Abstract
Thermobifida fusca is a high-G+C-content, thermophilic, Gram-positive soil actinobacterium with high cellulolytic activity. In T. fusca, CelR is thought to act as the primary regulator of cellulase gene expression by binding to a 14-bp inverted repeat [5′-(T)GGGAGCGCTCCC(A)] that is upstream of many known cellulase genes. Previously, the ability to study the roles and regulation of cellulase genes in T. fusca has been limited largely by a lack of established genetic engineering methods for T. fusca. In this study, we developed an efficient procedure for creating precise chromosomal gene disruptions and demonstrated this procedure by generating a celR deletion strain. The celR deletion strain was then characterized using measurements for growth behavior, cellulase activity, and gene expression. The celR deletion strain of T. fusca exhibited a severely crippled growth phenotype with a prolonged lag phase and decreased cell yields for growth on both glucose and cellobiose. While the maximum endoglucanase activity and cellulase activity were not significantly changed, the endoglucanase activity and cellulase activity per cell were highly elevated. Measurements of mRNA transcript levels in both the celR deletion strain and the wild-type strain indicated that the CelR protein potentially acts as a repressor for some genes and as an activator for other genes. Overall, we established and demonstrated a method for manipulating chromosomal DNA in T. fusca that can be used to study the cellulolytic capabilities of this organism. Components of this method may be useful in developing genetic engineering methods for other currently intractable organisms.
Thermobifida fusca (formerly Thermomonospora fusca) is an aerobic, moderately thermophilic, filamentous soil bacterium (18). T. fusca is a major degrader of plant cell walls in heated organic materials, such as compost piles, rotting hay, and manure piles (2). The extracellular enzymes produced by T. fusca, including a host of cellulases, have been studied extensively because of their high activity, thermostability, and functionality at a broad pH range (pH 4 to 10) (18).
The widely accepted mechanism for enzymatic hydrolysis of cellulose involves the synergistic activity of endoglucanases (EC 3.2.1.4), exoglucanases (or cellobiohydrolase) (EC 3.2.1.91), and β-glucosidases (EC3.2.1.21). Six different cellulases have been identified in T. fusca and purified. Three of these cellulases are endocellulases (Cel9B, Cel6A, and Cel5A) (1, 9, 14); two are exocellulases (Cel6B and Cel48A) (13, 27); and one is a novel processive endocellulase (Cel9A) (33). In addition to the cellulases, T. fusca also produces and secretes hemicellulases. Two of the hemicellulases found in T. fusca are xylanases (Xyl11A and Xyl10B), one is a xyloglucanase (Xg74A), and one is a mannanase (ManB) (3, 15, 28). The cellulases in T. fusca act synergistically to convert insoluble cellulose to cellobiose and/or glucose.
In T. fusca, CelR (Tfu_0938) has previously been identified as a regulator of several cellulase genes, is a member of the lactose repressor family, and binds cellobiose with a millimolar dissociation constant (Kd) (24). Initially, CelR was thought to act as a repressor of cellulase gene expression by binding to a 14-bp inverted repeat [5′-(T)GGGAGCGCTCCC(A)] that is upstream of many cellulase genes. It has also been found that physiological concentrations of cellobiose (the major end product of cellulases) cause dissociation of the CelR-DNA complex (6). However, recent observations have suggested that CelR is not only a repressor but also a putative activator (28). Our previous research also suggested that CelR may not act solely as a repressor of cellulase gene expression (8). However, there is little direct experiment evidence that indicates the regulatory function of CelR in cellulase gene expression, and there is no direct evidence showing that CelR may act as an activator. Directed disruption of the celR gene in T. fusca is one of the most direct methods to address this regulatory role of CelR (10, 12, 25, 32), but such disruption has not been accomplished prior to this study.
No method for disruption of chromosomal genes in T. fusca has been reported previously. Here, we describe development of a PCR-based gene disruption protocol for T. fusca that was adapted from previously described methods (7) and permits deletion of the celR gene, resulting in a genetically stable T. fusca deletion strain. Our approach was to use the λ Red (γ, β, exo) system (7) to construct a cassette which contains neo flanked at both ends with nucleotides homologous to the desired region of the chromosome of T. fusca (7, 20, 31, 32) in Escherichia coli and then transform the cassette into T. fusca protoplasts to replace the celR gene by double-crossover homologous recombination (12). The celR deletion strain that was constructed was then characterized using measurements for cellular growth, cellulase activity, and expression of 18 cellulase-related genes (6).
MATERIALS AND METHODS
Strains and culture conditions.
T. fusca ATCC BAA-629 was grown in Hagerdahl medium (11) containing 1.0% cellobiose. The strains used and generated in this study are listed in Table 1. E. coli strain BW25113 was used to propagate the recombination plasmid pIJ790 (7). E. coli strain ET12567 carrying pUZ8002 was used as the nonmethylating plasmid donor strain (21). All E. coli strains were grown in SOB or LB medium.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Rita Shiang, Virginia Commonwealth University |
| E .coli BW25113 | K-12 derivative, ΔaraBAD ΔrhaBAD | 7, 12 |
| E. coli ET12567 | dam dcm hsdS cat tet | 5 |
| T. fusca(celR::neo) transformant | celR::neo | This study |
| Plasmids | ||
| pGEM-T | bla lacZ | Promega |
| pGEM2816 | bla celR lacZ | This study |
| pGEM2948neo | bla celR::neo lacZ | This study |
| pIJ790 | l RED (gam, bet, exo) cat araC, rep101ts | 12 |
| pET-30a(+) | neo lacI, T7 | Novagen |
Plasmids.
