Abstract
A total of 227 isolates of Aeromonas obtained from different geographical locations in the United States and different parts of the world, including 28 reference strains, were analyzed to determine the presence of various virulence factors. These isolates were also fingerprinted using biochemical identification and pulse-field gel electrophoresis (PFGE). Of these 227 isolates, 199 that were collected from water and clinical samples belonged to three major groups or complexes, namely, the A. hydrophila group, the A. caviae-A. media group, and the A. veronii-A. sobria group, based on biochemical profiles, and they had various pulsotypes. When virulence factor activities were examined, Aeromonas isolates obtained from clinical sources had higher cytotoxic activities than isolates obtained from water sources for all three Aeromonas species groups. Likewise, the production of quorum-sensing signaling molecules, such as N-acyl homoserine lactone, was greater in clinical isolates than in isolates from water for the A. caviae-A. media and A. hydrophila groups. Based on colony blot DNA hybridization, the heat-labile cytotonic enterotoxin gene and the DNA adenosine methyltransferase gene were more prevalent in clinical isolates than in water isolates for all three Aeromonas groups. Using colony blot DNA hybridization and PFGE, we obtained three sets of water and clinical isolates that had the same virulence signature and had indistinguishable PFGE patterns. In addition, all of these isolates belonged to the A. caviae-A. media group. The findings of the present study provide the first suggestive evidence of successful colonization and infection by particular strains of certain Aeromonas species after transmission from water to humans.
Aeromonas species cause both intestinal and extraintestinal infections (25, 33, 78), and the latter include septicemia, cellulitis, wound infections, urinary tract infections, hepatobiliary tract infections, soft tissue infections, and, occasionally, meningitis and peritonitis (25, 30, 78). In immunocompromised children, these pathogens can cause even more severe forms of infections, such as hemolytic-uremic syndrome (HUS) and necrotizing fasciitis (3, 23), although detailed studies are needed to establish such associations. Worldwide, the rate of isolation of Aeromonas from diarrheic stools has been reported to be as high as 10.8%, compared to 2.1% for healthy controls (25, 37, 78). An increased rate of isolation of Aeromonas species was reported in flood water samples during Hurricane Katrina in New Orleans (58), and skin and soft tissue infections caused by Aeromonas species were among the most common infections in the survivors of the 2004 tsunami in southern Thailand (28). In particular, Aeromonas salmonicida causes fish infections that result in huge economical losses in the fishing industry (6, 22). The ability of aeromonads, as well as other bacteria, to survive in chlorinated water when they are in biofilms and their resistance to multiple antibiotics are major public health concerns (46).
Aeromonas-related gastroenteritis remains somewhat controversial (24, 36). There have been a number of well-described cases and a few documented outbreaks, but whether all aeromonad fecal isolates from symptomatic persons are the actual causes of diarrheal disease is still questionable. One theory for this conundrum was posed in 2000 by two of us, who suggested that only specific subsets of Aeromonas strains within and between species are actually pathogenic for humans (38). This highlights the importance of developing accurate biotyping, molecular fingerprinting, and virulence factor analysis methods for differentiating environmental and clinical aeromonads from one another and for comparing them (38).
Of the 19 currently recognized Aeromonas species, A. hydrophila, A. caviae, and A. veronii biovar sobria are the most common species known to cause the majority of human infections, and they account for more than 85% of all clinical isolates (34). The pathogenesis of Aeromonas infections is multifactorial, as aeromonads produce a wide variety of virulence factors, including hemolysins, cytotonic and cytotoxic enterotoxins, proteases, lipases, leucocidins, endotoxin, adhesions, and an S layer, that act in concert to cause disease in the host (12-14, 50, 51). The cytotoxic enterotoxin Act, which has some similarities to aerolysin (31), is one of the most significant virulence factors in diarrheal isolate SSU of A. hydrophila and was first characterized in our laboratory (12). Act is secreted by the type II secretion system (T2SS) and has hemolytic, cytotoxic, and enterotoxic activities (12). In addition, our laboratory recently sequenced and characterized two other secretion systems, T3SS and T6SS, that were found to contribute to the virulence of A. hydrophila SSU (66, 67, 72). We recently characterized an effector of the T3SS, which was designated AexU, and found that it was associated with ADP ribosylation of host cell proteins, a rounded phenotype in HeLa cells, inhibition of phagocytosis, induction of apoptosis, and mouse mortality (66, 67). In recent studies, we also investigated the role of two T6SS-associated effectors, the valine-glycine repeat G (VgrG) family of proteins and hemolysin-coregulated protein (Hcp), in the virulence of A. hydrophila (71, 72). We demonstrated that VgrG1 of A. hydrophila had actin-ADP ribosylation activity that induced host cell cytotoxicity (71). Based on the model for T6SS, the VgrG1 protein must assemble with the highly homologous VgrG2 and VgrG3 proteins to form a cell-puncturing device to deliver effector proteins into the host cells (59). We also obtained evidence that expression of the hcp gene in HeLa cells led to their apoptosis, and animals immunized with recombinant Hcp were protected from subsequent challenge with a lethal dose of wild-type A. hydrophila SSU (72).
In addition, cytotonic enterotoxins (e.g., Alt [heat labile] and Ast [heat stable]) were identified in a diarrheal A. hydrophila SSU isolate (14, 63) that induced fluid secretion in the ligated small intestinal loops of animals (47). More recently, we identified some additional virulence factor-encoding and regulatory genes, such as the enolase, hlyA (hemolysin), gidA (glucose-inhibited division A), vacB (virulence-associated protein B), dam (DNA adenine methyltransferase), and tagA (ToxR-regulated lipoprotein) genes, which modulated the virulence of A. hydrophila SSU (19-21, 57, 62, 64). The production of such a wide array of virulence factors by Aeromonas species is indicative of their potential to cause severe diseases in humans. These virulence factor-encoding genes might be differentially expressed in Aeromonas species depending on the environmental conditions, such as water or the human host.
A cell-to-cell signaling system, known as quorum sensing (QS), might play an important role in sensing physiological conditions and helping bacteria express the virulence genes at an appropriate time under the appropriate conditions. Thus far, at least three QS circuits have been identified in Gram-negative bacteria, and they were designated LuxRI (autoinducer 1 [AI-1]), LuxS (AI-2), and AI-3 (epinephrine/norepinephrine). All of these QS systems were detected in our SSU clinical strain of A. hydrophila, and we recently demonstrated that N-acyl homoserine lactone (AHL) (AI-1) and AI-2-mediated QS controlled the virulence of A. hydrophila SSU (40, 43). Further, we observed decreased production of N-acyl homoserine lactones when we deleted two major virulence factor-encoding genes, the act gene and the gene encoding an outer membrane protein (aopB), an important component of the T3SS (65), from A. hydrophila SSU. Likewise, we observed that N-acyl homoserine lactone production was also modulated by regulatory genes, such as dam and gidA, in A. hydrophila SSU (18). Thus, differential expression of genes might also be an important factor in the pathogenesis of Aeromonas species.
The presence of any virulence gene in strains of Aeromonas isolated from water should be carefully scrutinized, as such genes could be expressed better in a human host, which could lead to devastating outcomes. In addition, it is possible that in the environment certain Aeromonas clones may predominate and cause human diseases more frequently than other clones. Thus, it is important to determine the clonal variation of a range of Aeromonas species isolated from various sources and identify predominant clones by a polyphasic approach that includes biochemical phenotyping, virulence marker detection, and molecular fingerprinting techniques.
In the present study, we compared 199 Aeromonas isolates, 146 of which were from water sources and 53 of which were from human patients with diarrhea in the Unites States. In addition, 28 reference and classical strains that were obtained from various culture collections and/or were isolated from specimens obtained in diverse geographical areas of the world, including water and clinical specimens, were also characterized. All isolates were biochemically identified to the phenospecies group level, examined for the presence of a set of 11 virulence factors by using DNA colony blot hybridization, and fingerprinted by using pulsed-field gel electrophoresis (PFGE). Some of the virulence factors selected, including T6SS effectors, were also examined by using functional assays. Our data provide the first suggestive evidence of water-to-human transmission, i.e., of successful colonization and infection by particular strains of certain Aeromonas species.
MATERIALS AND METHODS
Bacterial strains.
The strains examined in this study originated from multiple sources and collections (see Tables S1 to S3 in the supplemental material). One set consisting of 83 strains (NM collection; originally 84 strains, but NM-59 did not grow and could not be used for biochemical studies) (see Table S1 in the supplemental material) was selected from the larger collection of one of us (N.P.M.) of clinical and water isolates resulting from his previous epidemiological studies related to Aeromonas and diarrhea that were identified using conventional biochemical tests and ribotyped (52, 54, 55). A second collection, consisting of 101 environmental strains, was obtained from the U.S. Environmental Protection Agency (EPA) with the species group identification “blinded,” and these strains originated from a variety of water sources (see Table S2 in the supplemental material). A third smaller set consisting of 17 clinical diarrheal stool isolates (MB collection) was obtained by one of us (M.A.B.), and these isolates originated from patients at the Marshfield Clinic in Marshfield, WI (see Table S3 in the supplemental material). The last set, which consisted of 28 reference and classical strains, was obtained from the collection of another one of us (A.J.H.) (Table 1). For the biochemical phenotypic tests, the strains were grown on sheep blood agar (SBA) (BDMS, Sparks, MD) plates and incubated at 37°C. Stock cultures were maintained frozen at −80°C in Trypticase soy broth (TSB) with 30% (vol/vol) glycerol.
TABLE 1.
