reactors R1 and R6. These biogas plants were fed with cattle liquid manure as the main substrate. However, the methanogenic community of reactor R5, also fed with cattle liquid manure, was nonconforming with the communities of reactors R1 and R6. Also the two reactors R2 and R3, which were operated with pig slurry as a main substrate, showed a lower similarity in the structure of their methanogenic biocoenosis.
FIG. 1.
MDS of CJ similarity values for archaeal diversity determined in biogas plants R1 to R6. The following symbols represent the main substrates of the biogas plants: ○, cattle liquid manure; •, pig liquid manure; and ▵, maize silage. CJ values were calculated on the basis of OTUs determined by ARDRA. The relative distance between the points correlates with their dissimilarity.
DISCUSSION
Several studies have been published regarding the community structure of methanogenic Archaea in biogas reactors. However, the structure of microbial biocoenosis in full-scale biogas reactors supplied with renewable energy crops was not sufficiently known. Thus, to enlarge the recently acquired knowledge of methanogenic consortia in this kind of biogas plant, a polyphasic approach was applied to study methanogenic biocoenosis in six agricultural biogas plants.
Each of the biogas plants analyzed showed an individual methanogenic community structure. However, the hydrogenotrophic order Methanomicrobiales was predominant in five of the six biogas plants as determined by FISH, Q-PCR, and ARDRA. In one biogas plant only (R6) the aceticlastic family Methanosaetaceae was determined to be predominant methanogenic group (Table 3).
This finding stands in contrast to the common opinion which favors aceticlastic methanogenesis as the dominant methane generation pathway also in agricultural biogas plants. Therefore, possible influencing factors on the structure of methanogenic consortia were included in the analysis. To compare the methanogenic communities within the six biogas plants with consideration of the main substrate supplied, CJ similarity indices were computed and visualized by MDS (Fig. 1). Analysis of the MDS illustration showed no direct dependence of methanogenic diversity on the main substrate supplied. Although R1, R5, and R6 were operated with cattle liquid manure as the main substrate, methanogenic biocoenoses only of R1 and R6 showed close similarities, as illustrated in Fig. 1. In addition R2 and R3, which were supplied with pig slurry, also showed low similarity in their methanogenic consortia (Fig. 1). Reactor R4, fed only with maize silage, showed no conformity in its structure of methanogenic diversity with the remaining biogas plants (Fig. 1).
Interestingly, reactors R2 and R5 possessed the highest total ammonia nitrogen (sum of NH3-N + NH4+-N) concentrations, with 4.3 and 4.0 g liter−1, respectively (Table 1). These high contents of total ammonia nitrogen concentrations might result from the supply of nitrogen-rich poultry feces as the substrate for the fermentative process in these reactors (Table 1). However, biogas plant R4, which was supplied with maize silage as the sole substrate, also showed high values of total ammonia nitrogen (Table 1). In these three biogas reactors high temperatures between 41 and 44.6°C were measured (Table 1), which in combination with high pH values influence the dissociation equilibrium of NH4+/NH3. The calculated NH3-N concentrations were 0.37 (R4), 0.45 (R5), and 1.15 g liter−1 (R2), the highest values of the analyzed biogas reactors. This could be an explanation for the absence of Methanosaetaceae in these three biogas plants (Table 3). It is well known that the growth of Methanosaeta spp. is inhibited by high total ammonia nitrogen concentrations, whereas Methanosarcina spp. and the hydrogenotrophic methanogens also prosper at high concentrations (3, 17, 32). The critical total NH4+-N concentrations may range between 2.5 and 8.0 g liter−1, depending on the supplied substrates (48). For pure culture of Methanosaeta concilii a maximum concentration of less than 1.1 g liter−1 NH4+ (corresponding to 0.047 g liter−1 NH3) at 35°C and pH 7.6 was determined (60). In full-scale biogas reactors supplied with wastewater sludge or manure, Methanosaetaceae were detected just at total ammonia concentrations below 1.5 g liter−1, whereas in reactors with total ammonia concentrations between 2.1 and 4.1 g liter−1, Methanosarcinaceae and not Methanosaetaceae were found (32).
Currently, the nature of the influence from ammonia and ammonium, on the anaerobic-digestion microbial community has not bee examined adequately. In the literature a variety of toxicity mechanisms and the possibility of adaptation to high total ammonia concentrations have been discussed (3, 22, 30, 36). Therefore, the effect of high ammonium or ammonia concentrations on methanogenic Archaea in full-scale biogas reactors requires further investigation.