The ampicillin resistance marker bla on pKD20 was replaced by cat, the chloramphenicol resistance (Cmr) gene, generating pIJ790 (Table 1). The relevant plasmid structure was confirmed by restriction analysis. The 2,816-bp DNA fragment containing celR flanked by 901 bp upstream and 892 bp downstream was amplified and cloned in the pGEM-T vector (Promega, United States) to obtain pGEM2816 (Fig. 1) . A new plasmid, pGEM2948neo, was then constructed for disruption of celR by replacing the majority of the coding region of celR with neo, a kanamycin resistance (Kanr) gene, using homologous recombination. The neo gene was cloned from pET-30a(+) (Novagen) and was flanked on both sides by 39 nucleotides homologous to celR nucleotides. pGEM2948neo was then transformed into E. coli strain ET12567 (a nonmethylating strain).
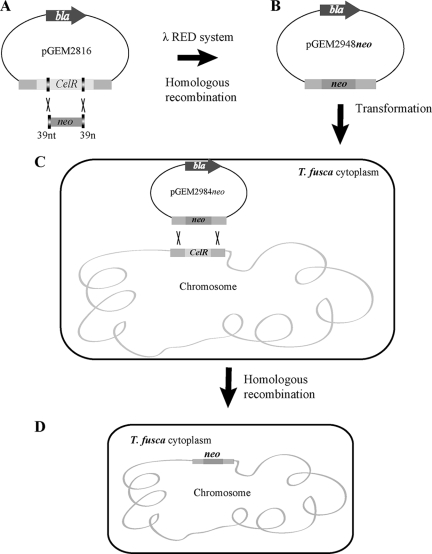
FIG. 1.
Overview of the strategy used for replacement of celR in T. fusca. (A) A gene disruption cassette containing neo was obtained by PCR by using primers containing 39-nucleotide homology extensions corresponding to regions of the celR gene. The PCR product was transformed into E. coli BW25113/pIJ790 (expressing the λ Red recombination functions) containing pGEM2816. (B) The celR gene was replaced by neo using homologous recombination with the λ Red system to obtain pGEM2948neo, which was introduced into the methylation-deficient host E. coli ET12567. (C) Nonmethylated pGEM2948neo was transformed into T. fusca protoplasts with the help of PEG 1000. After overnight regeneration, soft R2YE agar with kanamycin was added to the plates, and they were incubated for two more days. (D) Double-crossover allelic exchange was confirmed by PCR and DNA sequencing. nt and n, nucleotides.
Gene replacement on plasmids.
Disruption of the celR gene was accomplished by first generating a linear celR DNA fragment with upstream and downstream flanking regions (fragment C2816) using primers 2816F and 2816R (Table 2) to PCR amplify the gene from T. fusca. Fragment C2816 was cloned into the pGEM-T vector using the manufacturer's instructions (Promega). E. coli strain BW25113 carrying pIJ790 was grown overnight at 30°C in 10 ml LB medium containing chloramphenicol (Cm) at a concentration of 25 μg/ml. One hundred microliters of an E. coli BW25113/pIJ790 overnight culture was inoculated into 10 ml SOB medium containing 20 mM MgSO4 (200 μl of a 1 M MgSO4 stock solution was added to 10 ml SOB medium) and Cm (25 μg/ml) and grown for 3 to 4 h at 30°C with agitation at 200 rpm to obtain an optical density at 600 nm (OD600) of 0.4. After this, the cells were recovered by centrifugation at 8,000 × g for 5 min at 4°C. The supernatant was decanted, and the cell pellet was resuspended in 10% glycerol. Fifty microliters of the cell suspension was mixed with 100 ng (1 to 2 μl) of purified DNA fragment C2816. Electroporation was carried out in a 0.2-cm ice-cold electroporation cuvette using a Bio-Rad GenePulser II set to 200 Ω, 25 μF, and 3.0 kV. The expected time constant was 4.5 to 4.9 ms. One milliliter of ice-cold SOC medium was immediately added to the electroporated cells, and the cells were incubated for 1 h at 30°C. Incubated cells were then spread onto LB agar plates containing Cm (25 μg/ml) and ampicillin (Amp) (100 mg/ml) and incubated overnight at 30°C. Positive transformants were selected using ampicillin resistance and were characterized by isolating plasmid DNA and verifying that the plasmid construction was correct by restriction analysis and sequencing of the plasmid by Sanger sequencing using SP6 and T7 as the forward and reverse primers, respectively. Only the plasmids on which C2816 had the same direction of expression as it had on pGEM-T were used in the next experiment. Thirty-nine nucleotides were homologous to 5′ and 3′ parts of celR whose positions started and ended at positions 1104577 and 1103895 in the genome, respectively. Primers KanknockF and KanknockR, which were used to obtain neo from pET-30a (+), were flanked by the 39-nucleotide sequences described above (5′-AGTCCGGCCACGCGAGAGGCCGTGAAGCGGGCGATCAAA and 5′-CGAGTCGTCGTAGCCGACCACCGCCACATCCTCGGGCAC, homologous to regions upstream and downstream of celR, respectively) (Table 2). PCR was performed using a 50-μl reaction mixture containing 100 ng of template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, 50 pmol each primer, and 3% dimethyl sulfoxide according to the manufacturer's instructions (high-fidelity polymerase; Finnzymes), as follows: denaturation at 98°C for 30 s and then 30 cycles of denaturation at 98°C for 10 s, annealing at 55.9°C for 30 s, and extension at 72°C for 1 min 30 s, followed by elongation at 72°C for 5 to 10 min. The neo gene flanked by 39-nucleotide sequences was transformed into E. coli strain BW25113 containing C2816, and the λ Red recombinase system was activated using 1 M l-arabinose.
TABLE 2.