Sources and identification of reference and classic historic strains
| Strain | Reference no. | Source | Serotype |
|---|---|---|---|
| AA-1 | ATCC 7966T | Canned milk | A. hydrophila HG 1 |
| AA-2 | ATCC 9071 | Frog | A. veronii biovar sobria HG 8Y |
| AA-3 | ATCC 15468T | Guinea pig | A. caviae HG 4 |
| AA-4 | ATCC 49568T | Stool | A. jandaei HG 9 |
| AA-5 | ATCC 35624T | Sputum | A. veronii biovar veronii HG 10 |
| AA-6 | ATCC 43700T | Abscess | A. schubertii HG 12 |
| AA-7 | ATCC 49657T | Stool | A. trota HG 14 |
| AA-8 | ATCC 51108T | Fish | A. bestiarum HG 2 |
| AA-9 | AH65 | Fish | A. bestiarum |
| AA-10 | CDC 0434-84 | Freshwater | A. salmonicida (motile) HG 3 |
| AA-11 | Altwegg/A63 | Stool | A. salmonicida (motile) |
| AA-12 | CDC0862-83 | Fish | A. media HG 5a |
| AA-13 | L149 | Well water | A. caviae-A. media group |
| AA-14 | L327 | Stool | A. caviae-A. media group |
| AA-15 | L321 | Stool | A. caviae-A. media group |
| AA-16 | L226 | Well water | A. caviae-A. media group |
| AA-17 | CIP 7433T | Fish | A. sobria HG 7 |
| AA-18 | Rumpf/AH | Leg | A. hydrophila group |
| AA-19 | X3 | Abscess | A. hydrophila group |
| AA-20 | VT1 | Osteomyelitis | A. veronii-A. sobria group |
| AA-21 | Camare | Heel wound | A. hydrophila group |
| AA-22 | SSU | Stool | A. hydrophila group |
| AA-23 | PM3 | Clinical | A. hydrophila group |
| AA-24 | PM25 | Clinical | A. hydrophila group |
| AA-25 | Dog | Veterinary | A. hydrophila group |
| AA-26 | W1 | Water distribution | A. hydrophila group |
| AA-27 | W2 | Water distribution | A. hydrophila group |
| AA-28 | W3 | Water distribution | A. hydrophila group |
Identification of the genus of isolates.
All strains were initially screened by using the following tests: Gram stain, oxidase activity (1% solution of N,N,N′,N′-tetramethyl-ρ-aminodimethylaniline oxalate), acid production from glucose using triple sugar iron (TSI) slants, motility using semisolid agar, and resistance to vibriostatic agent O/129 (150 μg/ml; Oxoid, Ogdensburg, NY). Only the strains that were motile and oxidase positive, produced acid from glucose, and were O/129-resistant Gram-negative rods were considered members of the genus Aeromonas.
“Species group” identification.
Each strain was then examined using a battery of biochemical tests from Aerokey II (10), the 2003 Abbott-Janda schema (1), and selected supplementary biochemical tests described in Bergey's Manual of Systematic Bacteriology (48), including the Voges-Proskauer test and tests for esculin hydrolysis; indole production (Kovács method); methyl red; citrate utilization; the presence of lysine decarboxylase, ornithine decarboxylase, and arginine dihydrolase; production of acid and gas from glucose; acid production from arabinose, cellobiose, lactose, mannitol, rhamnose, salicin, sorbitol, and sucrose; hydrolysis of urea; o-nitrophenyl-β-d-galactopyranoside (ONPG) production; and the ability to grow in the presence of 6.5% NaCl.
Additionally, all isolates were tested to determine their susceptibilities to ampicillin (10 μg) and cephalothin (30 μg) by using the latest Clinical and Laboratory Standards Institute (CLSI) charts and guidelines. All of the tests were performed using conventional methods described previously (1, 10, 30, 48). Using the test results for the isolates and the combined reference strains, each isolate was assigned to a “species group,” either the A. hydrophila group, the A. caviae-A. media group, or the A. veronii-A. sobria group. Each “species group” includes at least three different DNA hybridization groups and phenotypic species (1, 35, 48). For example, the A. hydrophila group includes A. hydrophila, A. bestiarum, and A. salmonicida (both motile and nonmotile species). Likewise, the A. caviae-A. media group includes A. caviae, A. media, and A. eucrenophila, and the A. veronii-A. sobria group includes A. veronii biovar sobria, A. jandaei, A. schubertii, and A. trota. “Species groups” were used because of the preponderance of isolates from water, where presumably all aeromonad species live. However, there is not sufficient data to support actual species identification of environmental isolates without additional uncommon biochemical tests. If an isolate could not be definitely placed in any one “species group,” it was considered an “atypical” aeromonad.
Colony blot hybridization.
Aeromonas cultures (clinical and water isolates) were inoculated onto nylon membrane filters in a grid pattern on Luria-Bertani (LB) agar plates. The plates were incubated at 37°C for 4 to 8 h, and then the filters were removed and processed as described previously (5). Briefly, after denaturation and neutralization, the filters were extensively washed with 1.5 M NaCl, 1 M Tris (pH 7.4) and chloroform under a vacuum to remove cell debris. Then the filters were dried, baked at 80°C for 2 h, and prewashed with 50 mM Tris-HCl (pH 8.0), 1 M NaCl, 1 mM EDTA, 0.1% SDS at 42°C to remove additional cell debris. For hybridization, filters were first prehybridized in the Quickhyb solution (Stratagene, La Jolla, CA) at 68°C for 2 h and then hybridized by using Quickhyb and [α-32P]dCTP-labeled gene probes under high-stringency conditions for 3 h at 68°C. The membranes were washed twice at 68°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate, pH 7.0) with 0.1% SDS for 20 min and then twice in 1× SSC with 0.1% SDS for 20 min at 68°C. Subsequently, the filters were dried, and X-ray film was exposed to the filters as described previously (57).
The probes encompassed coding regions of the following genes: act, alt, ast, and ascV (which encode members of the T3SS); aexU (which encodes an effector protein secreted via the T3SS); gidA; the enolase gene; dam; tagA; hlyA (which encodes hemolysin); and ahyRI (which encode an autoinducer synthase-transcription regulator of AHL). The probes were prepared by performing PCR using specific primers for each of the genes and the appropriate plasmid DNA as the template (see Table S4 in the supplemental material). The conditions used for PCR included an initial denaturation step at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing of the primers at 55 to 68°C for 1 min, and extension at 72°C for 1 min. After final extension at 72°C for 10 min, the reaction mixtures were kept at 4°C until the PCR products were analyzed by agarose gel electrophoresis. Probes were labeled with [α-32P]dCTP (MP Biomedicals, Solon, OH) by using a random primer kit (Invitrogen, Carlsbad, CA) (20). The sizes and descriptions of the probes are given in Table S4 in the supplemental material. For each colony blot DNA hybridization assay, A. hydrophila SSU and Escherichia coli DH5α were used as a positive control and a negative control, respectively.
Measurement of hemolytic activity.
To examine the lysis of rabbit red blood cells (RBCs), culture filtrates from various Aeromonas cultures (grown for 18 h in LB medium at 37°C with shaking [180 rpm]) were first treated with trypsin (final concentration, 0.05%) at 37°C for 1 h and then subjected to a hemolysis assay as described previously (63). We defined the hemolytic activity titer as the reciprocal of the highest dilution of a culture filtrate that caused 50% lysis of the red blood cells. The hemolytic activity titer was divided by the optical density of the culture to obtain the hemolytic activity per unit of growth. The results were expressed per ml of supernatant. Treatment of culture filtrates with trypsin was required to activate Act and HlyA for measuring hemolytic activity, as RBCs do not produce any of their own proteases (20, 21).
The limitation of the hemolytic activity assay used was that various hemolysins produced by Aeromonas species might have different affinities for rabbit erythrocytes. However, because of the number of Aeromonas isolates that were examined in this study, it was not possible to employ RBCs from different animal species. Since most Aeromonas hemolysins can be detected by using rabbit erythrocytes, we used only these RBCs. We successfully detected hemolytic activity of Act and HlyA (in the act mutant strain) of A. hydrophila SSU after trypsin treatment using rabbit erythrocytes (21). However, a previous study demonstrated that trypsin treatment might inactivate the biological activity of some hemolysins produced by Aeromonas species (56). In our analyses of the Aeromonas isolates, we used culture filtrates both before and after trypsin treatment and detected only increased hemolytic activity after trypsin treatment associated with Act and HlyA. These data suggested that none of the Aeromonas isolates used in this study had a trypsin-sensitive hemolysin(s).
Measurement of cytotoxic activity.
The RAW 264.7 murine macrophage cell line (American Type Culture Collection, Manassas, VA) was used for the cytotoxicity assay, as described previously (63). Briefly, culture filtrates from various Aeromonas cultures were serially diluted (2-fold) and added to macrophages. The cytotoxicity was measured by examining cells with a microscope after 18 h of incubation. The dead cells were rounded and detached from the monolayer. Alternatively, the adherent cells in the monolayer were stained with Giemsa stain and then examined with the microscope. We defined the cytotoxicity titer as the reciprocal of the highest dilution of the culture filtrate that caused destruction of 50% of the RAW 264.7 cells. The cytotoxicity titer was divided by the optical density of the bacterial culture to obtain the cytotoxic activity per unit of growth. The results were expressed per ml of supernatant.
Measurement of protease activity.
The protease activities in the culture filtrates of various Aeromonas strains were measured using the procedure that we described previously (20). The protease activity was calculated per ml of culture filtrate and was divided by the optical density of the culture to obtain the protease activity per unit of growth. The hide azure powder substrate (Calbiochem, La Jolla, CA) was used for measuring protease activity because of the sensitivity and rapidity of the assay. Further, this substrate could detect both metalloproteases and serine proteases, which are the two major classes of proteases produced by Aeromonas species. The substrate incubated with Dulbecco phosphate-buffered saline (DPBS) alone served as a negative control (57).
AHL production.
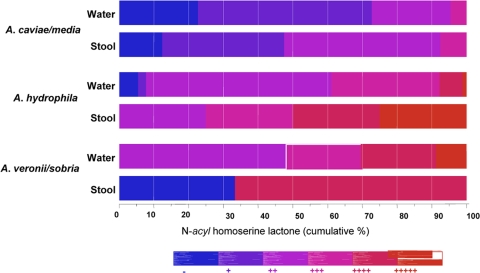
N-Acyl homoserine lactone (AHL) production was detected by cross-streaking various Aeromonas strains with the biosensor strain Chromobacterium violaceum CV026 on LB agar (49). The lactones produced by Aeromonas isolates diffused through the agar and induced production of the pigment violacein in the biosensor strain (73), resulting in various intensities of purple depending on the amounts of lactones produced by Aeromonas isolates after overnight incubation at 30°C. The levels of lactone production were expressed as follows: −, no lactone production; +, very weak production; ++, weak production; +++, moderate production; ++++, high level of production; and +++++, very high level of production (Fig. 1). An isogenic ahyRI mutant of A. hydrophila SSU was used as a negative control, as this mutant was negative for lactone production (40). Although several reporter strains are available for measuring AHL production, C. violaceum CV026 is particularly sensitive for detection of N-acyl chains ranging in length from C4 to C8. Previous studies have shown that Aeromonas species produce C4 to C6 homoserine lactones (HSLs) as the major AI-1 autoinducer molecules (9, 73); consequently, in this study we used C. violaceum for measuring AHLs. However, some of the Aeromonas isolates might produce HSLs with longer acyl chains, and in such cases Agrobacterium tumefaciens A136 (82) can be used for detection.
