Abstract
Many cyanobacteria are known to tolerate environmental extremes. Motivated by an interest in selecting cyanobacteria for applications in space, we launched rocks from a limestone cliff in Beer, Devon, United Kingdom, containing an epilithic and endolithic rock-dwelling community of cyanobacteria into low Earth orbit (LEO) at a height of approximately 300 kilometers. The community was exposed for 10 days to isolate cyanobacteria that can survive exposure to the extreme radiation and desiccating conditions associated with space. Culture-independent (16S rRNA) and culture-dependent methods showed that the cyanobacterial community was composed of Pleurocapsales, Oscillatoriales, and Chroococcales. A single cyanobacterium, a previously uncharacterized extremophile, was isolated after exposure to LEO. We were able to isolate the cyanobacterium from the limestone cliff after exposing the rock-dwelling community to desiccation and vacuum (0.7 × 10−3 kPa) in the laboratory. The ability of the organism to survive the conditions in space may be linked to the formation of dense colonies. These experiments show how extreme environmental conditions, including space, can be used to select for novel microorganisms. Furthermore, it improves our knowledge of environmental tolerances of extremophilic rock-dwelling cyanobacteria.
The surface and interior of rocks is a ubiquitous environment for microorganisms. Comprehensive culturing and culture-independent analyses of endolithic (interior of rocks) and epilithic (on the surface of rocks) microbial communities have been conducted. The primary producers in these environments are phototrophs, such as cyanobacteria, that are either free living or endosymbionts in lichens (16).
Epilithic microorganisms are often an important part of rock-dwelling communities. The characterization of the epilithic cyanobacteria from natural environments, such as beach rock and caves, and from human-made environments, such as hypogea and buildings, has identified a variety of cyanobacteria. This includes both unicellular and filamentous forms, for example, Lyngbya-related species and Chroococcidiopsis (5, 14, 37, 47).
Many microorganisms also inhabit the interior of rocks as endoliths. The endolithic environment provides protection from environmental stresses such as desiccation, extreme temperature, UV radiation, and high photosynthetically active radiation (400 to 700 nm) (6, 16, 25, 32). Endolithic communities are often the dominant form of life in extreme environments such as hot and cold deserts (15-17), savannahs, and semideserts (3, 6, 15, 48). In these extreme environments, the endolithic cyanobacteria are mainly unicellular cyanobacteria, for example, Chroococcidiopsis, Myxosarcina, and Gloeocapsa species (11, 46, 50). Conversely, in nondesert environments, such as dolomitic rocks in Switzerland (41), the limestone of the Niagara Escarpment (19, 20), and travertine deposits in Yellowstone National Park, the endolithic communities are more diverse and include both filamentous and unicellular types of cyanobacteria, such as Leptolyngbya, Nostoc, and Synechocystis (34).
Although rock-dwelling cyanobacteria communities are diverse, there has been limited, if any, use of artificial extreme conditions to select for novel extremophilic cyanobacteria from these environments. Such an investigation could have implications for understanding the physiological requirements of life in extreme environments.
The work described in this paper was motivated by an interest in understanding the physiological tolerance of cyanobacteria to space conditions and their potential use in space applications, such as oxygen and feedstock provision, which are crucial for extraterrestrial settlements (23, 29). In this work, we exposed samples of a coastal limestone cliff in Beer, Devon, United Kingdom, which is inhabited by a diverse cyanobacterial community, to low Earth orbit (LEO) to isolate novel extreme-tolerant cyanobacteria.
MATERIALS AND METHODS
Sample collection.
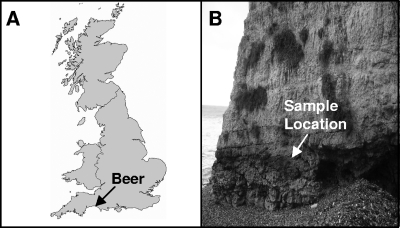
The sample site for this work was a cliff face in Beer, Devon, United Kingdom (50°41.50′N, 3°08.19′W) (Fig. 1A), that is made of Cretaceous nodular chalk limestone (21). Rocks were collected from the upper greensand layer, where limestone is predominant, with various amounts of the mineral glauconite. During high tide, the sampling site is submerged in seawater. The cyanobacteria inhabit the surface and interior of the rocks and form a homogenous epilithic covering (Fig. 1B). For the exposure experiments, the rocks were cut into blocks with an upper surface area of 1 cm2.