PCR primers used in this studya
| Primer | Sequence (5′-3′) | Sequence generated |
|---|---|---|
| 2816F | CCCAAGCTTTGCCGTGATGCGGGACTT | celR and upstream and downstream sequences |
| 2816R | CACCGTGAACGGGAAAACC | |
| KanknockF | AGTCCGGCCACGCGAGAGGCCGTGAAGCGGGCGATCAAAATGAGCCATATTCAACGGGA | neo flanked by 39 nucleotides homologous to celR |
| KanknockR | CGAGTCGTCGTAGCCGACCACCGCCACATCCTCGGGCACTTAGAAAAACTCATCGAGCA | |
| CelRF | GGGTTGGGGGAACACATATGGAGCGTC | celR |
| CelRR | TCCGCCTGCCTCCCGTTGTCCTC | |
| KanF | ATGAGCCATATTCAACGGGA | neo |
| KanR | TTAGAAAAACTCATCGAGCA | |
| NestKanF | AAACATGGCAAAGGTAGCGT | neo, nested PCR |
| NestKanR | GAGAAAACTCACCGAGGCAG |
The 39 nucleotides homologous to celR nucleotides are underlined. Bold type indicates restriction enzyme sites.
Preparation of T. fusca protoplasts.
A 5% spore suspension with 0.5% glycine was added to a shaking flask and incubated for 12 h at 55°C in an orbital incubator shaker in the first step of the procedure used to prepare T. fusca protoplasts for transformation. The culture was poured into a 20-ml screw-cap bottle and precipitated using a benchtop centrifuge (10,000 × g, 10 min). After centrifugation, the culture supernatant was discarded. The pellet was resuspended in 15 ml 10.3% sucrose and precipitated using a benchtop centrifuge as described above, and the supernatant was discarded. Mycelia were resuspended in 4 ml of a lysozyme solution (2 mg/ml P buffer) and incubated at 37°C for 2 h. Protoplasts were filtered through cotton wool and transferred to a plastic tube. Protoplasts were obtained by gentle centrifugation with a benchtop centrifuge (1,000 × g, 5 min), and then protoplasts were stored in 1 ml phosphate buffer.
Transformation of T. fusca and allelic exchange.
The Kanr mutagenized pGEM2948neo construct was introduced into E. coli ET12567/pUZ8002 by electroporation as described above and transformed into T. fusca protoplasts. Fifty-microliter samples of protoplasts were used for all transformations. Up to 5 μl of a DNA solution was added to protoplasts and mixed immediately by tapping the tube. Two hundred microliters of a 25% polyethylene glycol 1000 (PEG 1000) solution was added to P buffer and mixed by pipetting. The resulting protoplast suspension (100 to 200 μl) was spread onto two dried R2YE agar plates (with 12% sucrose). The plates were incubated at 45°C for 14 to 20 h. After incubation, the plates were flooded with kanamycin (50 μg/ml) after the R2YE agar was melted, and they were incubated for another 2 days. Replica plating onto DNA agar plates with kanamycin (50 μg/ml) was performed to screen for kanamycin-resistant transformants. Genomic DNA of positive transformants was extracted, and the cassette used for genomic integration was amplified using the 2816F and 2816R primers. PCR products were sequenced by Sanger sequencing using the CelRF and CelRR primers (Table 2) to verify that the target gene sequence was deleted.
Enzyme activity assays.
Two assays were used to measure the overall cellulase and endoglucanase activities of culture supernatants using a previously described protocol (29). Filter paper was used as the starting material to measure the overall cellulase activity, and endoglucanase activity was assayed by measuring the reducing sugars generated from 0.5% medium-viscosity carboxymethyl cellulose (CMC). Assays were conducted using 60-μl mixtures as follows. Aliquots (20 μl) of raw enzyme were added to wells of PCR plates containing 40 μl of 50 mM sodium acetate buffer and a filter paper disk (diameter, 7 mm) for the cellulase activity assay or containing 40 μl of 50 mM sodium acetate buffer with 0.5% CMC for the endoglucanase activity assay. After 60 min of incubation at 50°C, 120 μl of a 3,5-dinitrosalicylic acid (DNS) solution was added to each reaction mixture and incubated at 95°C in a thermal cycler (iCycler thermocycler; Bio-Rad, Hercules, CA) for 5 min. Finally, 36-μl aliquots of each sample were transferred to wells of a flat-bottom plate containing 160 μl of H2O, and the absorbance at 540 nm was measured with a VersaMax EXT microplate reader (Molecular Devices, Sunnyvale, CA). One enzyme unit was defined as release of an average of 1 μmol of cellobiose equivalent per min in the assay reaction. All of the enzyme activity values presented below are averages for triplicate measurements.
Cell density measurement.
Due to the growth physiology of T. fusca (filamentous cells that aggregate), the culture density of T. fusca was determined by measuring the cytoplasmic protein content. One milliliter of a culture was centrifuged at 10,000 × g for 5 min. The pellet was resuspended in fresh medium and centrifuged at 10,000 × g for 5 min again. The sediment was dissolved in 200 μl 50 mM Tris-HCl buffer (pH 6.8) containing 2% SDS, 0.1 M dithiothreitol, and 50% glycerol. Samples were then pulsar sonicated at 80% strength for 10 min in an ice bath. After centrifugation at 10,000 × g for 5 min, the protein content of the supernatant was measured using the Bradford protein assay (5). The cell dry weight was proportionally related to the overall protein content.
RNA preparation and real-time PCR (8).
To study molecular-level differences in the cultures of T. fusca, gene expression was studied using real-time PCR and cells harvested at the mid-log growth phase and early stationary phase, corresponding to times when different cellulase activities were exhibited. The probes and primers (see Table S1 in the supplemental material) were designed using Primer Express 3.0. The levels of expression of 18 different cellulase-related genes were measured along with the level of expression of one housekeeping gene (Tfu_02001404) that was used as a control, and all transcript levels were normalized to the level of this housekeeping gene.
RESULTS
Adaptation of PCR targeting for gene disruption in T. fusca.