FIG. 1.
Hue diagram showing levels of N-acyl homoserine lactones (AHLs) produced by different species groups of Aeromonas. AHL production was detected using the C. violaceum CV026 biosensor strain. The different numbers of plus signs indicate the different levels of AHLs produced as described in Materials and Methods.
PFGE for DNA fingerprinting of Aeromonas isolates.
The PFGE procedure used for Aeromonas was a modification of methods described previously (74). Isolates were grown overnight in 5 ml of brain heart infusion broth at 37°C, harvested by centrifugation, and washed with 1 ml PIV buffer (10 mM Tris-HCl [pH 7.6], 1 M NaCl). The concentration of pelleted cells in PIV buffer was adjusted to 1 × 109 CFU/ml by using a Vitek colorimeter (Hach Co., Loveland, CO). The cells were mixed with an equal volume of 2% low-melting-temperature agarose (FMC Bioproducts, Rockland, ME), dispensed into plug molds (Bio-Rad Laboratories, Hercules, CA), and allowed to solidify for 10 min at room temperature. Plugs were incubated in 3 ml of lysis buffer (6 mM Tris-HCl, 1.0 M NaCl, 0.1 M EDTA, 0.5% Brij 58, 0.5% Sarkosyl, 0.2% deoxycholate, 1 mg/ml lysozyme) at 37°C for 4 h. Lysis buffer was replaced with a proteinase K solution (0.5 M EDTA, 1% N-lauroyl sarcosine, 1 mg/ml proteinase K), which was followed by incubation at 55°C overnight. The plugs were washed four times in Tris-EDTA buffer (10 mM Tris-HCl, 0.1 M EDTA [pH 8.0]) and stored at 4°C.
The genomic DNA (gDNA) was digested with 30 U XbaI (Promega, Madison, WI) at 37°C overnight. Electrophoresis was performed in 1% Seakem Gold agarose (FMC Bioproducts) by using the CHEF-DRIII system (Bio-Rad) with 0.5× Tris-borate-EDTA buffer (45 mM Tris, 45 mM boric acid, 1 mM and EDTA [pH 8.0]) at 14°C. The running parameters were 6 V/cm for 20 h with 0.5- to 20-s pulses. Salmonella serotype Braenderup strain H9812 digested with XbaI was run in multiple lanes of each gel as a DNA global reference for standardizing runs and to determine DNA band sizes. DNA banding patterns were visualized with 0.1% ethidium bromide and digitally photographed. BioNumerics software (version 4.01; Applied Maths, Austin, TX) was used to compare the genetic similarity of isolates and to construct a similarity dendrogram using the Dice coefficient and the unweighted-pair group method with arithmetic mean (UPGMA) algorithm with a position tolerance of 1.0% (8).
Western blot analysis.
The T6SS effectors Hcp and VgrG2-VgrG3 in the culture supernatants of selected 7 Aeromonas isolates were detected by using Western blot analysis and specific antibodies according to the procedure described previously (40). We used recombinant VgrG2 for antibody production, and the immune sera obtained did not differentiate between the VgrG2 and VgrG3 proteins of A. hydrophila SSU due to their high level homology (∼90%) and their similar sizes on Western blots (71).
Statistical analysis.
The number of isolates in each of the three Aeromonas species groups was summarized by isolate source (stool, water, and water source). Standard descriptive statistics were used to summarize virulence factors organized by Aeromonas species group and isolate source. The prevalence of Aeromonas species groups and the presence of virulence factors were compared by isolate source, using Fisher's exact test. The activity levels for the virulence factors were compared by Aeromonas species group with the Wilcoxon rank-sum test (analogous to the t test or analysis of variance [ANOVA] with two groups). Results were considered statistically significantly different if the P value was <0.05 with no correction for multiple comparisons. Adjustment for multiple comparisons is always potentially controversial, because adjustment to reduce false-positive results always increases the false-negative result rate, thus reducing the power.
RESULTS
Biochemical properties of Aeromonas isolates.
All of the Aeromonas strains produced medium-size tan to buff colonies on Trypticase soy agar (TSA) with 5% sheep blood when they were incubated at 35°C for 2 to 5 days. The overwhelming majority of the strains also displayed beta-hemolysis on the TSA plates with blood; the beta-hemolysis was significantly stronger (3+ or 4+) with the A. hydrophila and A. veronii-A. sobria group strains, and with the A. caviae-A. media group strains the beta-hemolysis was somewhat “weaker” (1+ or 2+) or there was alpha-hemolysis or gamma-hemolysis.
The breakdown of the numbers of members of the different “species groups” for each set of isolates was as follows. The NM collection (81 strains, including water isolates and 2 isolates obtained from food) included 41 A. caviae-A. media group strains, 17 A. hydrophila group strains, 12 A. veronii-A. sobria group strains, and 11 “atypical aeromonad” strains. The MB collection (n = 17) included 12 A. caviae-A. media group strains, 3 A. hydrophila group strains, 2 atypical aeromonad strains, and no A. veronii-A. sobria group strains. The largest group, the EPA collection (n = 101), included 6 A. caviae-A. media group strains, 74 A. hydrophila group strains, 14 A. veronii-A. sobria group strains, and 7 atypical aeromonad strains (see Tables S1 to S3 in the supplemental material).
Of the 28 reference strains (Table 1), 7 were “bona fide” type strains of species, and 6 strains were additional reference strains of specific species in a hybridization group (HG). The remaining classical, historical strains included 10 strains identified as members of the A. hydrophila group, 4 strains belonging to the A. caviae-A. media group, and 1 strain belonging to the A. veronii-A. sobria group.
Sources of species groups.
A total of 227 Aeromonas isolates were used in this study, including 53 clinical stool isolates, 146 water isolates, and 28 reference and classical strains from environmental, clinical, and veterinary sources. The majority of the water and clinical stool isolates were collected at different geographical locations in the United States, and the remainder originated from locations in different parts of the world. In water samples, the A. hydrophila group was the most prevalent and accounted for 59.5% of the isolates (n = 87), followed by the A. veronii-A. sobria group (15.7%; n = 23) and the A. caviae-A. media group (14.3%; n = 21). The least prevalent organisms were the atypical aeromonads (10.2%; n = 15) (Table 2).
TABLE 2.
Identification of Aeromonas isolates obtained from water and clinical samples
| Species group | No. (%) of isolates from: |
|
|---|---|---|
| Water | Stools | |
| A. hydrophila group | 87 (59.5) | 7 (13.2) |
| A. veronii-A. sobria group | 23 (15.7) | 3 (5.6) |
| A. caviae-A. media group | 21 (14.3) | 38 (71.6) |
| Atypical aeromonads | 15 (10.2) | 5 (9.4) |
| Total | 146 | 53 |
Water isolates were obtained from different sources, such as surface water (SW), groundwater (GW), well water (WW), wellhead hydrant water (WHHW), kitchen tap water (KTW), and municipal drinking water distribution systems (DW). The A. hydrophila and A. veronii-A. sobria groups were found more commonly in SW or DW and GW, whereas members of the A. caviae-A. media groups were commonly isolated from DW, GW, and WW. Atypical aeromonads were more common in GW and SW (Table 3).
TABLE 3.
Water sources from which Aeromonas isolates were obtained
| Species group | No. of isolates obtained froma: |
|||||
|---|---|---|---|---|---|---|
| SW | GW | DW | WW | WHHW | KTW | |
| A. hydrophila group | 36 | 34 | 15 | 0 | 0 | 2 |
| A. veronii-A. sobria group | 13 | 8 | 1 | 1 | 0 | 0 |
| A. caviae-A. media group | 0 | 6 | 6 | 5 | 2 | 2 |
| Atypical aeromonads | 4 | 6 | 2 | 2 | 1 | 0 |
Abbreviations: SW, surface water; GW, groundwater; DW, distribution water; WW, well water; WHHW, wellhead hydrant water; KTW, kitchen tap water.
In clinical samples, the A. caviae-A. media group was the predominant group, accounting for 71.6% (n = 38) of the total isolates, followed by the A. hydrophila group (13.2%; n = 7), the atypical aeromonads (9.4%; n = 5), and the A. veronii-A. sobria group (5.6%; n = 3) (Table 2).
Hemolytic activity.
The hemolytic activity associated with Aeromonas isolates was measured by using rabbit RBCs. The percentages of clinical and water isolates of Aeromonas that were found to be positive in the hemolytic assay are shown in Table 4. For the water isolates, the highest percentage of positive strains (97%) belonged to the A. hydrophila group, whereas the lowest percentage of positive strains (10%) belonged to the A. caviae-A. media group. Similarly, for the clinical isolates, the percentages were 100% for the A. hydrophila group and 3% for the A. caviae-A. media group. Importantly, for the A. veronii-A. sobria group, all of the stool isolates (100%) were hemolytic, while only 61% of the water isolates were positive for hemolytic activity. For atypical aeromonads, 67% of the water isolates were positive for hemolytic activity, while 40% of the stool isolates had this activity.
TABLE 4.
Hemolytic, cytotoxic, and protease activities and lactone production for isolates of Aeromonas from water and stool
| Species group | % of positive isolates |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hemolytic activity |
Cytotoxic activity |
Protease activity |
Lactone production |
|||||
| Water | Stool | Water | Stool | Water | Stool | Water | Stool | |
| A. hydrophila group | 97 | 100 | 98 | 100 | 98 | 100 | 94 | 100 |
| A. veronii-A. sobria group | 61 | 100 | 61 | 100 | 100 | 100 | 100 | 67 |
| A. caviae-A. media group | 10 | 3 | 29 | 3 | 86 | 97 | 81 | 92 |
| Atypical aeromonads | 67 | 40 | 73 | 40 | 93 | 100 | 93 | 100 |
The levels of hemolytic activity detected for the water isolates belonging to the A. hydrophila group ranged from very low to very high (4 to 7,314 U). The levels of hemolytic activity were low and in a narrow range (19 to 33 U) for members of the A. caviae-A. media group isolated from water samples. On the other hand, for the clinical (stool) strains, members of the A. veronii-A. sobria group exhibited the highest levels of hemolytic activity (750 to 1,765 U), followed by members of the A. hydrophila group (20 to 1,406 U), the atypical aeromonad group (320 to 457 U), and the A. caviae-A. media group (50 U) (data not shown).