Besides total ammonia, acetate is another factor limiting the growth of aceticlastic methanogens: Methanosaeta spp. have a lower acetate threshold and, thus, a lower growth rate at high acetate concentrations than Methanosarcina spp. (16, 33, 59). Karakashev and coworkers (32) showed that not only acetate but also volatile fatty acids (VFA) have a strong influence on the methanogenic consortia in biogas plants. In this study, a correlation between high VFA (HAc eq.) concentration and the presence of Methanosaetaceae was not observed (Fig. 2b). In R4 no aceticlastic Archaea were detected although this reactor showed comparatively low VFA concentrations. Hence, high ammonium concentrations might have a higher impact than high VFA concentrations on the aceticlastic Archaea (52).
FIG. 2.
Influence of total ammonia nitrogen (NH4+-N +NH3-N) and calculated ammonia nitrogen (NH3-N) (A) and volatile fatty acids (acidic acid equivalents) (B) on methanogenic community composition in biogas reactors R1 to R6. White bars, only hydrogenotrophic methanogens were detected; gray bars, hydrogenotrophic methanogens dominated aceticlastic methanogens; black bars, aceticlastic methanogens dominated hydrogenotrophic methanogens. In panel A, plain bars represent data of total ammonia nitrogen, and striped bars represent data of calculated NH3-N.
The underrepresentation of aceticlastic methanogens in five of six analyzed biogas reactors supports the conclusion that aceticlastic methanogenesis plays a minor role in the biomethanation process. The fact that aceticlastic methanogens have a lower rate of substrate conversion (8.4 g CODs g CODBM−1 day−1; COD is chemical oxygen demand, S is substrate, and BM is biomass) than hydrogenotrophic methanogens (37.0 g CODs g CODBM−1 day−1) fortify this thesis (6). Moreover, hydrogenotrophic methanogens posses higher growth rates, up to 2.0 day−1 than aceticlastic methanogens with 0.4 day−1 (6).
This assumption raises the question, which microorganisms utilize the acetate derived from bacterial hydrolysis of organic compounds if aceticlastic methanogens are not present. Under these conditions potential candidates for acetate degradation to CO2/H2 are the syntrophic acetate oxidizers (27). As this process is thermodynamically extremely unfavorable (ΔG0′, 104.6 kJ mol−1) the acetate oxidizers need partner organisms such as hydrogenotrophic methanogens (39, 53). Up to now, only a few microbial species which are able to degrade acetate to H2 and CO2 in syntrophy with hydrogenotrophic methanogens have been identified: an acetate-oxidizing, rod-shaped (AOR) bacterium (39), Clostridium ultunense strain B (53), Thermacetogenium phaeum strain PB (31), Thermotoga lettingae strain TMO (5), and Acetobacterium woodii (63). Other bacteria also present in biogas reactors (37) oxidize acetate by parallel reduction of sulfur. Examples are members of the orders Desulfovibrionales, Desulfobacterales, and Desulfomonadales. Earlier studies following acetate utilization processes by 14C isotope-labeled acetate revealed that in the absence of aceticlastic methanogens due to high ammonium concentrations, nonmethanogenic acetate oxidation took place (33, 54). Accordingly, in biogas plants with nonmethanogenic acetate oxidation, the hydrogenotrophic genus Methanoculleus was observed to be the predominant group of methanogens (54). Members of this genus were also detected to be most prevalent in several lab-scale and full-scale biogas reactors (35, 37, 45).
In this study, members the genus Methanoculleus (order Methanomicrobiales) were also found to be the predominant methanogen as determined by ARDRA (see data at http://www.atb-potsdam.de/veroeffent/AEM_Nettmann_et_al_2010_Supplement.pdf). This might point to the occurrence of nonmethanogenic acetate oxidation by syntrophic acetate oxidizers in the analyzed biogas plants. However, to solve this question, further investigations of the syntrophic relationships within microbial communities, their influence on the carbon fluxes, and, finally, their influence on the efficiency of the biogas formation process in full-scale biogas reactors are required.