T. fusca is a cellulolytic bacterium that has been shown to be a difficult host for genetic manipulation, largely due to high exonuclease activity. The only previously documented success was the transformation of plasmid pIJ702 into T. fusca protoplasts (22). Recently, allelic exchange in the E. coli chromosome was achieved using homologous recombination with a PCR-generated selectable marker flanked on both ends by nucleotides with homology to the desired region of the chromosome. Recombination with chromosomal DNA was induced using the λ Red system, where the Redα (exo), Redβ (bet), and Redγ (gam) proteins of phage λ were present in the targeted strain (12) and RecBCD, which is responsible for exonuclease activity, was inactivated. Although the RecBCD complex is not present in T. fusca, there is a high level of exonuclease activity in T. fusca due to the RecF, RecO, RecR, and RecN proteins (18). To address the problem of degradation of extracellular DNA by nucleases in T. fusca, long nucleotide regions (>1 kb) homologous to the target gene were used; 2,816 bp of celR flanked by upstream and downstream fragments was obtained by high-fidelity PCR and cloned into the pGEM-T vector, generating pGEM2816 (Table 1), which was transformed into E. coli BW25113 competent cells. After transformation, seven positive transformants were determined to contain pGEM2816. To obtain a plasmid whose inserted DNA fragment had the same direction of expression as the pGEM-T vector, all of transformants were sequenced, and in three of the seven the insert was in the right direction. To disrupt celR, the neo gene from pET-30a(+) was amplified and used as an insertion cassette for in-frame disruption of celR. Because pET-30a(+) can make E. coli BW25113 resistant to kanamycin, the PCR product should have been purified using gel purification to eliminate any trace pET-30a(+). The purified PCR product could then be transformed into DH5α. Through homologous recombination with the λ Red system, most of the celR gene in pGEM2816 was replaced by the neo gene, generating the new plasmid pGEM2948neo. The neo gene was flanked by 1,038-bp and 1,095-bp sequences homologous to sequences upstream and downstream of celR, respectively.
Transformation of T. fusca.
The transformation of Streptomyces spp. has been studied intensively (4, 12, 21, 25), and methods for transforming protoplasts of mycelia are improving. Since T. fusca is closely related to Streptomyces coelicolor (18), transformation of T. fusca protoplasts should be possible. One complicating factor is that T. fusca is thermophilic, so use of common selecting agents, such as antibiotic resistance, can be problematic. In particular, the kanamycin resistance protein (kanamycin nucleotidyltransferase) is stable and functional at temperatures under 47°C (17). Although the optimal temperature for T. fusca growth is 55°C, we found that T. fusca was able to grow well at temperatures as low as 45°C. To avoid degradation of kanamycin nucleotidyltransferase, all of the T. fusca cultures were incubated at 45°C.
Because the genes coding for a methyl-directed mismatch repair system (MutSLH proteins) are not present in T. fusca (18), homologous recombination could not occur unless extracellular DNA fragments were not methylated. The pGEM2948neo construct was transformed into the nonmethylating strain E. coli ET12567 containing the RP4 derivative pUZ8002 to be demethylated. Because T. fusca protoplasts are very fragile, after pGEM2948neo was transformed into them, plates without kanamycin were used to regenerate protoplasts (incubation at 45°C for 14 to 20 h) before the R2YE agar was melted and 50 μg/ml kanamycin was added to the plates for selection. After further incubation with kanamycin selection for 2 days, 11 colonies were found, which were replica plated onto a new R2YE agar plate with 50 μg/ml kanamycin. Three of the 11 transformants were determined to be resistant to kanamycin, and their sequences were validated by DNA sequencing. After these three transformants were grown on kanamycin plates for 20 generations, only one of them remained resistant, and it was designated the T. fusca (celR::neo) transformant. Overall, use of this method resulted in a transformation efficiency of ∼1 × 10−3 transformant/μg DNA, which is comparable to previously reported transformation efficiencies for the anaerobic, cellulolytic bacterium Clostridium thermocellum (26).
Characteristics of the T. fusca (celR::neo) transformant in culture.
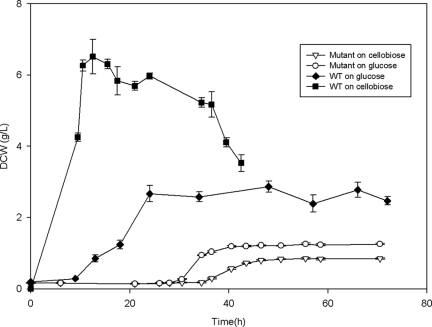
After successful construction of a T. fusca celR deletion mutant, cell growth and cellulase specific activity were measured to characterize phenotypic changes resulting from the disruption of celR. The mutant strain was cultured on cellobiose and glucose to determine if glucose repressed T. fusca growth. When growth was examined, the celR deletion strain exhibited severely decreased growth on both cellobiose and glucose compared to the wild-type strain (Fig. 2). One characteristic of the celR deletion strain was that it exhibited a very long lag phase on both cellobiose and glucose (28 h and 34.5 h, respectively) compared to the wild-type strain, which typically had a lag phase of less than 9.5 h. In addition, the cell yield (dry weight) of the mutant strain at stationary phase was much lower than that of the wild-type strain on both cellobiose and glucose (6.51 ± 0.49 g/liter for the wild type versus 0.85 ± 0.023 g/liter for the mutant on cellobiose and 2.87 ± 0.16 g/liter for the wild type versus 1.25 ± 0.027 g/liter for the mutant on glucose).
FIG. 2.
Growth phenotypes of wild-type and celR deletion strains. Dry cell weights (DCW) of the celR deletion strain (open symbols) of T. fusca grown on glucose and on cellobiose and of the wild-type strain (WT) (filled symbols) of T. fusca grown on glucose and on cellobiose were determined during growth in a bioreactor.