We performed a statistical analysis of the prevalence of a hemolysin(s) (based on the results of a functional biological assay) in water and stool Aeromonas isolates across all three groups (A. hydrophila, A. caviae-A. media, and A. veronii-A. sobria). The mean hemolytic activity of isolates from water samples was 336.5 U, and the standard deviation and median were 850.4 and 80 U, respectively. The corresponding values for the stool isolates were 133.7, 364.5, and 0 U, respectively. The water isolates had statistically significantly higher hemolytic activity than the stool isolates (P < 0.001) (data not shown).
However, the trend in the data was reversed when we considered different groups of Aeromonas separately. For example, the mean hemolytic activity of the A. hydrophila group isolates from water samples was 376.1 U, and for the clinical isolates of the A. hydrophila group it was 525.5 U (Table 5). Likewise, for the A. veronii-A. sobria group, the mean hemolytic activities were 854.5 U for the clinical isolates and 501.0 U for the water isolates (Table 5). Although the differences were not statistically significant, there was clearly an upward trend in the hemolytic activity for the clinical isolates. For the A. caviae-A. media group, however, the mean hemolytic activities of the clinical and water isolates were 1.3 and 2.4 U, respectively. Thus, the values were very low for both the water and stool isolates, which suggested that the members of the A. caviae-A. media group either produce low levels of hemolysins or do not produce hemolysins (Table 5). The mean hemolytic activities of reference Aeromonas strains were 1,160.8 U for strains clinical samples and 279.5 U for strains from water samples (data not shown).
TABLE 5.
Statistical analysis of hemolytic, cytotoxic, and protease activities of isolates of Aeromonas species from water and stool
| Species group | Source of isolates | n | Hemolytic activity |
Cytotoxic activity |
Protease activity |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activity (optical density units/ml) |
P valuea | Activity (optical density units/ml) |
P valuea | Activity (optical density units/ml) |
P valuea | |||||||||
| Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | ||||||
| A. hydrophila group | Water | 87 | 376.1 | 911.9 | 116.7 | 0.105 | 13,716 | 29,717 | 2,382.7 | 0.021 | 7.7 | 6.7 | 4.9 | 0.907 |
| Stool | 7 | 525.5 | 487.3 | 405.0 | 29,820 | 32,572 | 15,437 | 7.3 | 5.3 | 6.2 | ||||
| A. veronii-A. sobria group | Water | 23 | 501.0 | 945.7 | 123.1 | 0.269 | 12,944 | 29,046 | 1,280.0 | 0.065 | 7.0 | 6.8 | 4.0 | 0.446 |
| Stool | 3 | 854.5 | 863.0 | 750.0 | 19,516 | 11,712 | 24,904 | 2.5 | 2.5 | 1.5 | ||||
| A. caviae-A. media group | Water | 21 | 2.4 | 8.0 | 0.0 | 0.268 | 254.8 | 876.6 | 0.0 | 0.037 | 2.9 | 4.1 | 0.8 | 0.079 |
| Stool | 38 | 1.3 | 7.9 | 0.0 | 640.1 | 4,047.7 | 0.0 | 5.4 | 7.1 | 1.6 | ||||
P values were generated by using the Wilcoxon rank-sum test.
Cytotoxic activity.
The cytotoxic activities of various Aeromonas isolates were measured by using the RAW 264.7 macrophage cell line. The distribution of cytotoxic activity in clinical and water sample isolates is shown in Table 4. For both water and stool samples, 98 to 100% of the isolates belonging to the A. hydrophila group produced cytotoxic activity. A small number of A. caviae-A. media group isolates (from water or stools) exhibited cytotoxicity in RAW 264.7 macrophages, and almost no cytotoxicity was detected for the stool isolates (3%) belonging to the A. caviae-A. media group. All of the A. veronii-A. sobria group stool isolates, on the other hand, exhibited toxicity, while only 61% of the members of this group of isolates from water samples exhibited cytotoxic activity (Table 4) and cytotoxic activity was exhibited by 73 and 40% of the atypical aeromonad water and stool isolates, respectively.
Like the prevalence of hemolytic activity, the prevalence of cytotoxic activity in Aeromonas isolates across the three species groups (A. hydrophila, A. caviae-A. media, and A. veronii-A. sobria) was significantly greater for water isolates than for stool isolates. For example, the mean cytotoxic activity for the water isolates was 11,391 U (standard deviation, 27,379 U; median, 1,190.7 U), compared to 6,328 U (standard deviation, 17,071 U; median, 0 U) for the stool isolates, and the difference was statistically significant (P < 0.001). However, when each group of Aeromonas species was examined separately, the opposite was observed. For instance, the mean cytotoxic activity of clinical isolates belonging to the A. hydrophila group (29,820 U) was significantly higher (P = 0.021) than that of the water sample isolates (13,716 U) (Table 5). For the A. veronii-A. sobria group, the mean cytotoxic activities for clinical and water isolates were 19,516 and 12,944 U, respectively (P = 0.065) (Table 5). Likewise, the mean cytotoxic activities associated with the A. caviae-A. media group water and stool isolates were 254.8 and 640.1 U, respectively, indicating that the activity of the latter isolates was significantly higher (P = 0.037) (Table 5). For reference strains, the mean cytotoxic activities of Aeromonas species clinical and water sample isolates were 32,393 and 21,497 U, respectively.
Protease activity.
The distributions of the strains that had protease activity were similar for the water and clinical strains (Table 4). Like the cytotoxic activity, for the A. caviae-A. media group the mean protease activity was greater for the clinical strains (7.1 U) than for the water isolates (2.9 U), but the difference was not statistically significant (P = 0.079). For the A. hydrophila and A. veronii-A. sobria groups, the mean protease activity for the water sample isolates was either similar to or greater than that for the clinical sample isolates (Table 5), but the difference was not statistically significant. The mean protease activities for the reference strains were 4.7 U for clinical isolates and 3.95 U for water isolates.
N-Acyl homoserine lactone production.
When the AHL production data for water and stool isolates belonging to the three Aeromonas species groups were compared using Fisher's exact test, higher percentages of the stool isolates than of the water isolates belonging to the A. hydrophila and A. caviae-A. media groups (P = 0.019 to 0.028) produced more AHLs, whereas the reverse was true for the A. veronii-A. sobria group, for which the percentage of water isolates producing AHLs was higher than the percentage of stool isolates producing AHLs (Table 4 and Fig. 1).
Distribution of virulence factors.
All Aeromonas isolates were examined to determine the presence of 11 virulence genes by using specific probes in colony blot DNA hybridization under high-stringency conditions (see Table S5 in the supplemental material). For water samples, 79% or more of the A. hydrophila group strains harbored six virulence factor genes, the alt, dam, tagA, gidA, ahyRI, and enolase genes, while for the clinical samples, all of the A. hydrophila group isolates contained these six virulence factor genes (Table 6). Approximately 45 to 57% of the A. hydrophila group isolates obtained from both water and clinical samples were positive for the hlyA, ascV, and aexU genes. However, the act gene was more prevalent in water isolates (83%) than in clinical isolates (57%) belonging to the A. hydrophila group.
TABLE 6.
Distribution of virulence factor-encoding genes in water and clinical Aeromonas isolates
| Species | Source of isolates | n | No. (%) of isolates containing the following virulence factor genes: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| act | alt | ast | dam | tagA | gidA | ahyRI | Enolase | hlyA | ascV | aexU | |||
| A. caviae-A. media group | Water | 21 | 4 (19) | 16 (76.2) | 0 (0) | 19 (90.5) | 4 (19) | 21 (100) | 20 (95.2) | 19 (90.5) | 3 (14.3) | 0 (0) | 0 (0) |
| Stool | 38 | 1 (2.6) | 35 (92.1) | 0 (0) | 37 (97.4) | 3 (7.9) | 36 (94.7) | 37 (97.4) | 34 (89.5) | 3 (7.9) | 0 (0) | 0 (0) | |
| A. hydrophila group | Water | 87 | 72 (82.8) | 84 (96.6) | 69 (79.3) | 74 (85.1) | 69 (79.3) | 87 (100) | 86 (98.9) | 82 (94.3) | 37 (44.5) | 39 (44.8) | 46 (52.9) |
| Stool | 7 | 4 (57.1) | 7 (100) | 6 (85.7) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 4 (57.1) | 4 (57.1) | 4 (57.1) | |
| A. veronii-A. sobria group | Water | 23 | 10 (39.1) | 13 (56.5) | 1 (4.3) | 18 (78.3) | 2 (8.7) | 20 (87) | 7 (30.4) | 20 (87) | 0 (0) | 14 (60.9) | 17 (73.9) |
| Stool | 3 | 3 (100) | 2 (66.7) | 0 (0) | 3 (100) | 0 (0) | 3 (100) | 0 (0) | 3 (100) | 0 (0) | 1 (33.3) | 1 (33.3) | |
| Atypical aeromonads | Water | 15 | 10 (66.7) | 13 (86.7) | 5 (33.3) | 14 (93.3) | 10 (66.7) | 15 (100) | 15 (100) | 14 (93.3) | 4 (26.7) | 7 (46.7) | 5 (33.3) |
| Stool | 5 | 2 (40) | 4 (80) | 1 (20) | 5 (100) | 1 (20) | 3 (60) | 5 (100) | 4 (80) | 1 (20) | 0 (0) | 0 (0) | |
Table 6 also shows the distribution of the 11 virulence genes in both water and clinical isolates belonging to the A. caviae-A. media and A. veronii-A. sobria groups and the atypical aeromonad species. For the A. caviae-A. media group, the alt, dam, and ahyRI genes were more prevalent in clinical isolates than in water isolates. In addition to the alt and dam genes in the A. veronii-A. sobria group, the act, gidA, and enolase genes were more common in clinical isolates than in water isolates. Further, T3SS genes (ascV and aexU) were found in both water and clinical isolates belonging to the A. hydrophila and A. veronii-A. sobria groups, while none of the water and stool isolates in the A. caviae-A. media group harbored these T3SS genes. For the reference strains, 61.5% of the clinical isolates and 16.7% of the water isolates harbored act, while 38.5% of the clinical isolates and 50% of water isolates contained both the ascV and aexU genes (data not shown).
PFGE analysis.
