Abstract
Thiopeptide antibiotics are an important class of natural products resulting from posttranslational modifications of ribosomally synthesized peptides. Cyclothiazomycin is a typical thiopeptide antibiotic that has a unique bridged macrocyclic structure derived from an 18-amino-acid structural peptide. Here we reported cloning, sequencing, and heterologous expression of the cyclothiazomycin biosynthetic gene cluster from Streptomyces hygroscopicus 10-22. Remarkably, successful heterologous expression of a 22.7-kb gene cluster in Streptomyces lividans 1326 suggested that there is a minimum set of 15 open reading frames that includes all of the functional genes required for cyclothiazomycin production. Six genes of these genes, cltBCDEFG flanking the structural gene cltA, were predicted to encode the enzymes required for the main framework of cyclothiazomycin, and two enzymes encoded by a putative operon, cltMN, were hypothesized to participate in the tailoring step to generate the tertiary thioether, leading to the final cyclization of the bridged macrocyclic structure. This rigorous bioinformatics analysis based on heterologous expression of cyclothiazomycin resulted in an ideal biosynthetic model for us to understand the biosynthesis of thiopeptides.
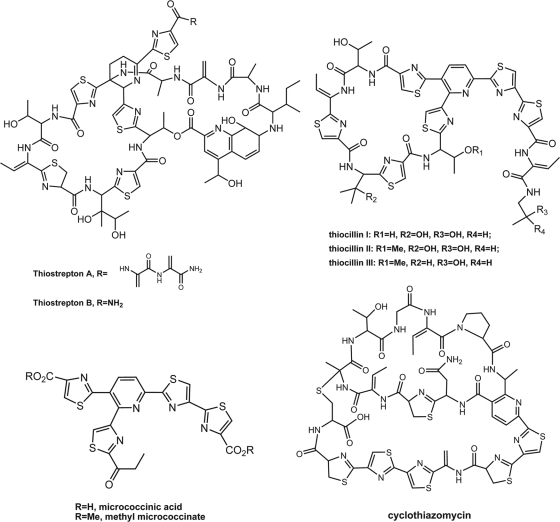
The thiopeptides are a family of highly modified, sulfur-containing macrocyclic peptides, such as thiostrepton, thiocillins, and micrococcinic acid (3, 11). Their structures have several common features: a tri- or tetrasubstituted nitrogen heterocycle central domain, a macrocyclic framework, and heavily modified amino acid residues, including thiazoles, oxazoles, and dehydroamino acids (Fig. 1) (3). Members of this family exhibit various biological properties, such as inhibition of ribosomal protein synthesis (24), rennin inhibitory activity (2), and induction of TipA (20). Moreover, many thiopeptide antibiotics show bioactivity against some bacterial strains resistant to most conventional treatments, including methicillin-resistant Staphylococcus aureus (MRSA), penicillin-resistant Streptococcus pneumoniae (PRSP), and vancomycin-resistant enterococci (VRE) (3, 17).
FIG. 1.
Structures of thiostrepton, thiocillin, microncoccinate, and cyclothiazomycin (3).
Early in vitro investigations of these special heterocycles of thiopeptides were performed by organic chemists and included stereoselective synthesis of a γ-lactam acidic hydrolysate of cyclothiazomycin (4) and total synthesis of thiostrepton (21). Extensive research on microccins (5, 19) and lantibiotics (6, 29) described biosynthesis of the thiazole- and dehydroamino acid-containing polypeptides that were derived from ribosomally synthesized prepeptides. Similarly, Lee et al. described a widely conserved gene cluster for toxin biosynthesis and suggested a ribosome biosynthetic pathway for the modified polypeptide containing thiazoles and oxazoles (16).
Recently, Wieland Brown et al. (28) and Kelly et al. (14) identified the gene clusters encoding thiocillin and thiostrepton, respectively, with a probe that targeted hypothetic prepeptide genes, whereas Liao and coworkers (17) took advantage of the conservation of one putative cyclodehydratase. Although most of the biochemical reactions involved in the biosynthetic pathway remain obscure, it is clear that thiopeptides are synthesized ribosomally and then there is a series of posttranslational modifications.
Cyclothiazomycin, which was isolated as a novel selective inhibitor of human plasma rennin, is a unique bridged macrocyclic thiopeptide (2) whose stereo structure was recently revealed by degradation experiments and spectroscopic analysis (10). It contains a dehydroserine, two dehydrothreonine residues, three thiazolines, three thiazoles, and a trisubstituted pyridine. Compared with common thiopeptides, it lacks the characteristic 2- and 3-azole substituent on the central pyridine domain; instead, it has an alanine-derived heterocyclic residue with the (R) configuration, a quaternary sulfide, and two macrocyclic peptide loops (Fig. 1). Moreover, a pair of convertible isomers of cyclothiazomycin B, the cyclothiazomycin analogues produced by Streptomyces sp. strain A307 (10), were identified as Z and E configurations caused by the tautomerization of the dehydrated threonine. These structural traits may indicate that some new genetic elements are likely involved in posttranslational modification.
Here we reported cloning and sequencing of the cyclothiazomycin biosynthetic gene cluster of Streptomyces hygroscopicus 10-22 (23). In addition, an analysis of heterologous expression in Streptomyces lividans 1326 and a deletion analysis were also performed, which indicated that a gene cluster at least 22.7 kb long is required for biosynthesis. A bioinformatics-based approach to analysis of this gene cluster postulated that there is a posttranslational modification pathway in which eight proteins are involved in the biosynthetic machinery.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Strains and plasmids used in this work are listed in Table 1. S. hygroscopicus 10-22 and S. lividans 1326, as well as derivatives of the latter strain, were grown at 30°C on SFM agar (20 g soy flour, 20 g mannitol, and 20 g agar in 1 liter tap water) plates for sporulation or in tryptic soy broth (TSB) that was supplemented with sucrose (10.3%, wt/vol) and yeast extract (1%, wt/vol) for growth of mycelia. Isolation of total DNA and manipulation of all Escherichia coli strains were performed as described by Sambrook et al. (26). For selection of Streptomyces transformants, the concentrations of apramycin, kanamycin, and thiostrepton that were used in agar were 30 mg ml−1, 10 mg ml−1, and 15 mg ml−1, respectively, and the concentrations that were used in liquid medium were 20 mg ml−1, 5 mg ml−1, and 5 mg ml−1, respectively. Heterologous expression mutations in S. lividans 1326 were obtained by interspecies conjugation by using E. coli ET12567::pUZ8002 as the donor and S. lividans 1326 as the recipient (Table 1) as described by Kieser et al. (15). Unless specified otherwise, restriction enzymes, T4 DNA ligase, Taq polymerase, and alkaline phosphatase were purchased from MBI Fermentas (Vilnius, Lithuania).