FIG. 1.
(A) Map of the United Kingdom showing the location of the sample site, Beer, Devon, United Kingdom. (Reprinted with permission of The Open University, Milton Keynes, United Kingdom.) (B) The rocks were collected from the upper greensand layer of the limestone cliffs, which is covered with a homogeneous epilithic covering of cyanobacteria.
Light transmission.
Light transmission measurements were used to determine the depth at which damaging UV radiation and photosynthetically active radiation could penetrate the rock. Transmission spectra were measured on rock sections that were 100 and 200 μm in thickness with an optical AvaSpec-1024 Spectrometer (Avantes, Netherlands) system. The UV/vis spectrometer measured the intensity of each wavelength of light in the UV and visible region (between 200 and 800 nm). For each of the rock sections, six positions were used to measure transmission at random locations on the sample. The means of these measurements were calculated. To determine the depth at which the minimal photon flux for photosynthesis would be reached, we used the theoretical value of 0.1 μmol photon/m2/s required for photosynthetic growth supported by O2-evolving photosynthesis (39).
Culturing of cyanobacteria.
To culture cyanobacteria, samples from the limestone cliff were incubated in 5 ml of modified BG-11 medium or filtered sterilized seawater (36). The filtered seawater was prepared by vacuum filtering 500 ml of seawater from Beer, Devon, through a 0.22-μm filter (Millipore, United Kingdom). The cultures were grown at 25°C under natural sunlight and day/night cycles.
16S rRNA gene analysis of isolates and whole-community genomic DNA.
For the analysis of the cyanobacteria community, rocks were collected from the upper greensand layer. To avoid contamination, the rocks were collected aseptically and stored in sterile plastic bags (Whirlpak; Fisher Scientific) at −80°C. For DNA extraction, three of the collected rocks were crushed, as previously described (24). The powdered rock was ground in a sterile pestle and mortar containing liquid nitrogen. DNA was extracted from 5 g of the crushed rock using the PowerMax Soil DNA Isolation kit (MoBio Laboratories, Cambridge, United Kingdom) according to the manufacturer's instructions.
Cyanobacterium-specific primers were used to amplify, by PCR, a partial region of the 16S rRNA gene, of between 613 and 618 nucleotides (35, 36). The 16S rRNA genes PCR product was extracted and purified from a 0.8% (wt/vol) agarose gel (Invitrogen, Paisley, United Kingdom) by means of the GenElute Gel Extraction kit (Sigma-Aldrich, Poole, United Kingdom) according to the manufacturer's instructions. The purified product was ligated at 4°C with the pCR4-TOPO vector. Chemical transformation was conducted with OneShot TOP10 chemically competent Escherichia coli from the TOPO-TA cloning kit (Invitrogen, Paisley, United Kingdom), as previously described (24).
The 16S rRNA gene inserts were sequenced using the T7 and T3 universal primers. The sequences were assembled with BioEdit and submitted to the CHECK_CHIMERA program of the Ribosomal Database Project (RDP; http://rdp8.cme.msu.edu/html) (10). The sequences were phylogenetically classified using the classifier in Ribosomal Data Project II, and the nearest 16S rRNA gene sequences were identified in the GenBank database using the BLASTN program (1). On the basis of these results, the sequences were aligned with representative cyanobacterial sequences from the GenBank database by ClustalX (45). The phylogenetic tree was constructed using the neighbor-joining method (40) and the Kimura two-parameter for distance correction (27) and visualized with NjPLOT (38). Rarefaction analysis was conducted with the FastGroupII program (51).
To identify the cyanobacteria isolates, a partial region of the 16S rRNA gene, of between 1,279 and 1,294 nucleotides, and the entire 16S-23S rRNA internal transcribed spacer (ITS) were sequenced. The 16S rRNA gene region was amplified with CYA106F and pH primers (4, 35). The ITS region was amplified with the 16S1407F and the 23S30R primers (26, 30). For both sets of primers, the PCR and amplification program were the same as those previously described (8). The PCR products were cleaned using the GenElute Gel Extraction kit (Sigma-Aldrich, Poole, United Kingdom), and the DNA sequencing of the PCR products was carried out directly. For phylogenetic analysis, only the region within the D1 and D4 conserved domains of the ITS region, of between 473 and 542 nucleotides, was used for the BLASTN analysis (26).