Of the 227 isolates studied, 226 were typeable by PFGE and are shown in a similarity dendrogram in Fig. S1 in the supplemental material. The one nontypeable isolate, EPA-3, was isolated from water and was identified as a member of the A. veronii-A. sobria group. XbaI digestion resulted in 15 to 24 well-resolved genomic DNA bands ranging in size from approximately 35 to 780 kb. Among the 226 isolates, there were 177 distinct pulsotypes exhibiting extensive genetic diversity. When all pairwise comparisons were analyzed, the median level of similarity for all isolates was 66.7% (range, 37.8% to 100%; n = 25,425), and for the three species groups the median levels of similarity were 66.7% for the A. hydrophila group (range, 43.9% to 100%; n = 5,460), 70.8% for the A. veronii-A. sobria group (range, 55.8% to 100%; n = 406), and 66.7% for the A. caviae-A. media group (range, 37.9% to 100%; n = 2,016).
To determine the clonal relationships between the water and clinical isolates, we compared the virulence clusters with the results of the cluster analysis of the PFGE data and found three sets of isolates from stool and water samples that were indistinguishable by PFGE and had the same or similar virulence signatures (Fig. 2 and Table 7). This is not surprising since one of us (N.P.M.) reported previously that for one pair of isolates, NM-22 and NM-33, the conventional biochemical identification was the same (A. caviae) and the isolates had the same ribotype pattern (53). Likewise, the isolate pair NM-14 and NM-35 also had the same conventional biochemical identification (A. caviae) and identical ribotype patterns, although that ribotype pattern differed from that of NM-22 and -33 (unpublished data).
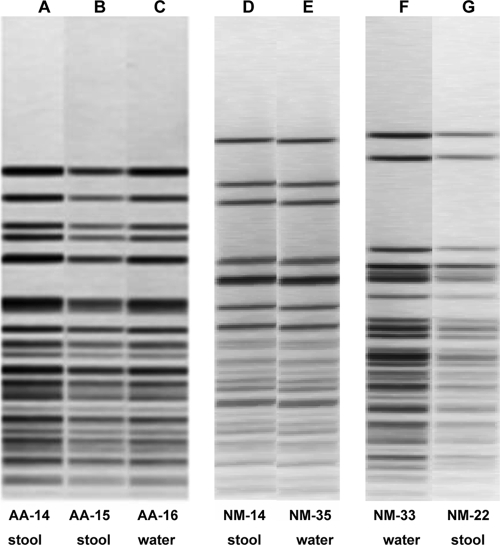
FIG. 2.
PFGE profile analysis of three sets of water and clinical Aeromonas isolates that had indistinguishable PFGE patterns. Lane A, AA-14 (A. caviae-A. media group); lane B, AA-15 (A. caviae-A. media group); lane C, AA-16 (A. caviae-A. media group); lane D, NM-14 (A. caviae-A. media group); lane E, NM-35 (A. caviae-A. media group); lane F, NM-33 (A. caviae-A. media group); lane G, NM-22 (A. caviae-A. media group).
TABLE 7.
Virulence patterns for three sets of Aeromonas isolates that are indistinguishable by PFGE
| Isolate | Source | Species group | Hemolytic activityb | Cytotoxic activityb | Proteolytic activityb | Lactone production | ahyRI | act | alt | ast | dam | tagA | gidA | Enolase gene | hlyA | ascV | aexU | T6SS effectorsa |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hcp | VgrG2- VgrG3 | ||||||||||||||||||
| AA-14 | Stool | A. caviae-A. media group | 0 | 0 | 3.4 | − | + | − | + | − | + | − | + | + | − | − | − | + | + |
| AA-15 | Stool | A. caviae-A. media group | 0 | 0 | 3.7 | − | + | − | + | − | + | − | + | + | − | − | − | + | + |
| AA-16 | Water | A. caviae-A. media group | 0 | 0 | 2.2 | − | + | − | + | − | + | − | + | + | − | − | − | + | + |
| NM-14 | Stool | A. caviae-A. media group | 0 | 0 | 1.2 | ++ | + | − | + | − | + | − | + | + | − | − | − | + | + |
| NM-35 | Water | A. caviae-A. media group | 0 | 0 | 0.3 | ++ | + | − | + | − | + | − | + | + | − | − | − | + | + |
| NM-22 | Stool | A. caviae-A. media group | 0 | 0 | 0.6 | − | + | − | + | − | + | − | + | + | − | − | − | − | − |
| NM-33 | Water | A. caviae-A. media group | 0 | 0 | 1.0 | + | + | − | + | − | + | − | + | + | − | − | − | − | − |
Abbreviations: Hcp, hemolysin-coregulated protein; VgrG, valine-glycine repeat G protein.
In optical density units/ml.
Epidemiological data.
NM-22 was isolated in 1988 from a stool from a child in Iowa that was ill with gastroenteritis and had presumably been exposed to water from the family well. NM-33 was isolated from the suspected infection source, the private household well used by the child's family. This epidemiologic linkage, the indistinguishable pulsotypes (Fig. 2), and the identical virulence patterns strongly suggest that these isolates are truly identical and that the child's Aeromonas infection was probably the result of waterborne transmission (Table 7).
The members of the second pulsotype and virulence group, NM-14 and NM-35, were not quite as tightly linked epidemiologically since NM-14 was obtained from a stool sample from a child in Iowa with diarrhea that was collected in early August 1987 and NM-35 was isolated from a different private farm well in another geographic area of Iowa in 1986, the year before NM-14 was isolated (Table 7). The most interesting fact is that a third strain of A. caviae (which was not available for this study) had the same ribotype as NM-14 and NM-35 and was submitted by a local hospital laboratory to a reference bacteriology laboratory in 1988 (N. Moyer, unpublished data). This suggests that genotypically identical strains, all belonging to the A. caviae-A. media group, were present in the environment and caused disease in patients between 1986 and 1988 (well sample collected in 1986, stool sample collected in 1987, and hospital laboratory referral in 1988). Interestingly, NM-2, -3, and -4 isolates were from the siblings with chronic diarrhea, and A. caviae was the only enteric pathogen isolated from the stools. They had similar ribotypes (53), and NM-2 and -3 isolates had identical PFGE patterns (see Fig. S1 in the supplemental material). NM-19 (also A. caviae with PFGE pattern identical to that of NM-2 and -3) was from the stool of an unrelated child with chronic diarrhea whose family had a private well approximately 10 miles from the well of the family with the above-mentioned siblings. Although NM-2, -3, and -4 were recovered from stools in 1984, NM-19 was isolated in 1996. Interestingly, all of these isolates had similar virulence signatures (see Table S5 in the supplemental material). These data might suggest that very similar strains were circulating in the population and environment over an extended period of time.
Three matching strains, AA-14, AA-15, and AA-16, were collected during a “dysentery” outbreak in rural villages in the Sudan in January 1984 (Table 7). AA-14 was isolated from a 1-year-old male with acute lymphocytic leukemia in the Kassala area (capital of the Sudan, south of Port Sudan) and with watery and bloody diarrheal stools (7 stools in the previous 24 h) that were also positive for fecal leukocytes and negative for other common enteric pathogens, including parasites and viruses. AA-15 was isolated from a 27-year-old female in the Port Sudan area with fever and diarrhea (less than 3 stools in the previous 24 h, no blood or white cells) who was also negative for other common enteric pathogens, including parasites and viruses. AA-16 was a well water isolate from the rural town of Sinkat, which is just outside Kassala on the road from Port Sudan. Again, although these three A. caviae-A. media group strains were not strictly linked epidemiologically, their indistinguishable PFGE pulsotypes (Fig. 2) combined with their identical or nearly identical virulence patterns (Table 7) suggest that they represent possible water-to-human transmission.
Detection of T6SS effectors.
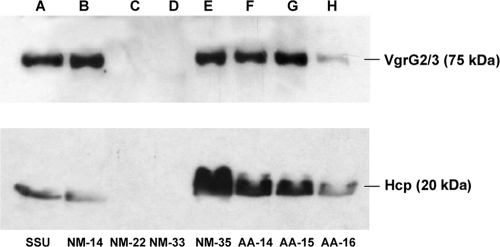
We then performed a Western blot analysis with 7 isolates belonging to the A. caviae-A. media group that exhibited similar DNA fingerprints and virulence signatures for detection of T6SS effectors (Hcp and VgrG2-VgrG3) in the culture supernatant. As shown in Fig. 3 and Table 7, 5 of these 7 isolates (AA-14, AA-15, AA-16, NM-14, and NM-35), which represented pairs of water and stool isolates, synthesized the Hcp and VgrG2-VgrG3 effectors of the T6SS and secreted them into the culture supernatant. We did not detect T6SS effectors in paired water and stool strains NM-22 and NM-33.
FIG. 3.
Western blot analysis for detection of the secreted T6SS effectors Hcp and VgrG2-VgrG3 in culture supernatants of seven water and clinical Aeromonas isolates. Lane A, A. hydrophila SSU (positive control); lane B, NM-14 (A. caviae-A. media group); lane C, NM-22 (A. caviae-A. media group); lane D, NM-33 (A. caviae-A. media group); lane E, NM-35 (A. caviae-A. media group); lane F, AA-14 (A. caviae-A. media group); lane G, AA-15 (A. caviae-A. media group); lane H, AA-16 (A. caviae-A. media group).
DISCUSSION
Aeromonas species inhabit various aquatic environments, and these pathogens can infect humans in several ways, including via recreational or occupational activities in water (e.g., fishing or swimming) and consumption of contaminated food or water. Previous studies have shown that Aeromonas species isolates from water possess various virulence factors and that these isolates can cause human diseases (11, 41, 80). However, the mode of transmission of these pathogens from water to humans and the bacterial virulence factors and host responses that eventually lead to Aeromonas-associated diseases are still not clearly understood. Many investigators, including us, have been seeking new virulence factors in Aeromonas species to better establish a relationship between the presence of virulence factors and the disease state in the host. Thus, it is important to examine the distribution of newly identified virulence factors in water and clinical Aeromonas isolates and to characterize their role in the virulence of Aeromonas species. In this study, we examined the presence of 11 virulence genes and clonal relatedness for a large number of water and clinical strains of different Aeromonas species isolated from various regions in the United States and other parts of the world. Further, our aim was to identify a subset of Aeromonas species that are prevalent in water and capable of causing human disease.