TABLE 1.
Bacterial strains, plasmids, and cosmids
| Strain, plasmid, or cosmid | Relevant propertiesa | Reference or source |
|---|---|---|
| Streptomyces hygroscopicus 10-22 | Producer of cyclothiazomycin | This study |
| Streptomyces lividans 1326 | Heterologous strain for cyclothiazomycin production | 8 |
| Botrytis cinerea Persoon | Plant-pathogenic fungus that is sensitive to cyclothiazomycin | 10 |
| Escherichia coli DH10B | F−recA lacZΔM15 | Gibco BRL |
| Escherichia coli ET12567/pUZ8002 | Strain used for conjugation between E. coli and Streptomyces spp.; recF dam dcm hsdS Cmlr Strr Tetr Kmr | 22 |
| Escherichia coli EPI300-T1R | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 lacX74 recA1 endA1 araD139 (ara-leu)7697 galU galK rpsL nupG trfA tonA dhfr | Epicentre Biotechnologies |
| Plasmids and cosmids | ||
| pHZ1358 | sti+oriT ori (pIJ101) bla tsr, 10.65 kb | 27 |
| Supercos1 | Ampr Kmr (aac) | Stratagene |
| 14E6 | pHZ1358-derived cosmid with insert from S. hygroscopicus 10-22 | This study |
| pJTU4891 | 5.7-kb XbaI-EcoRI fragment from pSET152 ligated with 26.6-kb XbaI-EcoRI fragment from 14E6 | This study |
| pJTU4892 | pSET152-derived plasmid containing 22.7-kb XbaI-NdeI fragment from 14E6 | This study |
| pJTU4893 | pSET152-derived plasmid containing 15.5-kb BclI-BclI fragment from 14E6 | This study |
| pJTU4894 | cltIMN in pJTU4891 replaced by aac by using the PCR targeting method | This study |
clt, genes for cyclothiazomycin production; oriT, origin of transfer of plasmid RK2; tsr, thiostrepton resistance gene; aac, kanamycin resistance gene; sti, origin for second-strand synthesis of multicopy plasmid pIJ101.
Production and analysis of cyclothiazomycin.
S. hygroscopicus 10-22 was grown at 30°C for 5 to 7 days on SFM agar plates for production of cyclothiazomycin. For a quick bioassay of heterologous production of cyclothiazomycin in an S. lividans 1326 mutant, agar patches were transferred to PDA agar (200 g diced potatoes, 20 g glucose, and 15 g agar in 1 liter tap water) that contained Botrytis cinerea Persoon (Table 1). Inhibition zones were observed after 48 h of incubation at 28°C. For liquid chromatography (LC)-mass spectrometry (MS) analysis, strains were cultured on SFM agar at 30°C for 7 days. The fermentation agar was harvested and extracted by using the same volume of methanol. After overnight soaking, the resulting suspension was dried with solid Na2SO4, filtered, and syringe filtered before injection. LC-electrospray (ES)-quadrupole time of flight (QTOF) analysis was performed with an Agilent 1200 system (Agilent Co., CA) using a 2.1- by 30-mm C18 reverse-phase column (Agilent Co., CA) at a flow rate of 0.3 ml/min. Solvent A was 10% aqueous acetonitrile with 0.1% HCOOH, and solvent B was 0.1% HCOOH in acetonitrile. The gradient used was a gradient from 0 to 90% solvent B over 10 min. Mass spectra were obtained with a 6530 accurate-mass QTOF mass spectrometer (Agilent Co., CA) equipped with an electrospray ionization source. Tandem MS (MS/MS) was performed in the target-dependent mode.
Construction of S. hygroscopicus 10-22 genomic library.
For generation of a cosmid library, high-molecular-weight chromosomal DNA was prepared using the protocol described by Kieser et al., partially digested with MboI, dephosphorylated with calf intestinal alkaline phosphatase (Fermentas), and size fractionated in low-melting-temperature agarose by using pulsed-field gel electrophoresis (15). The chromosome DNA fragments that were between 35 and 45 kb long were recovered from the low-melting-temperature agarose gel digested with β-agarase (New England Biolabs). The DNA recovered was ligated with BamHI-digested cosmid vector pHZ1358 (Table 1) at a 1:1 molar ratio. Packaging and transfection into E. coli EPI300-T1 (Table 1) were done with λ-packaging mixtures prepared as described by Sambrook et al. (26). Ampicillin (Amp)-resistant colonies were picked individually and inoculated into LB (100 μl) supplemented with Amp in each well of 22 96-well plates. The plates were incubated at 37°C for 18 h, and then 50% glycerol (100 μl) was added to each well before storage at −80°C.
PCR primers and cosmid library screening.