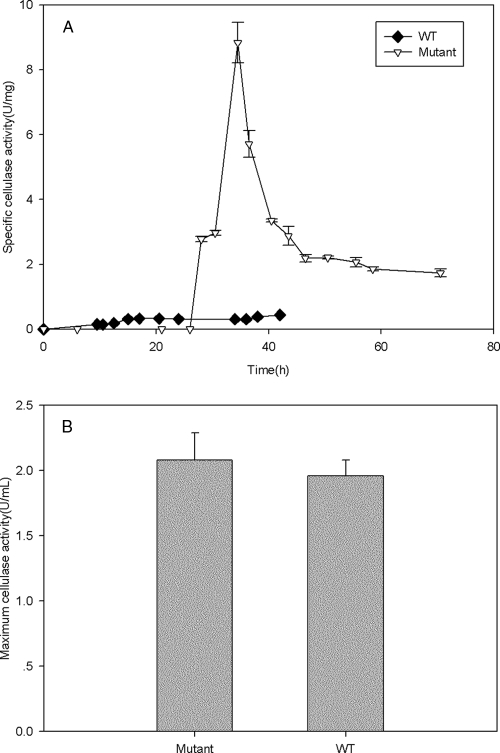
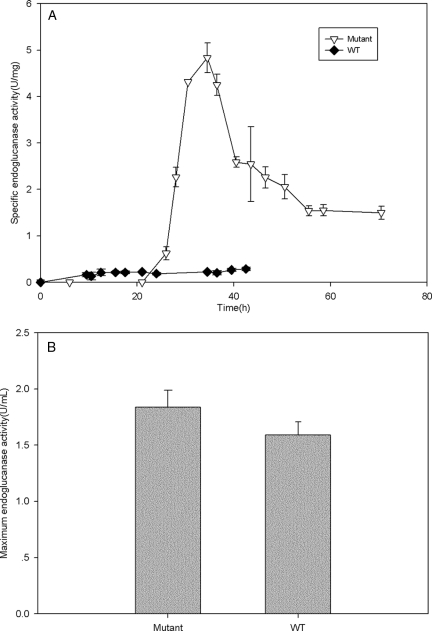
In addition to measuring the basic growth characteristics of the celR deletion strain, we conducted assays to measure the composite cellulase activity (combined endoglucanase, exoglucanase, and β-glucosidase activities) (Fig. 3) and the endoglucanase activity (Fig. 4) in the mutant and wild-type strains. The maximum cellulase activities of the wild-type and mutant strains did not differ appreciably (Fig. 3B), but when the large difference in the cell yields was taken into account, it was found that the celR deletion strain had a much higher cellulase specific activity (8.84 ± 0.63 U/mg [dry weight]) than the wild-type strain (0.441 ± 0.004 U/mg), as shown in Fig. 3A. When the endoglucanase activity was examined, a similar trend was found. There was no appreciable difference in the maximum endoglucanase activity between the mutant and wild-type strains (Fig. 4B), but the mutant strain had much higher activity per cell (4.84 ± 0.32 U/mg for the mutant strain and 0.29 ± 0.04 U/mg for the wild-type strain).
FIG. 3.
Cellulase activities of the celR deletion and wild-type strains. (A) Specific cellulase activities (U/mg [dry weight]) of the mutant and wild-type (WT) T. fusca strains. (B) Maximum cellulase activities (U/ml) of the mutant and wild-type strains.
FIG. 4.
Endoglucanase activities of the celR deletion and wild-type strains. (A) Specific endoglucanase activities (U/mg [dry weight]) of the mutant and wild-type (WT) T. fusca strains. (B) Maximum endoglucanase activities (U/ml) of the mutant and wild-type strains.
Cellulase gene expression in wild-type T. fusca and in T. fusca with celR disrupted.
To study the role of CelR in the regulation of expression of cellulase genes in T fusca, 18 cellulase-related genes were examined by real-time PCR for both wild-type and celR deletion strains of T. fusca (Table 3). While there is conflicting evidence concerning the role of CelR, it is commonly thought that CelR acts as a repressor of cellulase genes (23, 24) by binding to either the perfect (TGGGAGCGCTCCCA) or imperfect (NGGGAGCGCTCCCN) 14-bp palindrome site upstream of a target gene (18, 19). Of the 18 cellulase-related genes identified, 9 were determined to have a perfect or imperfect 14-bp palindrome upstream of the celR gene coding region (Table 3) (6, 16, 18, 23, 24, 30).
TABLE 3.