The diversity analysis by ARDRA also resulted in the detection of members of the hitherto unclassified Euryarchaeota ARC-I cluster (R6). Additionally, putative new species detected by ARDRA showed closest nucleotide similarity (83%) to members of the CA-11 cluster (R4 and R5). The CA-11 cluster was first determined by Godon et al. (25) in a reactor supplied with wine distillation waste. Members of the ARC-I and CA-11 clusters were also found in lab-scale and full-scale biogas reactors supplied with triticale (35), in sludge samples of a mesophilic wastewater reactor, and in a reactor supplied with wine waste (25). The role of these putative methanogen groups within the microbial consortia and their effect on the biogas formation process in biogas plants remain unclear and require further investigations.
This study provides new insights into the methanogenic community structure present within biogas plants supplied with energy crops and liquid manure. The results imply that hydrogenotrophic methanogenesis is favored for methane formation. However, this study is a snapshot of the microbial community at one moment in time. Further studies will be needed to monitor the microbial population dynamics during ongoing biogas fermentation.
Acknowledgments
E. Nettmann and I. Bergmann were supported by research grants from the German Federal Ministry of Food, Agriculture and Consumer Protection (BMELV)/Agency of Renewable Resources (FNR) (grant 22011804) and the Federal Ministry of Education and Research (BMBF)/Project Management Jülich (PTJ) (grant 03SF0317M).
We are very thankful to W. B. Herppich for critical reading of the manuscript. Moreover, we gratefully acknowledge most valuable discussions with U. Szewzyk and E. Grohmann, their advice on FISH experiments, and their critical reading of the manuscript.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Ahring, B. K. 2003. Perspectives for anaerobic digestion. Adv. Biochem. Eng. Biotechnol. 81:1-30. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chrisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angelidaki, I., and B. K. Ahring. 1993. Thermophilic anaerobic digestion of livestock waste: the effect of ammonia. Appl. Microbiol. Biotechnol. 38:560-564. [DOI] [PubMed] [Google Scholar]
- 4.Anthonisen, A. C., R. C. Loehr, T. B. S. Prakasam, and E. G. Srinath. 1976. Inhibition of nitrification by ammonia and nitrous acid. J. Water Pollut. Control Fed. 48:835-852. [PubMed] [Google Scholar]
- 5.Balk, M., J. Weijma, and A. J. M. Stams. 2002. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilick anaerobic reactor. Int. J. Syst. Evol. Microbiol. 52:1361-1368. [DOI] [PubMed] [Google Scholar]
- 6.Batstone, D. J., J. Keller, I. Angelidaki, S. V. Kalyuzhnyi, S. G. Pavlostathis, A. Rozzi, W. T. M. Sanders, H. Siegrist, and V. A. Vavilin. 2002. Anaerobic digestion model no. 1, p.1-77. IWA scientific and technical report no. 13. IWA Publishing, London, United Kingdom. [PubMed]
- 7.Bauer, C., M. Korthals, A. Gronauer, and M. Lebuhn. 2008. Methanogens in biogas production from renewable resources—a novel molecular population analysis approach. Water Sci. Technol. 58:1433-1439. [DOI] [PubMed] [Google Scholar]
- 8.Böckelmann, U., U. Szewzyk, and E. Grohmann. 2003. A new enzymatic method for the detachment of particle associated soil bacteria. J. Microbiol. Methods 55:201-211. [DOI] [PubMed] [Google Scholar]
- 9.Bundesforschungsanstalt für Landwirtschaft (FAL). 2005. Ergebnisse des Biogas-Messprogramms. Fachagentur für Nachwachsende Rohstoffe e. V. (FNR), Gülzow, Germany.
- 10.Chan, O. C., P. Claus, P. Casper, A. Ulrich, T. Lueders, and R. Conrad. 2005. Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment. Environ. Microbiol. 7:1139-1149. [DOI] [PubMed] [Google Scholar]
- 11.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 12.Chao, A., R. L. Chazdon, R. K. Colwell, and T. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159. [Google Scholar]
- 13.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, G., S. Kavanagh, S. McHugh, S. Connaughton, A. Kearney, O. Rice, C. Carrigg, C. Scully, N. Bhreathnach, T. Mahony, P. Madden, A. Enright, and V. O'Flaherty. 2006. Accessing the black box of microbial diversity and ecophysiology: recent advances through polyphasic experiments. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 41:897-922. [DOI] [PubMed] [Google Scholar]
- 16.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 17.Conrad, R., and M. Klose. 2006. Dynamics of the methanogenic archaeal community in anoxic rice soil upon addition of straw. Eur. J. Soil Sci. 57:476-484. [Google Scholar]
- 18.Crocetti, G., M. Murto, and L. Bjornsson. 2006. An update and optimisation of oligonucleotide probes targeting methanogenic Archaea for use in fluorescence in situ hybridisation (FISH). J. Microbiol. Methods 65:194-201. [DOI] [PubMed] [Google Scholar]
- 19.Daims, H., K. Stoecker, and Wagner, M. 2005. Fluorescence in situ hybridisation for the detection of prokaryotes. p. 213-239. In A. M. Osborn and C. J. Smith (ed.), Advanced methods in molecular microbial ecology. Bios-Garland, Abingdon, United Kingdom.