Degenerate PCR primers designed using homologs of putative cyclodehydratases (17) were used to amplify a cyclodehydratase gene fragment from S. hygroscopicus 10-22. PCR products were purified from agarose gels (0.8%) by using a DNA gel extraction kit (V-gene Biotechnology Ltd.) and were subsequently inserted into the pMD18-T vector (TaKaRa, Dalian, China) for sequencing. Based on the fragment sequence, one pair of specific primers (CYCP primers) (Table 2) were designed and used to screen the genomic cosmid library of S. hygroscopicus 10-22. Each of the 22 96-well plates was screened by combining equal aliquots of cells from every well and using the resulting mixtures as templates for the first batch of PCR. For plates that yielded the desired PCR products, aliquots of cells from all wells in a row were combined and screened by a second PCR. Finally, each well in the positive rows was separately screened to identify individual clones that carried the target gene.
TABLE 2.
Primers used in this study
| Primer typea | Sequence |
|
|---|---|---|
| Forward | Reverse | |
| aac-T | TATTCCAGAAGTAGTGAGG | CTGGATGCCGACGGATTTG |
| CYCP | CGATGAGGTCGTCCCGCAGAT | CGCACTTCGATCCCGAACAGG |
| KNde-T | TGAGTTCGTTTACGGCGGCAC | TCTGCAAGTACATCTCCGCGTCA |
| KT-IMN | GGGCGTACAGCACAGGACGGAGGCACTCGAT | GGCCAGGCCCGTCATACCTCGACTGCGGCCT |
| TGACCACATTATTCCAGAAGTAGTGAGG | GCTCCTCACTGGATGCCGACGGATTTG | |
| KT-IMN-T | TACAGCACAGGACGGAGGCACT | TCATACCTCGACTGCGGCCTG |
aac-T, primers used to test insertion of kanamycin resistance gene; KNde-T, primers used to confirm successful construction of pJTU4892; KT-IMN, primers used to construct pJTU4894; KT-IMN-T, primers used to test the correctness of pJTU4894.
Construction of shuttle plasmids for heterologous production of cyclothiazomycin.
Shuttle plasmids pJTU4891 to pJTU4893 were constructed from restricted enzyme-digested cosmid 14E6 and pSET152 (Table 1), and pJTU4894 was constructed based on pJTU4891 using the PCR-targeting method (9) with the KT-IMN primers (Table 2). To confirm successful construction of the plasmids, restriction enzyme digestion was performed, and two pairs of primers (Table 2) were tested.
Sequence analysis.
DNA sequencing was done at Shanghai Ding'an Ltd. (Shanghai, China), and a multiple-sequence alignment was constructed with BioEdit 7.0. Open reading frames (ORFs) and ribosome binding sites (RBS) were predicted by using Frame-Plot 4.0 (12). Comparisons of nucleotide or amino acid sequences with sequences in public databases were performed by using the BLAST program and the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (1).
Nucleotide and protein sequence accession number.
The DNA and deduced protein sequences described in this paper have been deposited in the GenBank database under accession number FJ472825.
RESULTS AND DISCUSSION
Cloning of the putative cyclothiazomycin biosynthetic gene cluster from S. hygroscopicus 10-22.
S. hygroscopicus 10-22, which was isolated by Zhou et al. in the 1980s, can produce three antifungal antibiotics, 5102-I, 5102-II, and 5102-III (23). One of these antibiotics, antibiotic 5102-I, has been identified and designated validamycin (13). Antibiotic 5102-II is a polypeptide containing Thr, Ser, and Cys and can protect corn plants against leaf spot (a disease caused by Cochliobolus heterostrophus). However, the lack of detailed structural information and an unsuccessful cloning strategy based on a nonribosomal peptide hypothesis have hampered cloning of the 5102-II biosynthesis gene cluster for years.
Previous failures raised the possibility that the polypeptide antibiotic 5102-II is synthesized by a ribosomal pathway, followed by posttranslational modification. Alignment of putative docking proteins encoded by the biosynthetic gene clusters of MccB17 (18) and streptolysin S, a ribosomally synthesized toxin secreted from the human pathogen Streptococcus pyogenes (16), suggested that there is a common domain belonging to the Ycao family that occurs in most putative toxin gene clusters (16). Accordingly, a pair of degenerate primers, ThioF and ThioR (17), was designed based on the common domain. Subsequently, a 700-bp fragment was obtained by performing PCR with the genomic DNA of S. hygroscopicus 10-22 as the template, and its sequence was found to be a segment of the YcaO family domain by BlastP analysis. Based on this nucleotide sequence, a pair of specific nested primers (CYCP primers [Table 2]) was designed to screen the S. hygroscopicus 10-22 genomic library constructed in pHZ1358. One of the positive cosmids, 14E6 with a 35-kb insertion, was sequenced (accession number FJ472825), and 27 ORFs were predicted in its sequence.
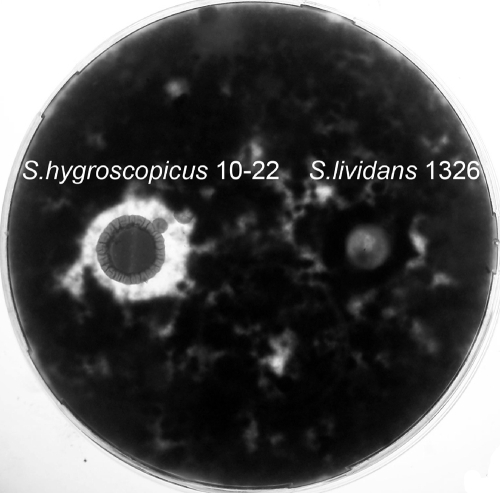
Significantly, a 180-nucleotide ORF, designated cltA, in the sequence was found to encode a 60-amino-acid precursor peptide (Fig. 2A) whose C-terminal sequence (SNCTSTGTPASCCSCCCC) from S43 to C60 is perfectly consistent with the structural peptide of cyclothiazomycin (Fig. 3C, from S1 to C18) (10), and the N-terminal 42-amino-acid sequence from M1 to A41 is likely to be the leader peptide (LP) (Fig. 4, compound 7). This putative cleavage site in the precursor is very similar to the cleavage sites in thiocillins (28) and two other putative thiopeptides, Sare_2569 and SGR_4418 (17), but very different from the cleavage site of another ribosomal synthesized polypeptide, where Gly is usually considered the initial residue recognized by some peptidase (16). In addition, the organization of all of the ORFs in this sequence (Fig. 2A) is also similar to that of thiostrepton (14, 17), thiocillin (28), and other putative ribosomally produced polypeptide gene clusters (16). Moreover, cyclothiazomycin was reported to be active against some important plant-pathogenic fungi (2, 10), including Cochliobolus miyabeanus, which belongs to the same genus as C. heterostrophus, and Botrytis cinerea Persoon, which was used as the indicator in a bioassay of the production by S. hygroscopicus 10-22 in this study (Fig. 5). These findings suggested that the polypeptide antibiotic 5102-II is likely to be cyclothiazomycin, and they indicated a successful strategy for cloning the cyclothiazomycin biosynthetic gene cluster.