Cellulase-related gene expression in wild-type and celR-disrupted T. fusca
| Gene no. | Locus | Gene product (designation) | No. of 14-bp palindromes | Gene expression |
||
|---|---|---|---|---|---|---|
| Wild-type activity (AU ± SD) | Mutant activity (AU ± SD) | P | ||||
| 1 | Tfu_1627 | β-1,4-Endoglucanase (Cel9B) | 1 perfect | 59.36 ± 1.68 | 98.14 ± 14.22 | 9.37E-03 |
| 2 | Tfu_1074 | β-1,4-Endoglucanase (Cel6A) | 1 perfect | 93.02 ± 5.28 | 29.27 ± 5.33 | 1.24E-04 |
| 3 | Tfu_0620 | β-1,4-Exocellulase (Cel6B) | 2 perfect, 1 imperfect | 34.94 ± 1.08 | 69.55 ± 5.47 | 4.24E-04 |
| 4 | Tfu_2176 | β-1,4-Endoglucanase (Cel9A), progressive | 1 perfect | 169.51 ± 17.82 | 456.57 ± 39.32 | 3.24E-04 |
| 5 | Tfu_0901 | Endo-1,4-β-glucanase (Cel5A) | 1 perfect | 51.85 ± 3.81 | 167.06 ± 22.29 | 9.10E-04 |
| 6 | Tfu_1959 | β-1,4-Exocellulase (Cel48A) | 1 perfect, 5 imperfect | 47.64 ± 3.27 | 86.48 ± 12.67 | 6.79E-03 |
| 7 | Tfu_1268 | Predicted cellulose-binding protein (E7) | None | 82.08 ± 10.07 | 151.16 ± 18.34 | 4.63E-03 |
| 8 | Tfu_1665 | Predicted cellulose-binding protein (E8) | 1 perfect | 50.19 ± 2.63 | 65.21 ± 5.37 | 1.21E-02 |
| 9 | Tfu_0934 | Predicted ABC-type sugar transport system, periplasmic component for cellobiose/cellotriose | 1 perfect | 138.12 ± 4.02 | 39.57 ± 12.34 | 1.93E-04 |
| 10 | Tfu_0937 | β-Glucosidase (BglC) | None | 335.90 ± 7.13 | 29.42 ± 5.35 | 4.76E-07 |
| 11 | Tfu_0938 | CelR | None | 83.23 ± 5.17 | ||
| 12 | Tfu_2923 | β-1,4-Endoxylanase (Xyl10A) | 1 imperfect | 19.53 ± 1.69 | ||
| 13 | Tfu_2788 | Putative β-1,4-endoxylanase | None | 80.57 ± 9.57 | 26.40 ± 4.11 | 8.41E-04 |
| 14 | Tfu_1213 | β-1,4-Xylosidase | None | 33.40 ± 2.49 | 19.56 ± 3.56 | 5.27E-03 |
| 15 | Tfu_2791 | β-1,4-Endoxylanase (Xyl10B) | None | 25.14 ± 1.04 | 24.26 ± 6.3 | 0.82 |
| 16 | Tfu_1612 | Putative secreted xyloglucanase | None | 31.02 ± 2.29 | 7.46 ± 0.99 | 8.18E-05 |
| 17 | Tfu_2990 | Putative secreted xylanase | None | 32.89 ± 1.19 | 8.26 ± 0.71 | 6.63E-06 |
| 18 | Tfu_0900 | Putative secreted β-mannanase | None | 5.35 ± 0.23 | 12.14 ± 0.89 | 2.15E-04 |
Of the nine genes with potential binding sites for CelR, six had exactly one perfect palindrome sequence upstream. The mRNA transcript levels for three of these six genes (Tfu_1627, Tfu_2176, and Tfu_0901) were statistically significantly (P < 0.01) higher in the celR deletion strain than in the wild-type strain, possibly indicating that CelR represses expression of these genes in the wild-type strain. For two genes (Tfu_1074 and Tfu_0934) the mRNA transcript levels were statistically significantly (P < 0.002) lower in the celR deletion strain than in the wild-type strain, indicating that CelR may activate expression of these genes in the wild-type strain. The mRNA transcript level for the remaining gene with exactly one perfect palindrome site (Tfu_1665) was slightly higher in the deletion strain than in the wild-type strain, but the level of statistical significance was not high. It should be noted that Tfu_0934, Tfu_0937, and Tfu_0938 (CelR) are predicted to be in the same operon (18) with a single promoter upstream of Tfu_0934. The mRNA transcript data are consistent with the predicted operon structure since the mRNA transcript levels for all three of these genes were significantly lower in the celR deletion strain (Tfu_0938 is the celR gene which was deleted, and mRNA transcripts were not detected in any sample).
Tfu_2923 with one imperfect 14-bp palindrome was not transcribed in the celR deletion strain as no mRNA transcripts were detected in any of the replicates. The mRNA transcript levels for Tfu_0620 (two perfect and one imperfect palindromes) and Tfu_1959 (one perfect and five imperfect 14-bp palindromes) were both higher (P < 0.007) in the celR deletion strain than in the wild-type strain. All of the other genes studied did not have a 14-bp palindrome site upstream, and the mRNA transcript levels for them were lower in the celR deletion strain, with the exception of Tfu_0900, which encodes a putative mannanase and for which the mRNA transcript levels were higher in the celR deletion strain than in the wild-type strain.
DISCUSSION
Because of the abundance of cellulose in nature and potential applications for converting cellulose to biofuels or other commodities, there has been increasing interest in studying cellulase enzymes. T. fusca is an interesting organism for studying cellulose degradation, as it is an aerobic, thermophilic organism with a variety of secreted cellulases that result in efficient cellulose degradation and utilization. To date, there have been a number of studies on the cellulases in T. fusca, but research has been hindered by the absence of a tractable genetic manipulation system. In this study, we developed a protocol for genetic manipulation of the chromosome of T. fusca using homologous recombination and demonstrated this protocol by disrupting the celR gene, the primary regulator of cellulase gene expression in T. fusca. Through construction and characterization of a celR deletion strain of T. fusca, we showed that (i) a protocol using carefully designed plasmids, intermediate host cells, and modified culture conditions allows genetic manipulation of the chromosome in T. fusca, (ii) a celR deletion strain has growth and cellulolytic capabilities that are significantly different from those of the wild-type strain, and (iii) CelR appears to function both as a repressor and as an activator when it regulates cellulase gene expression.
To create a protocol for manipulation of chromosomal DNA in T. fusca, multiple steps in methods used for other organisms had to be altered. Due to the high exonuclease activity in T. fusca, atypically long flanking regions of homology (>1 kb) were required in the disruption cassette that was designed, and the disruption cassette was introduced as a plasmid (with a replication origin that is not recognized by T. fusca) rather than as linear DNA. Since T. fusca does not possess a functional methyl-directed mismatch repair system, plasmid preparation and propagation were conducted using a nonmethylating host strain (E. coli strain ET12567). Adjustments to the culture conditions for T. fusca were also necessary. For transformation, T. fusca protoplasts must be prepared; however, these protoplasts are very fragile. Due to this fragility, it was necessary to include a period of protoplast regeneration in nonselective medium after transformation before a selecting agent (kanamycin) was added to screen for positive transformants. In addition, the culture temperature for T. fusca transformants was decreased to 45°C (T. fusca normally grows optimally at 55°C) so that the kanamycin nucleotidyltransferase was not heat inactivated. Employing these steps resulted in isolation of a single transformant that had a stable disruption in chromosomal celR.