- 20.Denhardt, D. T. 1966. A membran-filter technique for the detection of complementary DNA. Biochem. Biophys. Res. Commun. 23:641-646. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez, A. S., S. Huang, S. Seston, J. Xing, R. F. Hickey, C. S. Criddle, and J. M. Tiedje. 1999. How stable is stable? Function versus community composition. Appl. Environ. Microbiol. 65:3697-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallert, C., and J. Winter. 1997. Mesophilic and thermophilic anaerobic digestion of source-sorted organic wastes: effect of ammonia on glucose degradation and methane production. Appl. Microbiol. Biotechnol. 48:405-410. [Google Scholar]
- 23.Garrity, G. M., and J. G. Holt. 2001. Phylum AII. Euryarchaeota, p. 211-345. In D. R. Boone, R. W. Castenholz, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, NY. [Google Scholar]
- 24.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 25.Godon, J.-J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 27.Hattori, S. 2008. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 23:118-127. [DOI] [PubMed] [Google Scholar]
- 28.Hofman-Bang, J., D. Zhang, P. Westermann, B. K. Ahring, and L. Raskin. 2003. Molecular ecology of anaerobic reactor systems. Adv. Biochem. Eng. Biotechnol. 81:151-203. [DOI] [PubMed] [Google Scholar]
- 29.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadam, P. C., and D. R. Boone. 1996. Influence of pH on ammonia accumulation and toxicity in halophilic, methylotrophic methanogens. Appl. Environ. Microbiol. 62:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamagata, Y., and E. Mikami. 1989. Diversity of acetotrophic methanogens in anaerobic digestion. p. 459-464. In T. Hattori, Y. Ishida, Y. Maruyana, R. Y. Morita, and A. Uccida (ed.), Recent advances in microbial ecology. Japan Scientific Societies Press, Tokyo, Japan.
- 32.Karakashev, D., D. J. Batstone, and I. Angelidaki. 2005. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microbiol. 71:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karakashev, D., D. J. Batstone, E. Trably, and I. Angelidaki. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 72:5138-5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 35.Klocke, M., E. Nettmann, I. Bergmann, K. Mundt, K. Souidi, J. Mumme, and B. Linke. 2008. Characterization of the methanogenic Archaea within two-phase biogas reactor systems operated with plant biomass. Syst. Appl. Microbiol. 31:190-205. [DOI] [PubMed] [Google Scholar]
- 36.Koster, I. W. 1986. Characteristics of the pH-influenced adaptation of methanogenic sludge to ammonium toxicity. J. Chem. Technol. Biotechnol. 36:445-455. [Google Scholar]
- 37.Krause, L., N. N. Diaz, R. A. Edwards, K.-H. Gartemann, H. Krömeke, H. Neuweger, A. Pühler, K. J. Runte, A. Schlüter, J. Stoye, R. Szczepanowski, A. Tauch, A. Goesmann, N. N. Diaz, and R. A. Edwards. 2008. Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 136:91-101. [DOI] [PubMed] [Google Scholar]
- 38.Leclerc, M., J.-P. Delgenes, and J.-J. Godon. 2004. Diversity of the archaeal community in 44 anaerobic digesters as determined by single strand conformation polymorphism analysis and 16S rDNA sequencing. Environ. Microbiol. 6:809-819. [DOI] [PubMed] [Google Scholar]
- 39.Lee, M. J., and S. H. Zinder. 1988. Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2/CO2. Appl. Environ. Microbiol. 54:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovley, D. R., and M. J. Klug. 1983. Methanogenesis from methanol and methylamines and acetogenesis from hydrogen and carbon dioxide in the sediments of a eutrophic lake. Appl. Environ. Microbiol. 45:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotides probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackie, R. I., and M. P. Bryant. 1981. Metabolic activity of fatty acid-oxidizing bacteria and the contribution of acetate, propionate, butyrate, and CO2 to methanogenesis in cattle waste at 40 and 60°C. Appl. Environ. Microbiol. 41:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McHugh, S., M. Carton, T. Mahony, and V. O'Flaherty. 2003. Methanogenic population structure in a variety of anaerobic bioreactors. FEMS Microbiol. Lett. 219:297-304. [DOI] [PubMed] [Google Scholar]