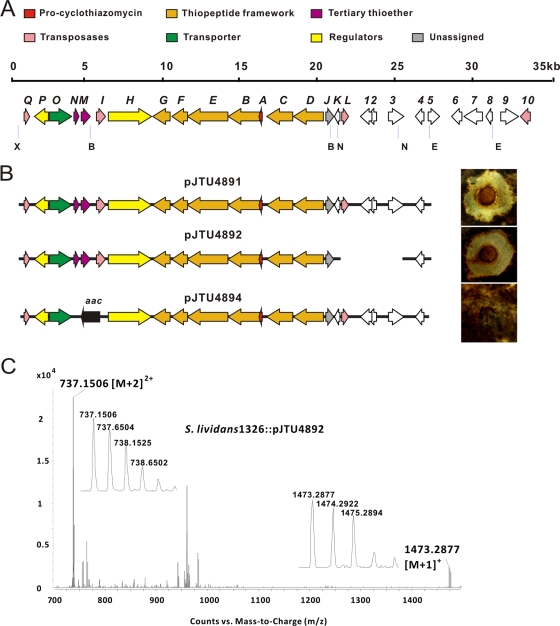
FIG. 2.
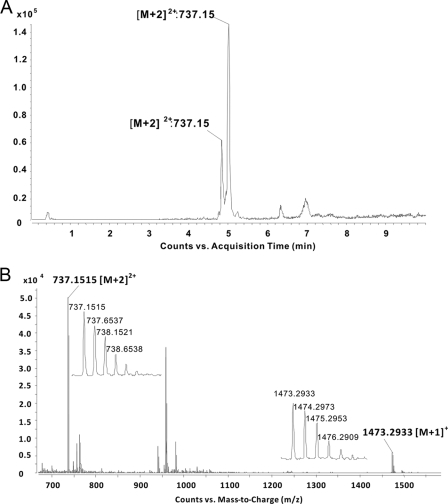
Heterologous expression of the cyclothiazomycin biosynthesis gene cluster cloned from S. hygroscopicus 10-22. (A) Organization of the ORFs in the gene cluster. The putative functions of the ORFs are indicated and are summarized in Table 3. The restriction enzymes used for gene deletion are indicated (B, BclI; E, EcoRI; X, XbaI; N, NdeI). (B) Heterologous expression profile of cyclothiazomycin in S. lividans 1326. The black bars represent the genomic fragments in the plasmids (see Table 1) corresponding to the regions in the gene cluster shown in panel A. The inhibition zones for the plasmids shown on the right are inhibition zones on the same bioassay agar plate (see Materials and Methods). (C) MS spectrum of the cyclothiazomycin produced by S. lividans 1326 harboring pJTU4892. Two characteristic peaks at m/z 737.1506 ([M+2]2+) and 1473.2877 ([M+1]+) with corresponding amplified isotopic peaks were extracted from the LC-ES-QTOF profile for heterologous production.
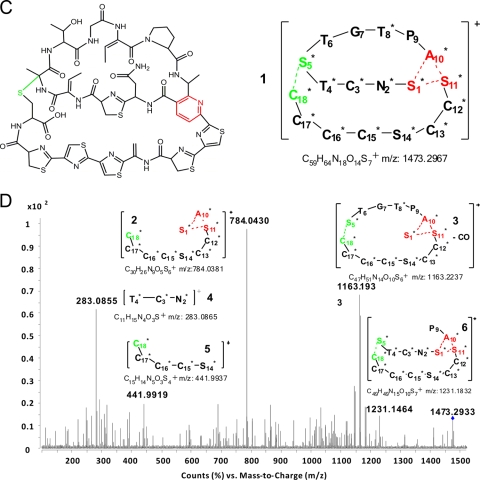
FIG. 3.
LC-ES-QTOF and MS/MS analysis of methanol extract of an S. hygroscopicus 10-22 culture. (A) LC-ES-QTOF analysis of methanol extract from an S. hygroscopicus 10-22 culture. Two distinct peaks were extracted with m/z 737.1515 ± 5 from total ion current chromatography, indicating the two isomers of cyclothiazomycin. (B) Accurate-mass QTOF spectrum of the active product obtained from S. hygroscopicus 10-22. The two signals at m/z 1473.2933 and 737.1515, corresponding to [M+1]+ and [M+2]2+, corresponded well to the theoretical value for cyclothiazomycin (m/z 1473.2967) within the error of the instrument (<2 ppm). The relative isotope abundances in the two isotopic graphs could be calculated, which resulted in measured average abundances of 88 and 70 for [M+2]+ and [M+3]+, respectively. (C) Cyclothiazomycin structure (left panel) and skeleton of the [M+1]+ ion indicated by its structural peptide (right panel). The dashed lines indicate the two cyclization sites: the substituted pyridine formed by S1*, A10*, and S11* (red) and the tertiary thioether formed by S5* and C18* (green). Asterisks indicate the amino acid residues that are posttranslationally modified. (D) MS/MS spectrum of cyclothiazomycin. Several fragments (fragments 2, 3, 4, 5, and 6) generated by double cleavage or multiple cleavage at an acrylamide bond or the tertiary C—S bond were assigned at m/z 784, 1163, 283, 441, and 1231. Differences between the theoretical values and the measurements, which were greater than the instrument errors for MS/MS (<5 ppm), might be caused by low abundances and impurities.