Isolation experiments.
To select for extremophilic cyanobacteria, samples from the limestone cliff were exposed to the combined environmental conditions of LEO (vacuum, 0.133 × 10−6 kPa; temperature, −20°C to +30°C; solar radiation, >170 nm). Furthermore, samples were exposed to the individual effects of vacuum (0.7 × 10−3 ± 0.01 × 10−3 kPa), desiccation, and UV radiation (325 to 400 nm) using ground-based experiments.
The ground-based experiments were conducted in triplicate, and a set of replicas was stored under ambient conditions in darkness as controls. Samples, including the controls, were collected aseptically after 1, 4, 7, 21, and 28 days (unless stated differently below). The LEO experiment did not include triplicate samples due to weight constraints, and the samples were exposed for 10 days.
Following exposure, each of the rocks was split aseptically and incubated in 5 ml of BG-11 medium or filtered and sterilized seawater. Microbial growth was monitored routinely using a Leica DMRP microscope (magnification, ×1,000) equipped with epifluorescence. Cell counts were conducted, after 10 weeks, by counting 35 fields of view, and the means and standard deviations associated with this value were determined. The cyanobacteria were isolated for morphological and molecular analysis by inoculating agar plates of BG-11 and seawater (1% agar; bacteriological agar no. 1; Oxoid, Basingstoke, United Kingdom).
Low Earth orbit.
Exposure to LEO was carried out as part of the European Space Agency (ESA)-funded Biopan VI mission. The samples were launched into LEO using a Soyuz launch vehicle (Russian Space Agency), which carries a Foton capsule into orbit. The Biopan, which is a space exposure facility, was installed on the external surface of the Foton capsule and opened in orbit to expose the samples to the combined environmental conditions of LEO for 10 days. The rocks were placed on two sample plates. The upper plate allowed for exposure to space conditions, extraterrestrial solar UV, and visible light. Cutoff filters, which permitted only the transmission of wavelengths that were longer than the specified values, also were used to expose the samples to the following wavelengths: >110 nm to simulate the full extraterrestrial spectrum, >200 nm to simulate UV on Mars, >290 nm to simulate UV on Earth, and >400 nm for the visible spectrum. The lower sample plate enabled the samples to be exposed to space conditions without exposure to UV or visible radiation as dark controls (36). An identical set of samples were kept for the same period in the laboratory at ambient conditions (21°C, atmospheric pressure) in darkness to serve as laboratory ground controls.
Ground-based exposure experiments.
To enhance our understanding of cyanobacterial survival in LEO, the samples from the limestone cliff were exposed to vacuum, desiccation, and UV radiation. The effect of desiccation was measured at room temperature in a desiccator containing silica gel beads, which were heated overnight at 80°C to eliminate moisture prior to use. Exposure to vacuum (0.7 × 10−3 ± 0.01 × 10−3 kPa) and UV radiation (325 to 400 nm) was conducted in a simulation chamber, as previously described (36). The samples were exposed to 1, 5, 10, 30, 45, and 60 min of UV radiation.
TEM analysis.
The isolate OU_20, obtained from the LEO exposure experiment, was examined by TEM. After 31 days of growth on BG-11 plates, colonies were transferred via an aseptic loop into 1 ml of fixative. Sections were prepared as previously described and examined with a JEM 1400 (JEOL) transmission electron microscope (TEM) at 80 kV, and digital images were acquired with an AMT XR60 camera of 11 megapixels (8).
Nucleotide sequence accession numbers.
The sequences obtained from the clone library and the isolates were deposited in the GenBank database under the accession numbers GQ162225 to GQ162317.
RESULTS
Light transmission of the rock.