- 44.Moreira, D. 1998. Efficient removal of PCR inhibitors using agarose-embedded DNA preparations. Nucleic Acids Res. 26:3309-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nettmann, E., I. Bergmann, K. Mundt, B. Linke, and M. Klocke. 2008. Archaea diversity within a commercial biogas plant utilizing herbal biomass determined by 16S rDNA and mcrA analysis. J. Appl. Microbiol. 105:1835-1850. [DOI] [PubMed] [Google Scholar]
- 46.Oude Elferink, S. J. W. H., R. van Lis, H. G. H. J. Heilig, A. D. L. Akkermans, and A. J. M. Stams. 1998. Detection and quantification of microorganisms in anaerobic bioreactors. Biodegradation 9:169-177. [DOI] [PubMed] [Google Scholar]
- 47.Pielou, E. C. 1966. The measurement of diversity in different types of biological colledions. J. Theor. Biol. 13:131-144. [Google Scholar]
- 48.Poggi-Varaldo, H. M., R. Rodriguez-Vázquez, G. Fernánez-Villagómez, and F. Esparza-Gracia. 1997. Inhibition of mesophilic solid-substrate anaerobic digestion by ammonia nitrogen. Appl. Microbiol. Biotechnol. 47:284-291. [Google Scholar]
- 49.Raizada, N. 2004. Application of molecular-biological method for optimization of anaerobic reactors. In Berichte aus Wassergüte- und Abfallwirtschaft. Technische Universität München, Garching, Germany.
- 50.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. Stahl. 1994. Group-specific 16S ribosomal-RNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Schnürer, A., and A. Nordberg. 2008. Ammonia, a selective agent for methane production by syntrophic acetate oxidation at mesophilic temperature. Water Sci. Technol. 57:735-740. [DOI] [PubMed] [Google Scholar]
- 53.Schnürer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 54.Schnürer, A., G. Zellner, and B. H. Svensson. 1999. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 29:249-261. [Google Scholar]
- 55.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana, IL.
- 56.Solera, R., L. I. Romero, and D. Sales. 2001. Analysis of the methane production in thermophilic anaerobic reactors: use of autofluorescence microscopy. Biotechnol. Lett. 23:1889-1892. [Google Scholar]
- 57.Souidi, K., J. Mumme, K. Mundt, E. Nettmann, I. Bergmann, B. Linke, and M. Klocke. 2007. Microbial diversity in a biogas-producing co-fermentation of maize silage and bovine manure. Agrartechnische Forsch. 13:197-205. [Google Scholar]
- 58.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics, John Wiley & Sons Ltd., Chichester, England.
- 59.Stams, A. J., S. J. Oude Elferink, and P. Westermann. 2003. Metabolic interactions between methanogenic consortia and anaerobic respiring bacteria. Adv. Biochem. Eng. Biotechnol. 81:31-56. [DOI] [PubMed] [Google Scholar]
- 60.Steinhaus, B., M. L. Garcia, A. Q. Shen, and L. T. Angenent. 2007. A portable anaerobic microbioreactor reveals optimum growth conditions for the methanogen Methanosaeta concilii. Appl. Environ. Microbiol. 73:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular evolutionary genetics analysis (Mega) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter, J. U., and R. S. Wolfe. 1980. Methane formation from fructose by syntrophic associations of Acetobacterium woodii and different strains of methanogens. Arch. Microbiol. 124:73-79. [DOI] [PubMed] [Google Scholar]
- 64.Yu, Y., J. Kim, and S. Hwang. 2006. Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: population dynamics and community structures. Biotechnol. Bioeng. 93:424-433. [DOI] [PubMed] [Google Scholar]
- 65.Yu, Y., C. Lee, J. Kim, and S. Hwang. 2005. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89:670-679. [DOI] [PubMed] [Google Scholar]