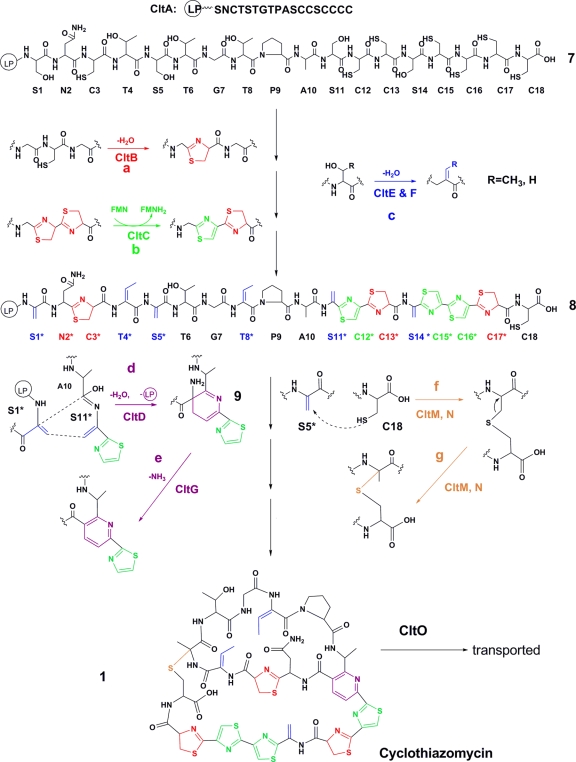
FIG. 4.
Proposed pathway for biosynthesis of cyclothiazomycin. Eight proteins, CltB, CltC, CltD, CltE, CltF CltG, CltM, and CltN, are the candidate enzymes involved in posttranslational modification, and one transport protein, CltO, is predicted to transport the final product. Reaction a, dehydration reaction to produce thiazolines (red); reaction b, selective dehydrogenation reaction to produce thiazoles (green); reaction c, dehydration reaction to produce dehydroamino acids (blue); reaction d, simultaneous hetero-Diels-Alder cyclization and LP cleavage to produce a dehydropiperidine intermediate (purple); reaction e, deamination of the amino group generated by LP cleavage to form a trisubstituted pyridine (purple); reaction f, Michael addition reaction to generate the putative thioether intermediate; reaction g, molecular rearrangement suggested to generate a tertiary thioether.
FIG. 5.
Bioassay for cyclothiazomycin production by S. hygroscopicus 10-22 with B. cinerea Persoon. An agar patch containing S. hygroscopicus 10-22 was placed on B. cinerea Persoon growing on medium to determine the zone of inhibition due to cyclothiazomycin production; S. lividans 1326 was used as a negative control (see Materials and Methods).
Characterization of the cyclothiazomycin produced by S. hygroscopicus 10-22.
To confirm production of cyclothiazomycin by S. hygroscopicus 10-22, the active components were extracted with methanol from a fermentation agar plate, and structural characterization was subsequently performed by using LC-ES-QTOF mass spectrometry analysis.
Two peaks were present in the extract ion current chromatogram extracted from the total ion current chromatogram of the methanol extract by m/z searching with the molecular weight of protonized cyclothiazomycin (m/z 1473.2967 ± 2) (Fig. 3B), suggesting that in solution cyclothiazomycin includes a pair of isomers, very similar to those of cyclothiazomycin B (10). The QTOF mass spectrum of each isomer had an accurate [M+1]+ signal at m/z 1473.2933 and an accurate [M+2]2+ signal at m/z 737.1515. Within the error of the instrument, these results matched the theoretical value for protonized cyclothiazomycin (C59H64N18O14S7+) (m/z 1473.2967) well (Fig. 3B). According to a Beynon table, isotope abundances could be calculated with the formulas [M+2]+ = 1.1x + 0.2z + 0.4S and [M+3]+ = (1.1x)2/200 + 4.4S, where x, z, and S are the numbers of C, O, and S atoms, respectively; these calculations resulted in abundances of 87 and 55, respectively (5a). The calculated results for [M+2]+ and [M+3]+ matched the measured results shown in Fig. 3D well. This isotope analysis, together with the high-resolution mass spectrum, allowed assignment of a unique formula to cyclothiazomycin using the Masshunter qualitative analysis software (Agilent Co., CA).
The role of cleavage of cyclothiazomycin, a bridged macrocyclic thiopeptide, in a mass spectrometer is difficult to determine, and the number of possible cleavage sites is much less than the number for a normal peptide. Most fragments were generated by double cleavage at the acrylamide bond or at the C—S bond between the modified S5 and C18 residues; three examples are fragments 4, 5, and 6 with [M+1]+ signals at m/z 283, 441, and 1231, respectively. Ions 2 and 3 generated from the neutral loss of ion 4 were observed at m/z 784 and 1163, respectively. Therefore, these important fragments in the tandem MS spectrum were characterized as fragments of cyclothiazomycin (Fig. 3C) and confirmed that this thiopeptide is produced by S. hygroscopicus 10-22.
Minimal biosynthetic gene cluster indicated by the heterologous expression study.
To define the functional gene cluster in the sequenced cosmid 14E6, a series of plasmids derived from pSET152 were constructed (Table 1; see Fig. S1 in the supplemental material). First, pJTU4891, containing a large XbaI-EcoRI fragment from 14E6 (Fig. 2B), was transformed into S. lividans 1326, resulting in a transformant with bioactivity against B. cinerea Persoon. Then a 4-kb NdeI fragment that is close to the 3′ end of the gene cluster was removed from pJTU4891 to create pJTU4892. Activity against B. cinerea Persoon confirmed that pJTU4892 contains all of the essential genes for producing cyclothiazomycin. Further deletion of cltIMN from the inserted sequence of pJTU4891 was obtained by a PCR targeting method to generate pJTU4894, which resulted in a failure to produce cyclothiazomycin (Fig. 2B).