To assess light absorbance inside the rock, spectra between 200 and 800 nm were measured by transmitting light through 100- and 200-μm-thick rock sections. No light transmission was measured through the 200-μm section. Through the 100-μm section, less than 0.5% of light was transmitted between wavelengths 250 and 400 nm. Below 250 nm, the low lamp transmission resulted in high background noise, and measurements were not reliable. The light transmission was heterogeneous and varied according to the presence of crystals, microfractures, and other heterogeneities in the rock content. At 680 nm, the transmission values varied from a minimum of 3.5% to a maximum of 4.5% (n = 6). The depth at which photosynthetically active radiation would be reduced to the minimum required for photosynthesis was calculated to be approximately 450 μm (n = 6).
Molecular diversity of the cryptoendolithic cyanobacterial community.
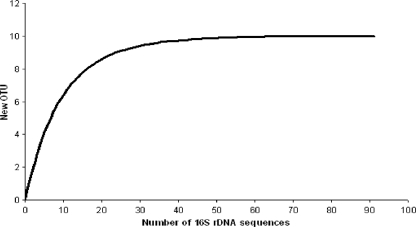
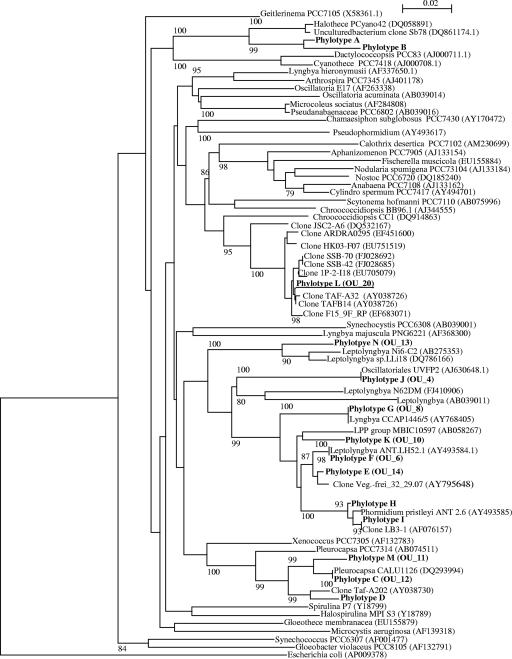
Approximately 0.9 μg of DNA per gram of rock was extracted from the limestone cliffs form Beer, United Kingdom. A total of 92 cyanobacterial clones were obtained. The rarefaction curve reached a plateau at a cutoff value of 97%, as shown in Fig. 2. The combined sequence data for the clone library and the isolates clustered to form 14 phylotypes, which have a threshold value of 97% similarity (42) (Fig. 3). BLASTN analysis revealed that 10 of the phylotypes had nearest-match identities of 97% or greater to sequences in the GenBank database. The largest phylotype group was A, with 33 clones, and the members were distantly related (92 to 94% identity) to a Halothece species from the marine environment of Shark Bay, Australia. The second largest group, E, included an uncultured sequence from an inland salt habitat. Of the remaining groups, five had nearest matches from extreme environments such as Antarctica and a cold sulfurous spring, while the remaining groups all had nearest matches to sequences identified from marine or freshwater environments.
FIG. 2.
Evaluation of the representation of the bacterial clones obtained from the limestone rock from Beer by rarefaction analysis. The appearance of new OTU strains was plotted as a function of the number of clones, which were analyzed randomly. The rarefaction curves were generated using FastGroupII (49). The curve was calculated with sequence similarity cutoff values of 97.0% (species level).
FIG. 3.
16S rRNA gene phylogenetic tree of the cyanobacterial community of the limestone rocks. The neighbor-joining tree was constructed using an approximately 700-nucleotide region of the 16S rRNA gene, which was amplified with cyanobacterium-specific primers (31). The phylogenetic tree was based on 92 sequences from the clone library, 9 sequences from the isolates, and 56 cyanobacterial sequences from GenBank, which were cut to the same length as the clone sequences. The phylotypes contained the following number of sequences: phylotype A, 33; phylotype B, 2; phylotype C, 10; phylotype D, 3; phylotype E, 13; phylotype F, 2; phylotype G, 9; phylotype H, 5; phylotype I, 9; phylotype J, 12; phylotype K, 1; phylotype L, 1; phylotype M, 1; and phylotype N, 1. The phylotypes are in boldface, and OU_20 is underlined. The scale bar corresponds to 0.02 changes per nucleotide. The percentage of bootstrap replicates (1,000 replicates) resulting in the same cluster is given near the respective nodes for bootstrap values higher than 80%.