Two of the earliest studies that presented somewhat compelling epidemiological data linking aeromonads to gastrointestinal disease were a study by Echeverria et al., who studied the causes of traveler's diarrhea in Peace Corp volunteers in Thailand in 1981 (17), and a study performed by Holmberg et al. in the United States in 1984 (29). The latter study was performed by investigators at CDC in Atlanta, GA, who evaluated 34 cases of gastroenteritis where Aeromonas species had been isolated from fecal specimens. Their conclusions were that some strains of Aeromonas could have caused diarrhea in previously normal hosts and that the organisms were most likely acquired by drinking untreated water. This study was followed in 1987 by a study by one of us (N.P.M.) of the clinical significance of Aeromonas species isolated from patients with diarrhea. This study examined 248 strains of Aeromonas isolated from 3,334 human fecal specimens submitted to a state public health laboratory over a 20-year period for culturing of enteric pathogens. The conclusions of this study were that some A. hydrophila, A. sobria (now A. veronii biovar sobria), and A. caviae strains were “capable of causing diarrhea and that antibiotic therapy and the drinking of untreated water were significant risk factors for susceptible hosts” (52). At the same time, researchers at the University of Maryland in College Park, MD, and at the Naval Medical Research Institute (NMRI) in Bethesda, MD, studied and reidentified isolates in previously described aeromonad strain collections from both clinical and environmental sources to promote and prove the value of accurate differential biotyping of aeromonads and plesiomonads in the environment (39).
In a later study in 1997, it was noted that only a small subset of Aeromonas strains belonging to specific hybridization groups (HG) can cause gastroenteritis in humans (45). Further, both Kirov et al. (42) and Hanninen et al. (26) indicated that HG 1 was associated with clinical specimens, while HG 3 and, to a lesser extent, HG 2 predominated in water and environmental samples. In 1997 and 1998, we (in collaboration with the International Centre for Diarrheal Disease Research, Bangladesh [ICDDR,B]) investigated the distribution of enterotoxin genes (alt, ast, act) in aeromonads isolated from children with diarrhea (n = 115), healthy matched controls (n = 27), and different water sources (n = 120) in Bangladesh in order to establish a link between virulence properties of aeromonads and the cause of gastroenteritis in human (5). In this study, 125 of 1,735 children with diarrhea were positive for aeromonads and other enteric pathogens (7.2%), and 28 (22%) of these children only had Aeromonas species in their stools. Only 27 of 830 (3.3%) control children enrolled in this study were positive for aeromonads. Of 2,120 environmental samples tested, 600 (28.3%) were positive for aeromonads and every fifth sample was subsequently examined to determine the distribution of enterotoxin genes. Our findings revealed that the number of isolates that were positive for both the alt and ast genes was significantly higher for children with diarrhea than for control children and the water sources. Further, patients from whom isolates positive for both the alt and ast genes were obtained had watery diarrhea, while patients from whom isolates positive for only the alt gene were obtained had loose stools (5). Another study reported that in the United States, 43, 70, and 30% of the Aeromonas isolates obtained by the U.S. EPA (a total of 205 isolates were examined) from drinking water from 16 utilities in four states harbored the alt, act, and ast genes, respectively (61). In addition, a recent study in Bangladesh showed that certain Aeromonas clones persisted in different sewage water treatment plants and that the same clones were found in fish and in children suffering from diarrhea (60). The authors concluded that certain genotypically and phenotypically stable clonal lineages of Aeromonas might spread from hospitalized children suffering from diarrhea to fish produced for human consumption through the sewage water treatment system (60).
Despite the association of virulence factors with aeromonads isolated from drinking water, there is increasing evidence that strains isolated from the environment generally belong to different hybridization groups than clinically associated strains. For example, Havelaar et al. (27) examined 187 Aeromonas strains from human diarrheal stools and 263 strains from drinking water in the Netherlands and observed little similarity between the two groups. Likewise, Borchardt et al. (8) noted that stool and water isolates were genetically unrelated. Therefore, it is still not clear whether the strains that are most prevalent and virulent in the environment are responsible for human diseases. Consequently, the current study was very important for resolution of this contradiction because it investigated the virulence potential and clonal relationship between water and clinical isolates of Aeromonas species isolated from different locations in the United States and different parts of the world.
In addition to the alt, act, and ast genes, we examined eight other virulence factor genes, including some of the newly identified genes, in Aeromonas isolates by using colony blot DNA hybridization. Since we used gene probes developed based on the sequences of one strain, A. hydrophila SSU, signals with different intensities for other strains and species of Aeromonas due to possible divergence in the sequences of the genes were expected. However, we obtained very clean and strong signals in our blots by using a technique that we modified and standardized in our laboratory. Although there have been arguments about the reproducibility of PCR-based typing methods (2, 76, 77), many investigators have used such techniques for detection of different virulence factor-encoding genes (61, 68). However, optimization of primers and PCR conditions is very crucial to obtain the best results (68), which is feasible only when the number of strains tested is small. Since in our study we examined more than 200 isolates, colony blot DNA hybridization was perhaps the best option to screen various virulence factor genes, although this approach has some limitations.
In the first step during statistical analysis of the data, we compared the prevalence of discriminating variables (i.e., the genotypic and phenotypic traits) one at a time for isolates collected from the two sources of interest, water and stools. We reasoned that the members of three species groups, the A. caviae-A. media, A. hydrophila, and A. veronii-A. sobria groups, could be grouped for analysis because these species are often found in water and together are responsible for 85% of clinical infections (34). Initially, we assumed that grouping across species would be the best method for statistical analysis because it would increase the sample size and improve the statistical power. However, we later obtained different outcomes when we analyzed subsets by species. To elaborate, when data were grouped across species, the stool and water isolates were statistically different for the prevalence of 11 genetic traits. For 10 of the genes, the prevalence was always much greater in the isolates collected from water. The prevalence was higher in stool isolates only for the dam gene. Likewise, the values for three phenotypic traits (protease activity, hemolytic activity, and cytotoxic activity) were much higher for the isolates from water, and the hemolysis and cytotoxicity values for these isolates were highly significantly different from those for the stool isolates.
However, when we analyzed the subsets of the species, a surprising result was obtained. Many of the genetic virulence traits were more prevalent in clinical isolates belonging to the three groups of Aeromonas isolates than in water isolates. Moreover, the values for phenotypic traits (e.g., cytotoxic activity) were significantly higher for the stool isolates belonging to the A. caviae-A. media and A. hydrophila groups, and they were also higher for stool isolates belonging to the A. veronii-A. sobria group, although the significance was borderline (Table 5). Other investigators have also reported that clinical isolates of A. hydrophila harbor a higher proportion of hemolysin genes (11) and exhibit greater cytotoxicity with CHO-K1 cells (44).
The fact that a pattern is the same for three species when they are analyzed separately but the pattern is reversed when the species are combined into one group is a little-known paradox called the reversal of inequalities or Simpson's paradox (75). This paradox arises when groups that are not the same size are combined and an unknown variable called a lurking variable is different in the different groups. The lurking variable skews the results in the direction opposite the direction observed when the groups are considered separately. In the present study, this was best illustrated by the cytotoxicity data. When the three Aeromonas species groups were examined separately, in each case the stool isolates produced substantially more cytotoxic activity than the water isolates. Even though this pattern was consistent for all three species groups, when the species groups were combined, the water isolates appeared to produce more cytotoxic activity than the stool isolates. Examining the cytotoxin levels by species group instead of by source, we noticed that members of the A. caviae-A. media group produced 2-orders-of-magnitude less cytotoxin than members of the two other species groups; moreover, the majority of the isolates from stools (72%) are members of the A. caviae-A. media group. Most isolates belonging to the A. hydrophila and A. veronii-A. sobria groups were obtained from water. This association between Aeromonas species group and isolate source is the lurking variable, and in the combined analysis the large number of A. caviae-A. media group isolates from stools that produced low levels of cytotoxin skewed the average downward so that it appeared that the water isolates produced more cytotoxin than the stool isolates.
In fact, the three species groups are significantly different in nearly all genotypic and phenotypic traits, and it is the differences and the association between species groups and source that could have led to the erroneous conclusion that virulence traits were more prevalent in water isolates than in stool isolates. The lesson is that it is important to correctly place isolates in the proper Aeromonas “species group” before any predictive model for discriminating stool and water isolates is applied.
The majority of the water (>78%) and stool (>90%) isolates belonging to the Aeromonas species groups (A. hydrophila, A. caviae-A. media, and A. veronii-A. sobria) possessed three virulence factor-encoding genes, the dam, gidA, and enolase genes. Based on these data, we hypothesized that these three genes are essential for the Aeromonas species to survive in the environment, as well as to cause human disease. Further, these three genes could be used as molecular identification markers of Aeromonas species if there are difficulties in serological and biochemical identification. A. caviae-A. media group strains were found to contain fewer virulence genes than the strains belonging to the other Aeromonas species groups, and these results agree with the data presented in a previous study (80). The alt and dam genes were more common in clinical strains than in water isolates in all three groups (Table 6). Further, for the clinical strains, the alt gene was found in 92% of the A. caviae-A. media group strains, 100% of the A. hydrophila group strains, and 67% of the A. veronii-A. sobria group strains, while the percentages were 76, 97, and 57% for the water isolates, respectively (Table 6). These results are in accordance with the results of our previous study (5) that implied that the alt gene encoding a heat-labile cytotonic enterotoxin is one of the major virulence factors of Aeromonas-associated diarrheal disease.
We and other workers (15, 65, 67, 81) have investigated the contribution of the T3SS to the virulence of the Aeromonas species. However, Silver et al. recently demonstrated that the functional T3SS of A. veronii was crucial for both beneficial and pathogenic colonization of the host (for example, in the leech and in the mouse model, respectively) (69). Other investigators also reported that the T3SS was required for intercellular beneficial associations, such as those in the insect endosymbiont Sodalis glossinidius (16) and Rhizobium-leguminous plant associations (79). Therefore, further studies to obtain a clear understanding of the role of bacterial T3SS in pathogenic and beneficial associations in different host environments are warranted.
In the present study, irrespective of the Aeromonas species, we found that the T3SS genes were more prevalent in water isolates (for aexU, 68 of 146 isolates were positive; for ascV, 60 of 146 isolates were positive) than in clinical isolates (for aexU, 5 of 53 isolates were positive; for ascV, 5 of 53 isolates were positive), a finding in agreement with data obtained in another recent study (4). However, these genes were more prevalent in clinical isolates than in water isolates specifically for the A. hydrophila group (Table 6). Based on these results, it is plausible that the T3SS might play a role in the ability of A. hydrophila group isolates to cause human diarrhea. However, we should interpret these data cautiously, as a limited number of clinical isolates were used in the study. In a study conducted in Thailand, 11% and 30% of the clinical isolates belonging to the A. caviae complex harbored the ascV and aexT (aexU-like) genes, respectively (80). However, in our study, none of the clinical and water isolates belonging to the A. caviae-A. media group contained these genes. These different results can be explained by the fact that the pathogenic mechanisms of the diseases caused by Aeromonas species may be different in different geographic regions. Nevertheless, a comparative analysis of isolates from different geographic locations would be important for understanding the pathogenesis of Aeromonas-associated infections.