To detect the cyclothiazomycin produced by S. lividans 1326::pJTU4892, a similar LC-ES-QTOF analysis was done to screen the transformant's product. Two peaks that were characterized as a pair of isomers of cyclothiazomycin were also detected at the same retention time (see Fig. S2 in the supplemental material), and the mass spectrum of each peak had two signals, one at m/z 737.1511 ([M+2]2+) and one at m/z 1473.2907 ([M+1]+) (Fig. 3C). Together with the isotope abundances of [M+2]+ and [M+3]+, these high-resolution mass spectra also allowed assignment of a unique formula to cyclothiazomycin.
Successful heterologous production of cyclothiazomycin by S. lividans 1326::pJTU4891 did not include expression of ORFs 5, 6, 7, 8, 9, and 10 in the cyclothiazomycin biosynthetic gene cluster. Since pJTU4892 lacks the 4-kb NdeI fragment that includes cltL, ORF 1, ORF 2, and most of cltK and ORF 3, expression of these genes was also not included, and ORF 4 at the 3′ end is unlikely to encode a functional protein involved in biosynthesis. Moreover, the failure of pJTU4894 to produce cyclothiazomycin suggested that at least one gene in the cltIMN cluster is essential for production of cyclothiazomycin (Fig. 2B). Thus, a 22.7-kb gene cluster containing 15 ORFs was the minimal cluster to govern biosynthesis of cyclothiazomycin.
Putative functional enzymes for the cyclothiazomycin biosynthesis model.
According to the bioinformatics analysis of the insert of pJTU4892 (Table 3), besides the 60-amino-acid precursor peptide, the products of 9 of the 15 ORFs in the biosynthetic cluster were candidates for molecules involved in biosynthesis of cyclothiazomycin. Several mutagenesis experiments were done using in-frame deletion of cltD, the C-domain of cltC, and cltA, but none of the mutants was able to generate cyclothiazomycin (see Fig. S2 in the supplemental material). Other genes, the cltHPOIQ genes, were likely to encode two regulators, a transporter, and two transposases, respectively.
TABLE 3.
Putative functions of ORFs in cyclothiazomycin biosynthesis gene cluster
| Gene | Size of product (amino acids) | Homolog |
Putative function | % Identity | % Similarity | ||
|---|---|---|---|---|---|---|---|
| Protein | Organism | Accession no. | |||||
| cltA | 60 | Structural prepeptide | |||||
| cltB | 680 | Hypothetical protein | Actinomadura melliaura | ABC02784 | Cyclodehydratase | 47 | 57 |
| cltC | 562 | NADH oxidase | A. melliaura | ABC02783.1 | NADH oxidase (containing two McbC-like domains) | 36 | 47 |
| cltD | 647 | Hypothetical protein | A. melliaura | ABC02785.1 | Peptidase M14(N) YcaO family (C) | 28 | 41 |
| cltE | 875 | Lantibiotic dehydratase domain protein | Salinispora arenicola CNS-205 | ABV98429.1 | Lantibiotic dehydratase domain protein | 26 | 37 |
| cltF | 341 | Lantibiotic biosynthesis protein | Bacillus cereus ATCC 14579 | AAP11952.1 | Putative lantibiotic biosynthesis protein | 32 | 41 |
| cltG | 375 | Unknown | A. melliaura | ABC02780.1 | Unknown | 37 | 51 |
| cltH | 932 | NysRI | Streptomyces noursei | AF263912_17 | LuxR family transcriptional regulator | 30 | 40 |
| cltI | 192 | Putative transposase | Streptomyces ambofaciens ATCC 23877 | CAJ87970.1 | Putative transposase | 63 | 67 |
| cltJ | 170 | Epoxide hydrolase 1 | Streptomyces sp. strain Mg1 | EDX23590.1 | Epoxide hydrolase | 47 | 62 |
| cltM | 196 | Hypothetical protein | Streptomyces sp. strain FR1 | ABC67445.1 | Hypothetical protein | 44 | 58 |
| cltN | 111 | Ethyl tert-butyl ether degradation protein EthD | Caulobacter sp. strain K31 | ABZ74391.1 | Ethyl tert-butyl ether degradation protein EthD | 34 | 58 |
| cltO | 477 | Transport protein | Streptomyces coelicolor A3(2) | SCO2502 | Transport protein | 37 | 53 |
| cltP | 297 | DNA-binding protein | S. coelicolor A3(2) | SCO2501 | XRE family transcriptional regulator | 43 | 54 |
| cltQ | 116 | Putative transposase | S. coelicolor A3(2) | SCP1.215 | Putative transposase | 65 | 74 |
Similar to the tsrJKLMNOS and sioJKLMNOS genes in thiostrepton and siomycin A gene clusters, respectively (17), six ORFs, cltBCDEFG flanking cltA, are likely to encode the components of the main framework of cyclothiazomycin: dehydramino acids, thiazolines or thiazoles, and the trisubstituted pyridine. Two of the gene products, CltE and CltF, which are homologous to the lantibiotic dehydratases, were candidates for dehydration of Ser and Thr residues (Fig. 4, reaction c). About 4.5 kb from the main structural genes, the cltMN genes were likely to encode the enzymes involved in production of the tertiary thioether (Fig. 4, reactions f and g). cltJ, which was located at the 3′ end of the gene cluster, encoded a homolog of an epoxide hydrolase with high levels of identity (47%) and similarity (62%). According to the model proposed for other thiopeptides (14, 17, 28) and the thioether synthetic pathway in lantibiotic biosynthesis (6), cltJ was unlikely to be a functional gene for either the main framework or tertiary thioether biosynthesis.
Specific dehydrogenation of the thiazolines in the biosynthetic precursor.