After successful construction and verification of a celR deletion strain, phenotypic measurements were obtained to characterize changes in the growth and cellulase activity of the mutant. Growth and cellulase activity are known to be different in T. fusca with different carbon substrates (e.g., glucose acts as a repressor of cellulase activity), so growth experiments were conducted both with cellobiose and with glucose. The two main observations when growth was monitored were that the celR deletion strain had a prolonged lag phase and a significantly decreased cell yield compared to those of the wild-type strain. The prolonged lag phase is not surprising because of the deletion of a regulatory gene and the potential need for the metabolic system to adjust to this. Of greater interest is the fact that the cell yield was significantly decreased for both growth on cellobiose and growth on glucose. While the cell yield of the celR deletion strain on cellobiose was decreased by a factor of approximately 8 (a large effect was expected), it was interesting that the cell yield of the celR deletion strain on glucose was less than one-half that of the wild-type strain. This was even more interesting considering that T. fusca does not explicitly need any of the cellulase genes regulated by CelR to grow on glucose, so the impact of celR deletion potentially should be minimal.
A plausible explanation for the decreased cell yield both on glucose and on cellobiose is linked to the disruption of CelR. In the absence of a regulator, the endoglucanse and cellulase activities per cell were roughly 17 and 20 times higher, respectively, in the celR deletion strain than in the wild-type strain for growth on cellobiose. This marked increase in cellulase activity implies that there is an increased metabolic burden to produce larger quantities of functional cellulases in the celR deletion strain. The increased metabolic burden of producing unregulated cellulases decreases the availability of metabolic resources to produce basic biomass components. This scenario would be true both in the case of cellobiose and in the case of glucose.
Further characterization of the celR deletion strain was performed using real-time PCR to study the transcriptional consequences of removing CelR. In T. fusca, for the genes encoding all of the known cellulases and one of the xylanases there are one or two copies of an upstream 14-bp DNA sequence, (T)GGGAGCGCTCCC(A), which was found to be the binding site for the transcriptional regulator CelR (19, 23). While it has been shown that CelR can repress expression of several cellulase genes, there is some indirect evidence that CelR may also act as a putative activator of cellulase gene expression (28). In this study, we examined the regulatory role of CelR by creating a celR deletion strain and measuring the changes in mRNA transcript levels. The simple case was to consider the changes in the expression of genes that contained only a single copy of the perfect 14-bp CelR recognition sequence (5′-TGGGAGCGCTCCCA). These genes were Tfu_1627, Tfu_1074, Tfu_2176, Tfu_0901, Tfu_1665, and Tfu_0934. Of the genes that showed a statistically significant change in the mRNA transcript level in the celR deletion strain, three showed an increase in expression (Tfu_1627, Tfu_2176, and Tfu_0901) and two showed a decrease in expression (Tfu_1074 and Tfu_0934) in the deletion strain compared to the wild-type strain. If CelR is the only regulator that acts on these genes, these data indicate that CelR acts as a repressor in some cases and as an activator in other cases.
Several of the cellulase genes in T. fusca also contain copies of the imperfect CelR binding site upstream of the coding region of the gene (5′-NGGGAGCGCTCCCN). CelR is also believed to bind to this recognition site (18, 19, 24). It has been predicted that binding to this imperfect recognition site leads to activation of gene transcription (28). For the single gene containing only one imperfect recognition site (Tfu_2923), there were no detectable mRNA transcripts in any of the samples of the celR deletion strain, supporting the idea that CelR may activate transcription of this gene.
A secondary question is, what happens if a gene has both perfect and imperfect CelR binding sites upstream? The Tfu_1959 gene has one perfect and five imperfect binding sites upstream, and the Tfu_0620 gene has two perfect and one imperfect binding sites upstream. If we assume that CelR binding to an imperfect binding site leads to activation of mRNA transcription based on the results for Tfu_2923, we can interpret the results for the genes with multiple binding sites upstream. For both Tfu_1959 and Tfu_0620, the mRNA transcript levels for the celR deletion strain were statistically significantly (P < 0.007) higher than those for the wild-type strain. If CelR binding to the imperfect site activates gene expression, then the mRNA transcript levels for the mutant strain should be lower. This implies that CelR binding to the perfect site for these two genes must repress gene expression with higher affinity than binding to the imperfect site (which is related to different binding strengths of CelR for the perfect and imperfect 14-bp palindromes) (16, 18, 23). While this is one possible explanation for the regulatory interactions of CelR with the perfect and imperfect binding sites, regulation of these genes may have additional influences outside CelR that have not been identified yet.
In another study (19), it was predicted that Tfu_1268 should be repressed by CelR because it was induced by cellobiose and repressed by glucose, and cellobiose was thought to deactivate CelR binding. Our data showed that the mRNA transcript level of Tfu_1268 was higher in the celR deletion strain than in the wild-type strain, which is consistent with the prediction of Moser et al. (19). However, in the upstream sequence of Tfu_1268 there are no known CelR binding sites. Thus, if regulation of Tfu_1268 is controlled by CelR, there may be more binding sites or modes of action that have not been discovered yet. Overall, we developed a method for manipulation of chromosomal DNA in T. fusca and demonstrated application of this method by creating a celR deletion strain. Characterization of the celR deletion strain showed that there were great changes in the growth and cellulase phenotypes of T. fusca, and by using real-time PCR analysis we showed that CelR can act either as a repressor or as an activator. The method that we developed and our results provide a framework for further study of the cellulolytic capabilities of T. fusca and may facilitate development of techniques for genetic manipulation of other intractable thermophilic organisms.