AHL is an autoinducer for QS regulation, and it has been reported that AHL-mediated QS is a global regulator and controller of a number of virulence factors in many pathogens (7, 32, 70). Like the data for cytotoxic activity, higher AHL levels were detected in stool isolates than in water isolates for both the A. hydrophila and A. caviae-A. media groups (Table 4 and Fig. 1). Our recently published data (40) indicated that deletion of the ahyRI genes from A. hydrophila SSU decreased biofilm formation and affected the production of metalloprotease and secretion of the T6SS effectors Hcp and VgrG2-VgrG3. Further, the ahyRI mutant was less virulent in a mouse septicemia model. Therefore, increased production of AHLs by stool isolates belonging to the A. caviae-A. media and A. hydrophila groups could directly or indirectly modulate bacterial virulence. In addition, pairs of isolates belonging to the A. caviae-A. media group obtained from water and stools (e.g., NM-14 and NM-35) synthesized and secreted the Hcp and VgrG2-VgrG3 effectors of T6SS (Fig. 3 and Table 7) and also synthesized AHLs, indicating that these lactones might be involved in regulating T6SS effectors in these strains.
The PFGE analysis and colony blot DNA hybridization data revealed that three sets of isolates from stool and water samples had indistinguishable PFGE patterns and had the same or similar virulence signatures (Fig. 2 and Table 7). Based on our biochemical identifications, the seven strains were identified as members of the A. caviae-A. media group. Further, these strains in the A. caviae-A. media group harbored only 4 of the 11 virulence or regulatory genes tested (the alt, dam, gidA, and enolase genes). These strains also did not produce cytotoxic and hemolytic activities, as they did not have the act gene (Table 7). Nevertheless, 5 of the 7 paired water and stool strains possessed a functional T6SS (Fig. 3 and Table 7), which is one of the major virulence mechanisms of A. hydrophila SSU characterized recently in our laboratory (71, 72). Therefore, it would be interesting to examine in detail the virulence mechanism(s) of these selected A. caviae-A. media group isolates.
Finally, in the present study, we observed that members of the A. hydrophila and A. veronii-A. sobria groups contained more virulence genes and had greater cytotoxic and hemolytic activities than members of the A. caviae-A. media group. Thus, it seems possible that the A. hydrophila and A. veronii-A. sobria groups have much greater potential to cause human diseases than the A. caviae-A. media group. One limitation of the present study was that there were fewer clinical isolates, particularly in the A. hydrophila and A. veronii-A. sobria groups. Therefore, further investigation with a large number of clinical and water isolates of Aeromonas species is crucial to better understand the pathogenic mechanism of Aeromonas species. In conclusion, our data suggest that there is water-to-human transmission, at least for the A. caviae-A. media group, and that exposure to aeromonads in water can lead to gastroenteritis in humans.
Supplementary Material
Acknowledgments
This research was supported by grants from the AwwaRF and the Environmental Protection Agency and by NIH/NIAID grant AI041611. NSF grant EF-0334227, which was a conduit for sequencing of the A. hydrophila ATCC 7966T genome, is also acknowledged. Bijay K. Khajanchi was a recipient of a J. W. McLaughlin predoctoral fellowship at UTMB.
We thank Phil Bertz for his technical assistance with performance of PFGE, Irina Moffitt for her technical assistance with performance of the biochemical phenotypic analyses, and Louis Bourgeois for donation of strains AA-13 to AA-16 from the Sudan. We also thank Mark Rogers, Keya Sen, and Jennifer Best of EPA, Cincinnati, OH, for providing Aeromonas isolates for this study. We thank Mardelle Susman for her careful editing of the manuscript.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbott, S. L., W. K. Cheung, and J. M. Janda. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41:2348-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdullah, A. I., C. A. Hart, and C. Winstanley. 2003. Molecular characterization and distribution of virulence-associated genes amongst Aeromonas isolates from Libya. J. Appl. Microbiol. 95:1001-1007. [DOI] [PubMed] [Google Scholar]
- 3.Abuhammour, W., R. A. Hasan, and D. Rogers. 2006. Necrotizing fasciitis caused by Aeromonas hydrophila in an immunocompetent child. Pediatr. Emerg. Care 22:48-51. [DOI] [PubMed] [Google Scholar]
- 4.Aguilera-Arreola, M. G., C. Hernandez-Rodriguez, G. Zuniga, M. J. Figueras, and G. Castro-Escarpulli. 2005. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 242:231-240. [DOI] [PubMed] [Google Scholar]
- 5.Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-244. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 7.Bertani, I., and V. Venturi. 2004. Regulation of the N-acyl homoserine lactone-dependent quorum-sensing system in rhizosphere Pseudomonas putida WCS358 and cross-talk with the stationary-phase RpoS sigma factor and the global regulator GacA. Appl. Environ. Microbiol. 70:5493-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borchardt, M. A., M. E. Stemper, and J. H. Standridge. 2003. Aeromonas isolates from human diarrheic stool and groundwater compared by pulsed-field gel electrophoresis. Emerg. Infect. Dis. 9:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruhn, J. B., I. Dalsgaard, K. F. Nielsen, C. Buchholtz, J. L. Larsen, and L. Gram. 2005. Quorum sensing signal molecules (acylated homoserine lactones) in gram-negative fish pathogenic bacteria. Dis. Aquat. Org. 65:43-52. [DOI] [PubMed] [Google Scholar]
- 10.Carnahan, A. M., S. Behram, and S. W. Joseph. 1991. Aerokey II: a flexible key for identifying clinical Aeromonas species. J. Clin. Microbiol. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chacon, M. R., M. J. Figueras, G. Castro-Escarpulli, L. Soler, and J. Guarro. 2003. Distribution of virulence genes in clinical and environmental isolates of Aeromonas spp. Antonie Van Leeuwenhoek 84:269-278. [DOI] [PubMed] [Google Scholar]
- 12.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 13.Chopra, A. K., C. W. Houston, J. W. Peterson, and G. F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 14.Chopra, A. K., J. W. Peterson, X. J. Xu, D. H. Coppenhaver, and C. W. Houston. 1996. Molecular and biochemical characterization of a heat-labile cytotonic enterotoxin from Aeromonas hydrophila. Microb. Pathog. 21:357-377. [DOI] [PubMed] [Google Scholar]
- 15.Dacanay, A., L. Knickle, K. S. Solanky, J. M. Boyd, J. A. Walter, L. L. Brown, S. C. Johnson, and M. Reith. 2006. Contribution of the type III secretion system (TTSS) to virulence of Aeromonas salmonicida subsp. salmonicida. Microbiology 152:1847-1856. [DOI] [PubMed] [Google Scholar]
- 16.Dale, C., S. A. Young, D. T. Haydon, and S. C. Welburn. 2001. The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc. Natl. Acad. Sci. U. S. A. 98:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echeverria, P., N. R. Blacklow, L. B. Sanford, and G. G. Cukor. 1981. Travelers' diarrhea among American Peace Corps volunteers in rural Thailand. J. Infect. Dis. 143:767-771. [DOI] [PubMed] [Google Scholar]
- 18.Erova, T. E., A. A. Fadl, J. Sha, B. K. Khajanchi, L. L. Pillai, E. V. Kozlova, and A. K. Chopra. 2006. Mutations within the catalytic motif of DNA adenine methyltransferase (Dam) of Aeromonas hydrophila cause the virulence of the Dam-overproducing strain to revert to that of the wild-type phenotype. Infect. Immun. 74:5763-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erova, T. E., V. G. Kosykh, A. A. Fadl, J. Sha, A. J. Horneman, and A. K. Chopra. 2008. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J. Bacteriol. 190:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erova, T. E., J. Sha, A. J. Horneman, M. A. Borchardt, B. K. Khajanchi, A. A. Fadl, and A. K. Chopra. 2007. Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol. Lett. 275:301-311. [DOI] [PubMed] [Google Scholar]
- 22.Faisal, M., A. E. Eissa, and E. E. Elsayed. 2007. Isolation of Aeromonas salmonicida from sea lamprey (Petromyzon marinus) with furuncle-like lesions in Lake Ontario. J. Wildl. Dis. 43:618-622. [DOI] [PubMed] [Google Scholar]
- 23.Figueras, M. J., M. J. Aldea, N. Fernandez, C. Aspiroz, A. Alperi, and J. Guarro. 2007. Aeromonas hemolytic uremic syndrome. A case and a review of the literature. Diagn. Microbiol. Infect. Dis. 58:231-234. [DOI] [PubMed] [Google Scholar]
- 24.Figueras, M. J., A. J. Horneman, A. Martinez-Murcia, and J. Guarro. 2007. Controversial data on the association of Aeromonas with diarrhoea in a recent Hong Kong study. J. Med. Microbiol. 56:996-998. (Author reply, 56:998). [DOI] [PubMed] [Google Scholar]
- 25.Galindo, C. L., C. Gutierrez, Jr., and A. K. Chopra. 2006. Potential involvement of galectin-3 and SNAP23 in Aeromonas hydrophila cytotoxic enterotoxin-induced host cell apoptosis. Microb. Pathog. 40:56-68. [DOI] [PubMed] [Google Scholar]
- 26.Hanninen, M. L., P. Oivanen, and V. Hirvela-Koski. 1997. Aeromonas species in fish, fish-eggs, shrimp and freshwater. Int. J. Food Microbiol. 34:17-26. [DOI] [PubMed] [Google Scholar]
- 27.Havelaar, A. H., F. M. Schets, A. van Silfhout, W. H. Jansen, G. Wieten, and D. van der Kooij. 1992. Typing of Aeromonas strains from patients with diarrhoea and from drinking water. J. Appl. Bacteriol. 72:435-444. [DOI] [PubMed] [Google Scholar]
- 28.Hiransuthikul, N., W. Tantisiriwat, K. Lertutsahakul, A. Vibhagool, and P. Boonma. 2005. Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin. Infect. Dis. 41:e93-e96. [DOI] [PubMed] [Google Scholar]
- 29.Holmberg, S. D., W. L. Schell, G. R. Fanning, I. K. Wachsmuth, F. W. Hickman-Brenner, P. A. Blake, D. J. Brenner, and J. J. Farmer III. 1986. Aeromonas intestinal infections in the United States. Ann. Intern. Med. 105:683-689. [DOI] [PubMed] [Google Scholar]