Thiazoles and thiazolines are known to be generated by nucleophilic attack on each Cys side chain on the adjacent carbonyl group, followed by dehydration and selective dehydrogenation (19, 25). CltB is likely to be a candidate for cyclodehydratase, governing the formation of thiazolines (Fig. 4, reaction a), since CltB has a putative C-terminal domain belonging to the YcaO superfamily and an N-terminal domain homologous to the uncharacterized protein DUF181. Similarly, in MicroB17 biosynthesis, McbD, which has a YcaO family domain, was proposed to dock protoxin with other subunits to form thiazolines and oxazolines (19). In addition, TclJ, also a YcaO-DUF181 double-domain protein, was suggested to be a cyclodehydratase that catalyzes the formation of a similar heterocycle in thiocillins (28).
Interestingly, in accordance with the prepeptide sequence, only the N-terminal thiazoline of a bithiazoline can be dehydrogenated to produce thiazole, resulting in the three thiazolines that are unchanged in cyclothiazomycin (compound 7) (Fig. 4, reaction b). Selective dehydrogenation of thiazolines to form thiazoles would be expected to be catalyzed by CltC, a putative NADH oxidase with two McbC-like domains, since it has been suggested that McbC catalyzes a similar dehydrogenation in pro-MccB17 and belongs to the nitro-flavin mononucleotide (FMN) reductase superfamily (19). Thus, with two “reductase” domains, CltC is likely to recognize a bithiazoline residue as a substrate but exhibit the dehydrogenation function only with the N-terminal thiazoline residue. The consecutive dehydrogenations of the two thiazoline residues on the prethiazoline chain illustrated by C15*-C16*-C17* (compound 8) (Fig. 4) suggest that CltC is capable of searching the bithiazoline residue from the N terminus and play a role as an LP-dependent enzyme.
Simultaneous reaction involved in LP cleavage and biosynthesis of a trisubstituted pyridine.
Cyclothiazomycin harbors a central trisubstituted pyridine, which is very different from that of thiocillin (Fig. 1). Since the central substituted pyridine and dehydropiperidine were considered a common feature of thiopeptides (3), the trisubstituted pyridine is likely to be synthesized via a piperidine intermediate, which was proposed to be formed by an intramolecular hetero-Diel-Alder reaction (14, 17) (Fig. 4, reaction d). According to the bioinformatics analysis, CltD can be deduced to be a double-domain protein involved in pyridine biosynthesis (Table 3). Significantly, the N-terminal domain of CltD is homologous to that of a putative M14peptidase, carboxypeptidase A (41% similarity and 28% identity), providing a clue for the timing of LP cleavage. Although LP cleavage of thiocillins and LP cleavage of thiostrepton were thought previously to be the final step in thiopeptide biosynthesis (17, 28), we believe that processing of the prepeptide precursor of cyclothiazomycin happens during pyridine formation. The C-terminal domain of CltD, a member of the YcaO family, may provide a docking site for the hetero-Diel-Alder reaction to form a piperidine intermediate (Fig. 4, reaction d). To complete the formation of pyridine, CltG is expected to participate in the deamination that is required to eliminate the amine group generated by LP cleavage, since it shows similarity to SioL and NosO (23% and 29% similarity, respectively), which were previously proposed to be involved in the six-member nitrogen heterocycle formation in siomycin (17) and nosiheptide (30) synthesis.
Tertiary thioether biosynthesis resulting in the final bridged macrocyclic structure of cyclothiazomycin.
An unusual tertiary thioether was found between the dehydrated S5 and C18 residues of cyclothiazomycin (Fig. 1) in the bridged macrocyclic structure. Intramolecular thioether cross-linking within a peptide through Michael addition of Cys to a dehydroamino acid originating from Ser or Thr was observed previously in lantibiotic biosynthesis (31). However, the tertiary thioether of cyclothiazomycin is very different from the primary thioether of a lantibiotic, implying that there is rearrangement after the intramolecular Michael addition reaction (Fig. 4, reactions f and g). With 34% sequence identity to EthD, which was proven to be an essential protein involved in ethyl tert-butyl ether degradation (7), CltN is likely to be one of the enzymes involved in the final formation of the tertiary thioether. Although no function can be determined from the bioinformatics analysis, CltM is the only unassigned candidate left for biosynthesis. It is likely to be involved in biosynthesis of the tertiary thioether rather than biosynthesis of the framework. Unlike CltD, neither CltM nor CltN possesses the docking subunit, implying that the cyclothiazomycin intermediate with a central pyridine can be properly folded, allowing the —SH group of C18 to approach the double bond of the dehydrated S5 residue.
Conclusion.
In this study, a novel and complete gene cluster involved in posttranslational modification of a ribosomally synthesized prepeptide was elucidated based on heterologous production. Based on the heterologous expression study and the bioinformatics analysis of the minimal intact gene cluster, we propose an eight-enzyme biosynthetic model for posttranslational modification of cyclothiazomycin (Fig. 4). In this model, three proteins, CltC, CltD, and CltN, might be important for biosynthesis of the characteristic structural elements: the selectively dehydrogenated thiazolines, trisubstituted central pyridine, and tertiary thioether. Importantly, LP cleavage was deduced to be catalyzed by the C-terminal domain of CltD during pyridine formation. Although this bioinformatics-based biosynthetic model needs to be substantiated, it may provide clues for further cyclothiazomycin bioengineering.
Supplementary Material
Acknowledgments
This work was supported by 863 Funds from the Ministry of Science and Technology, the National Science Foundation of China, the Ph.D. Training Fund of the Ministry of Education, and the Shanghai Municipal Council of Science and Technology.