Supplementary Material
Acknowledgments
We thank David B. Wilson of Cornell University for kindly providing plasmid pUZ8002, Rita Shiang of Virginia Commonwealth University for providing a stock of E. coli DH5α, and the John Innes Centre in the United Kingdom for providing plasmid pIJ790.
Footnotes
Published ahead of print on 22 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ai, Y. C., and D. B. Wilson. 2002. Mutation and expression of N233C-D506C of cellulase Cel6B from Thermobifida fusca in Escherichia coli. Enzyme Microb. Technol. 30:804-808. [Google Scholar]
- 2.Bachmann, S. L., and A. J. Mccarthy. 1991. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microbiol. 57:2121-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beki, E., S. Nagy, J. Vanderleyden, S. Jager, L. Kiss, L. Fulop, L. Hornok, and J. Kukolya. 2003. Cloning and heterologous expression of a beta-d-mannosidase (EC 3.2.1.25)-encoding gene from Thermobifida fusca TM51. Appl. Environ. Microbiol. 69:1944-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principal of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. L., and D. B. Wilson. 2007. Proteomic and transcriptomic analysis of extracellular proteins and mRNA levels in Thermobifida fusca grown on cellobiose and glucose. J. Bacteriol. 189:6260-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, Y., and S. S. Fong. 2009. Influence of culture aeration on the cellulase activity of Thermobifida fusca. Appl. Microbiol. Biotechnol. 85:965-974. [DOI] [PubMed] [Google Scholar]
- 9.Devillard, E., D. B. Goodheart, S. K. R. Karnati, E. A. Bayer, R. Lamed, J. Miron, K. E. Nelson, and M. Morrison. 2004. Ruminococcus albus 8 mutants defective in cellulose degradation are deficient in two processive endocellulases, Cel48A and Cel9B, both of which possess a novel modular architecture. J. Bacteriol. 186:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Karoui, M., S. K. Amundsen, P. Dabert, and A. Gruss. 1999. Gene replacement with linear DNA in electroporated wild-type Escherichia coli. Nucleic Acids Res. 27:1296-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferchak, J. D., and E. K. Pye. 1983. Effect of cellobiose, glucose, ethanol, and metal-ions on the cellulase enzyme complex of Thermomonospora fusca. Biotechnol. Bioeng. 25:2865-2872. [DOI] [PubMed] [Google Scholar]
- 12.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 14.Jung, H., D. B. Wilson, and L. P. Walker. 2003. Binding and reversibility of Thermobifida fusca Cel5A, Cel6B, and Cel48A and their respective catalytic domains to bacterial microcrystalline cellulose. Biotechnol. Bioeng. 84:151-159. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. H., D. Irwin, and D. B. Wilson. 2004. Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50:835-843. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y. C., D. C. Irwin, and D. B. Wilson. 2007. Processivity, substrate binding, and mechanism of cellulose hydrolysis by Thermobifida fusca Cel 9A. Appl. Environ. Microbiol. 73:3165-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao, H., T. McKenzie, and R. Hageman. 1986. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc. Natl. Acad. Sci. U. S. A. 83:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykidis, A., K. Mavromatis, N. Ivanova, I. Anderson, M. Land, G. DiBartolo, M. Martinez, A. Lapidus, S. Lucas, A. Copeland, P. Richardson, D. B. Wilson, and N. Kyrpides. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser, F., D. C. Irwin, S. Chen, and D. B. Wilson. 2008. Regulation and characterization of Thermobifida fusca carbohydrate-binding module proteins E7 and E8. Biotechnol. Bioeng. 100:1066-1077. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 21.Paget, M. S. B., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pidcock, K. A., B. S. Montenecourt, and J. A. Sands. 1985. Genetic recombination and transformation in protoplasts of Thermomonospora fusca. Appl. Environ. Microbiol. 50:693-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiridonov, N. A., and D. B. Wilson. 2000. A celR mutation affecting transcription of cellulase genes in Thermobifida fusca. J. Bacteriol. 182:252-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spiridonov, N. A., and D. B. Wilson. 1999. Characterization and cloning of CelR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J. Biol. Chem. 274:13127-13132. [DOI] [PubMed] [Google Scholar]
- 25.Stephan, J., V. Stemmer, and M. Niederweis. 2004. Consecutive gene deletions in Mycobacterium smegmatis using the yeast FLP recombinase. Gene 343:181-190. [DOI] [PubMed] [Google Scholar]
- 26.Tyurin, M. V., S. G. Desai, and L. R. Lynd. 2004. Electrotransformation of Clostridium thermocellum. Appl. Environ. Microbiol. 70:883-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson, D. L., D. B. Wilson, and L. P. Walker. 2002. Synergism in binary mixtures of Thermobifida fusca cellulases Cel6B, Cel9A, and Cel5A on BMCC and Avicel. Appl. Biochem. Biotechnol. 101:97-111. [DOI] [PubMed] [Google Scholar]
- 28.Wilson, D. B. 2004. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4:72-82. [DOI] [PubMed] [Google Scholar]
- 29.Xiao, Z., R. Storms, and A. Tsang. 2004. Microplate-based filter paper assay to measure total cellulase activity. Biotechnol. Bioeng. 88:832-837. [DOI] [PubMed] [Google Scholar]
- 30.Yang, C. H., S. F. Yang, and W. H. Liu. 2007. Production of xylooligosaccharides from xylans by extracellular xylanases from Thermobifida fusca. J. Agric. Food Chem. 55:3955-3959. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, X., and J. Huang. 2003. Combination of overlapping bacterial artificial chromosomes by a two-step recombinogenic engineering method. Nucleic Acids Res. 31:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, Y., J. P. Muyrers, G. Testa, and A. F. Stewart. 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18:1314-1317. [DOI] [PubMed] [Google Scholar]
- 33.Zhou, W. L., D. C. Irwin, J. Escovar-Kousen, and D. B. Wilson. 2004. 2004. Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry 43:9655-9663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.