- 30.Horneman, A. J., A. Ali, and S. Abbott. 2007. Aeromonas, p. 716-722. In P. R. Murray, E. J. Baron, J. H. Jorgenson, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 31.Howard, S. P., and J. T. Buckley. 1985. Activation of the hole-forming toxin aerolysin by extracellular processing. J. Bacteriol. 163:336-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain, M. B. B. M., H.-B. Zhang, J.-L. Xu, Q. Liu, Z. Jiang, and L.-H. Zhang. 2008. The acyl-homoserine lactone-type quorum-sensing system modulates cell motility and virulence of Erwinia chrysanthemi pv. zeae. J. Bacteriol. 190:1045-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janda, J. M. 2001. Aeromonas and Plesiomonas, p. 1237-1270. In M. Sussman (ed.), Molecular medical microbiology, vol. 2. Academic Press, London, United Kingdom. [Google Scholar]
- 34.Janda, J. M. 1991. Recent advances in the study of the taxonomy, pathogenicity, and infectious syndromes associated with the genus Aeromonas. Clin. Microbiol. Rev. 4:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janda, J. M., and S. L. Abbott. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45:2761-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 37.Joseph, S. W. 1996. Aeromonas gastrointestinal disease: a case study in causation?, p. 311. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas, 1st ed. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 38.Joseph, S. W., and A. M. Carnahan. 2000. Update on the genus Aeromonas. ASM News 66:218-223. [Google Scholar]
- 39.Joseph, S. W., A. Carnahan, D. Rollins, and R. I. Walker. 1988. Aeromonas and Plesiomonas in the environment: value of differential biotyping of aeromonads. J. Diarrhoeal Dis. Res. 6:80-87. [PubMed] [Google Scholar]
- 40.Khajanchi, B. K., J. Sha, E. V. Kozlova, T. E. Erova, G. Suarez, J. C. Sierra, V. L. Popov, A. J. Horneman, and A. K. Chopra. 2009. N-Acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155:3518-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kingombe, C. I., G. Huys, M. Tonolla, M. J. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirov, S. M., E. K. Ardestani, and L. J. Hayward. 1993. The growth and expression of virulence factors at refrigeration temperature by Aeromonas strains isolated from foods. Int. J. Food Microbiol. 20:159-168. [DOI] [PubMed] [Google Scholar]
- 43.Kozlova, E. V., V. L. Popov, J. Sha, S. M. Foltz, T. E. Erova, S. L. Agar, A. J. Horneman, and A. K. Chopra. 2008. Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 45:343-354. [DOI] [PubMed] [Google Scholar]
- 44.Krovacek, K., V. Pasquale, S. B. Baloda, V. Soprano, M. Conte, and S. Dumontet. 1994. Comparison of putative virulence factors in Aeromonas hydrophila strains isolated from the marine environment and human diarrheal cases in southern Italy. Appl. Environ. Microbiol. 60:1379-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhn, I., M. J. Albert, M. Ansaruzzaman, N. A. Bhuiyan, S. A. Alabi, M. S. Islam, P. K. Neogi, G. Huys, P. Janssen, K. Kersters, and R. Mollby. 1997. Characterization of Aeromonas spp. isolated from humans with diarrhea, from healthy controls, and from surface water in Bangladesh. J. Clin. Microbiol. 35:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeChevallier, M. W., T. M. Babcock, and R. G. Lee. 1987. Examination and characterization of distribution system biofilms. Appl. Environ. Microbiol. 53:2714-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ljungh, A., and T. Kronevi. 1982. Aeromonas hydrophila toxins—intestinal fluid accumulation and mucosal injury in animal models. Toxicon 20:397-407. [DOI] [PubMed] [Google Scholar]
- 48.Martin-Carnahan, A., and S. W. Joseph. 2005. Aeromonadaceae, p. 556-578. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 49.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 50.Merino, S., A. Aguilar, M. M. Nogueras, M. Regue, S. Swift, and J. M. Tomas. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merino, S., X. Rubires, S. Knochel, and J. M. Tomas. 1995. Emerging pathogens: Aeromonas spp. Int. J. Food Microbiol. 28:157-168. [DOI] [PubMed] [Google Scholar]
- 52.Moyer, N. P. 1987. Clinical significance of Aeromonas species isolated from patients with diarrhea. J. Clin. Microbiol. 25:2044-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moyer, N. P. 2002. The quest for understanding: a history of Aeromonas research, p. 26. 7th Int. Symp. Aeromonas and Plesiomonas, 8 to 10 September 2002, Orihuela (Alicante), Spain.
- 54.Moyer, N. P., G. M. Luccini, L. A. Holcomb, N. H. Hall, and M. Altwegg. 1992. Application of ribotyping for differentiating aeromonads isolated from clinical and environmental sources. Appl. Environ. Microbiol. 58:1940-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moyer, N. P., G. Martinettim, J. Luthy-Hottenstein, and M. Altwegg. 1992. Value of rRNA gene restriction patterns of Aeromonas spp. for epidemiological investigations. Curr. Microbiol. 24:15-21. [Google Scholar]
- 56.Nord, C. E., L. Sjoberg, T. Wadstrom, and B. Wretlind. 1975. Characterization of three Aeromonas and nine Pseudomonas species by extracellular enzymes and haemolysins. Med. Microbiol. Immunol. 161:79-87. [DOI] [PubMed] [Google Scholar]
- 57.Pillai, L., J. Sha, T. E. Erova, A. A. Fadl, B. K. Khajanchi, and A. K. Chopra. 2006. Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect. Immun. 74:3742-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Presley, S. M., T. R. Rainwater, G. P. Austin, S. G. Platt, J. C. Zak, G. P. Cobb, E. J. Marsland, K. Tian, B. Zhang, T. A. Anderson, S. B. Cox, M. T. Abel, B. D. Leftwich, J. R. Huddleston, R. M. Jeter, and R. J. Kendall. 2006. Assessment of pathogens and toxicants in New Orleans, LA following Hurricane Katrina. Environ. Sci. Technol. 40:468-474. [DOI] [PubMed] [Google Scholar]
- 59.Pukatzki, S., A. T. Ma, A. T. Revel, D. Sturtevant, and J. J. Mekalanos. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508-15513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahman, M., G. Huys, M. Rahman, M. J. Albert, I. Kuhn, and R. Mollby. 2007. Persistence, transmission, and virulence characteristics of Aeromonas strains in a duckweed aquaculture-based hospital sewage water recycling plant in Bangladesh. Appl. Environ. Microbiol. 73:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sen, K., and M. Rodgers. 2004. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J. Appl. Microbiol. 97:1077-1086. [DOI] [PubMed] [Google Scholar]
- 62.Sha, J., C. L. Galindo, V. Pancholi, V. L. Popov, Y. Zhao, C. W. Houston, and A. K. Chopra. 2003. Differential expression of the enolase gene under in vivo versus in vitro growth conditions of Aeromonas hydrophila. Microb. Pathog. 34:195-204. [DOI] [PubMed] [Google Scholar]
- 63.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sha, J., E. V. Kozlova, A. A. Fadl, J. P. Olano, C. W. Houston, J. W. Peterson, and A. K. Chopra. 2004. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 72:1084-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sha, J., S. F. Wang, G. Suarez, J. C. Sierra, A. A. Fadl, T. E. Erova, S. M. Foltz, B. K. Khajanchi, A. Silver, J. Graf, C. H. Schein, and A. K. Chopra. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb. Pathog. 43:127-146. [DOI] [PubMed] [Google Scholar]
- 67.Sierra, J. C., G. Suarez, J. Sha, S. M. Foltz, V. L. Popov, C. L. Galindo, H. R. Garner, and A. K. Chopra. 2007. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—part II. Microb. Pathog. 43:147-160. [DOI] [PubMed] [Google Scholar]
- 68.Silver, A. C., and J. Graf. 2009. Prevalence of genes encoding the type three secretion system and the effectors AexT and AexU in the Aeromonas veronii group. DNA Cell Biol. 28:383-388. [DOI] [PubMed] [Google Scholar]
- 69.Silver, A. C., Y. Kikuchi, A. A. Fadl, J. Sha, A. K. Chopra, and J. Graf. 2007. Interaction between innate immune cells and a bacterial type III secretion system in mutualistic and pathogenic associations. Proc. Natl. Acad. Sci. U. S. A. 104:9481-9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 71.Suarez, G., J. C. Sierra, T. E. Erova, J. Sha, A. J. Horneman, and A. K. Chopra. 2010. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192:155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talon, D., M. J. Dupont, J. Lesne, M. Thouverez, and Y. Michel-Briand. 1996. Pulsed-field gel electrophoresis as an epidemiological tool for clonal identification of Aeromonas hydrophila. J. Appl. Bacteriol. 80:277-282. [DOI] [PubMed] [Google Scholar]
- 75.Tu, Y. K., D. Gunnell, and M. S. Gilthorpe. 2008. Simpson's paradox, Lord's paradox, and suppression effects are the same phenomenon—the reversal paradox. Emerg. Themes Epidemiol. 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyler, K. D., G. Wang, S. D. Tyler, and W. M. Johnson. 1997. Factors affecting reliability and reproducibility of amplification-based DNA fingerprinting of representative bacterial pathogens. J. Clin. Microbiol. 35:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Belkum, A. 1997. Amplification-based DNA fingerprinting: from artifactual to definitive typing and in between. J. Clin. Microbiol. 35:3008-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vila, J., J. Ruiz, F. Gallardo, M. Vargas, L. Soler, M. J. Figueras, and J. Gascon. 2003. Aeromonas spp. and traveler's diarrhea: clinical features and antimicrobial resistance. Emerg. Infect. Dis. 9:552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]
- 80.Wu, C. J., J. J. Wu, J. J. Yan, H. C. Lee, N. Y. Lee, C. M. Chang, H. I. Shih, H. M. Wu, L. R. Wang, and W. C. Ko. 2007. Clinical significance and distribution of putative virulence markers of 116 consecutive clinical Aeromonas isolates in southern Taiwan. J. Infect. 54:151-158. [DOI] [PubMed] [Google Scholar]
- 81.Yu, H. B., P. S. Rao, H. C. Lee, S. Vilches, S. Merino, J. M. Tomas, and K. Y. Leung. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72:1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu, H., S. J. Thuruthyil, and M. D. Willcox. 2000. Invasive strains of Pseudomonas aeruginosa are able to cause epithelial cell cytotoxicity that is dependent on bacterial cell density. Clin. Exp. Ophthalmol. 28:201-204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.