Exposure experiments. (i) Isolation of cyanobacteria using LEO.
The samples from the limestone cliff were exposed to 10 days of LEO to select for resistant cyanobacterial strains. The environmental parameters during the experiment were the following: vacuum, 0.133 × 10−6 kPa; temperature, −20°C to +30°C; solar radiation, >170 nm. The incubation of the rocks, after exposure, resulted in the isolation of a cyanobacterium, OU_20. The growth of this isolate was detected on the control rocks (no UV exposure) after 160 days. This was followed by the growth of the same isolate on the rocks exposed to UV at >290 and >400 nm, as shown in Table 1.
TABLE 1.
Survival of cyanobacterium OU_20 from the rock-dwelling community after exposure to low Earth orbit
| Wavelength (nm) | Survivala (days) |
|
|---|---|---|
| With UV exposure | Without UV exposure | |
| >110 | 206 | |
| >200 (simulating UV on Mars) | 160 | |
| >290 (simulating UV on Earth) | 196 | |
| >400 (visible spectrum) | 198 | 201 |
The number of days before growth was detected after incubation.
(ii) Ground-based exposure experiments.
Ground-based experiments were conducted to isolate cyanobacteria from the samples after exposure to the individual effects of vacuum, desiccation, and UV radiation. Nine cyanobacteria were isolated after exposure to 1 day of desiccation, while only four were isolated after 28 days of exposure, as shown in Table 2. Furthermore, the cyanobacterial cell counts decreased from 4.0 × 105 ± 0.8 × 105 ml−1 after 1 day to 2.1 × 103 ± 0.5 × 103 ml−1 after 28 days. Vacuum was more detrimental than desiccation, and only three cyanobacterial strains were isolated after 1 day of exposure, as shown in Table 2. The total number of cells decreased from 2.0 × 102 ± 0.9 × 102 ml−1 after 1 day to 5.1 × 101 ± 0.6 × 101 ml−1 after 28 days.
TABLE 2.
Survival of cyanobacteria from the rock-dwelling community after exposure to UV radiation, desiccation, vacuum, and low Earth orbita
| Isolate | UV (min) | Desiccation (days) | Vacuum (days) | LEO (days) |
|---|---|---|---|---|
| OU_4 | 1 | 1 | ||
| OU_6 | 60 | 28 | ||
| OU_8 | 7 | |||
| OU_10 | 60 | 7 | 1 | |
| OU_11 | 1 | |||
| OU_12 | 60 | 1 | ||
| OU_13 | 60 | 28 | 28 | |
| OU_14 | 28 | |||
| OU_20 | 28 | 14 | 10 |
Shown are the maximum exposure times that the isolates could survive.
Moreover, when the samples were exposed to UV radiation (325 to 400 nm) with a total dose of 54.6 W/m2, the number of isolates decreased dramatically. After 5 min of exposure, five cyanobacterial strains were isolated (the total number of cyanobacteria cells was 2.1 × 101 ± 0.8 × 101 ml−1), and only four were isolated after 60 min of exposure (4.0 × 101 ± 1.1 × 101 ml−1), as shown in Table 2.
Morphological and molecular analyses of the isolated cyanobacteria.
The isolates were compared morphologically to the taxonomy descriptions of Anagnostidis and Komárek as well as Tatton et al. (2, 28, 44, and www.CyanoDB.cz). The isolates OU_6, OU_8, OU_13, and OU_14 were similar to the descriptions and the photomicrographs of the Leptolyngbya strains (order Oscillatoriales) described by Taton et al. (43). In addition, the isolates OU_4 and OU_10 were comparable to the Phormidium strains (order Oscillatoriales) (43). The remaining isolates were nonfilamentous. The isolates OU_12 and OU_11 formed spherical cells that were slightly elongated in shape and were similar to the description of the order Pleurocapsales by Komárek et al. (www.CyanoDB.cz). A detailed description of the morphology of the isolates is available in Table S1 in the supplemental material.