We thank Jie Chen for the gift of B. cinerea Persoon.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, M., T. Ohtsuka, M. Yamada, Y. Ohba, H. Yoshizaki, H. Yasuno, T. Sano, J. Watanabe, K. Yokose, and H. Seto. 1991. Cyclothiazomycin, a novel polythiazole-containing peptide with renin inhibitory activity. Taxonomy, fermentation, isolation and physico-chemical characterization. J. Antibiot. (Tokyo) 44:582-588. [DOI] [PubMed] [Google Scholar]
- 3.Bagley, M. C., J. W. Dale, E. A. Merritt, and X. Xiong. 2005. Thiopeptide antibiotics. Chem. Rev. 105:685-714. [DOI] [PubMed] [Google Scholar]
- 4.Bagley, M. C., and X. Xiong. 2004. Stereoselective synthesis of the gamma-lactam hydrolysate of the thiopeptide cyclothiazomycin. Org. Lett. 6:3401-3404. [DOI] [PubMed] [Google Scholar]
- 5.Baquero, F., D. Bouanchaud, M. C. Martinez-Perez, and C. Fernandez. 1978. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J. Bacteriol. 135:342-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Beynon, J. H., and A. E. Williams. 1960. Mass and abundance tables for use in mass spectrometry. Elsevier Publishing Co., New York, NY.
- 6.Bierbaum, G., and H. G. Sahl. 2009. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 10:2-18. [DOI] [PubMed] [Google Scholar]
- 7.Chauvaux, S., F. Chevalier, C. Le Dantec, F. Fayolle, I. Miras, F. Kunst, and P. Beguin. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert, M., S. Ostiguy, D. Kluepfel, and R. Morosoli. 1996. Cloning of a secA homolog from Streptomyces lividans 1326 and overexpression in both S. lividans and Escherichia coli. Biochim. Biophys. Acta 1296:9-12. [DOI] [PubMed] [Google Scholar]
- 9.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto, M., T. Murakami, K. Funahashi, T. Tokunaga, K. Nihei, T. Okuno, T. Kimura, H. Naoki, and H. Himeno. 2006. An RNA polymerase inhibitor, cyclothiazomycin B1, and its isomer. Bioorg. Med. Chem. 14:8259-8270. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, R. A., and C. J. Moody. 2007. From amino acids to heteroaromatics—thiopeptide antibiotics, nature's heterocyclic peptides. Angew. Chem. Int. Ed. Engl. l46:7930-7954. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 13.Jian, X., X. Pang, Y. Yu, X. Zhou, and Z. Deng. 2006. Identification of genes necessary for jinggangmycin biosynthesis from Streptomyces hygroscopicus 10-22. Antonie Van Leeuwenhoek 90:29-39. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, W. L., L. Pan, and C. Li. 2009. Thiostrepton biosynthesis: prototype for a new family of bacteriocins. J. Am. Chem. Soc. 131:4327-4334. [DOI] [PubMed] [Google Scholar]
- 15.Kieser, T., M. J. Bibb, K. F. Chater, M. J. Butter, and D. A. Hopwood. 2000. Practical Streptomyces genetics. A laboratory manual. John Innes Foundation, Norwich, United Kingdom.
- 16.Lee, S. W., D. A. Mitchell, A. L. Markley, M. E. Hensler, D. Gonzalez, A. Wohlrab, P. C. Dorrestein, V. Nizet, and J. E. Dixon. 2008. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. U. S. A. 105:5879-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao, R., L. Duan, C. Lei, H. Pan, Y. Ding, Q. Zhang, D. Chen, B. Shen, Y. Yu, and W. Liu. 2009. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem. Biol. 16:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClerren, A. L., L. E. Cooper, C. Quan, P. M. Thomas, N. L. Kelleher, and W. A. van der Donk. 2006. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U. S. A. 103:17243-17248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne, J. C., R. S. Roy, A. C. Eliot, N. L. Kelleher, A. Wokhlu, B. Nickels, and C. T. Walsh. 1999. Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry 38:4768-4781. [DOI] [PubMed] [Google Scholar]
- 20.Murakami, T., T. G. Holt, and C. J. Thompson. 1989. Thiostrepton-induced gene expression in Streptomyces lividans. J. Bacteriol. 171:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolaou, K. C., B. S. Safina, M. Zak, S. H. Lee, M. Nevalainen, M. Bella, A. A. Estrada, C. Funke, F. J. Zecri, and S. Bulat. 2005. Total synthesis of thiostrepton. Retrosynthetic analysis and construction of key building blocks. J. Am. Chem. Soc. 127:11159-11175. [DOI] [PubMed] [Google Scholar]
- 22.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pang, X., X. Zhou, Y. Sun, and Z. Deng. 2002. Physical map of the linear chromosome of Streptomyces hygroscopicus 10-22 deduced by analysis of overlapping large chromosomal deletions. J. Bacteriol. 184:1958-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porse, B. T., I. Leviev, A. S. Mankin, and R. A. Garrett. 1998. The antibiotic thiostrepton inhibits a functional transition within protein L11 at the ribosomal GTPase centre. J. Mol. Biol. 276:391-404. [DOI] [PubMed] [Google Scholar]
- 25.Roy, R. S., A. M. Gehring, J. C. Milne, P. J. Belshaw, and C. T. Walsh. 1999. Thiazole and oxazole peptides: biosynthesis and molecular machinery. Nat. Prod. Rep. 16:249-263. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. “Streptomyces nanchangensis,” a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 28.Wieland Brown, L. C., M. G. Acker, J. Clardy, C. T. Walsh, and M. A. Fischbach. 2009. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc. Natl. Acad. Sci. U. S. A. 106:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willey, J. M., and W. A. van der Donk. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477-501. [DOI] [PubMed] [Google Scholar]
- 30.Yu, Y., L. Duan, Q. Zhang, R. Liao, Y. Ding, H. Pan, E. Wendt-Pienkowski, G. Tang, B. Shen, and W. Liu. 2009. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem. Biol. 4:855-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, X., W. Ni, and W. A. van der Donk. 2007. On the regioselectivity of thioether formation by lacticin 481 synthetase. Org. Lett. 9:3343-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.