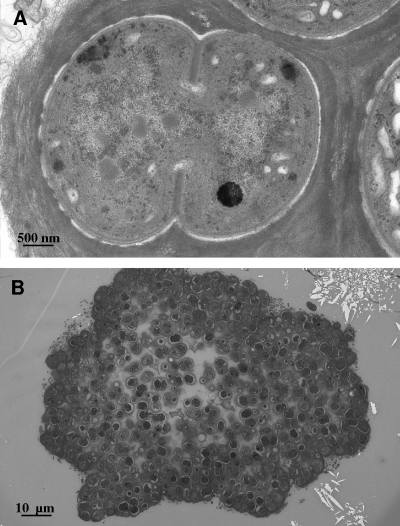
Due to the significance of isolate OU_20 as the only isolate that survived space conditions, TEM analysis also was conducted to investigate characteristics that may have improved its extreme tolerance. The isolate OU_20 formed large clumps, which were surrounded by a mucilaginous sheath, as shown in Fig. 4. The cells divided by a local thickening of the cell wall and the formation of a septum. The morphology of OU_20 is similar to that of the order Chroococcales (49).
FIG. 4.
TEM images of the cyanobacterium OU_20 that survived exposure to low Earth orbit.
In addition to the morphological analysis, we amplified a partial region of the 16S rRNA gene of between 1,279 and 1,294 nucleotides and the total ITS region. The results from the BLASTN analysis of the sequences are shown in Table 3. Where the nearest-matched sequences were from cultured cyanobacteria, the sequences were in agreement with the morphological descriptions of the isolates. Five of the isolates clustered with phylotype groups identified in the clone library, while the remaining sequences formed new phylotype groups, as shown in Table 3.
TABLE 3.
BLASTN analysis of the partial 16S rRNA gene and the 16S-23S rRNA ITS sequences for the isolates
| Isolate | Phylotype | 16S rRNA gene |
Internal transcribed rRNA genea |
||
|---|---|---|---|---|---|
| Nearest match | % | Nearest match | % | ||
| OU_4 | J | Oscillatoriales sp. UVFP2 (AJ630648.1) | 99 | Leptolyngbya fragilis OL 03 (AM398972.1) | 82 |
| OU_6 | F | Leptolyngbya sp. ANT.LH52.1 (AY493584.1) | 100 | Leptolyngbya sp. ANT.LH52.1 (AY493637.1) | 95 |
| OU_8 | G | Leptolyngbya sp. ANT.LH52.1 (AY493584.1) | 97 | Leptolyngbya sp. ANT.LH52.1 | 86 |
| OU_10 | K | Phormidium pristleyi sp. ANT.ACEV5.1 (AY493586.1) | 99 | Uncultured cyanobacterium R8-R79 (DQ181763.1) | 87 |
| OU_11 | M | Pleurocapsa sp. CALU 1126 (DQ293994.1) | 96 | Unicellular cyanobacterium LLi67 (DQ786165.1) | 88 |
| OU_12 | C | Pleurocapsa sp. CALU 1126 (DQ293994.1) | 97 | Unicellular cyanobacterium LLi67 (DQ786165.1) | 81 |
| OU_13 | Nb | Leptolyngbya sp. VRUC198/Albertano 1992 (X84809.1) | 96 | Leptolyngbya sp. VRUC198/Albertano 1992 (EF560653.1) | 99 |
| OU_14 | Eb | Leptolyngbya sp. ANT.LH52.1 (AY493584.1) | 98 | Leptolyngbya sp. ANT.LH52.1 (AY493637.1) | 77 |
| OU_20 | Lb | Bacterium clone JSC2-A6 (DQ532167.1) | 95 | Antarctic cyanobacterium clone S334-20 (EU032386.1) | 86 |
Conserved domains D1 to D4 were described by Iteman et al. (26).
Phylotypes are not identified in the clone library.
DISCUSSION
The objective of this study was to investigate the effects of LEO on a rock-dwelling cyanobacterial community. The epilithic and endolithic community used in this investigation was from a limestone cliff in Beer, Devon, United Kingdom. Although the environmental conditions at Beer are not as extreme as those experienced by rock-dwelling communities in the Atacama Desert or Antarctica, the cliff experiences periods of desiccation, temperature fluctuations, and cyclical exposure to freshwater and seawater, as well as exposure to UV radiation. This sample location was selected because we predicted that the cyanobacterial community would be diverse and therefore would yield useful results on the relative survivability of different cyanobacteria in space conditions. Furthermore, we hypothesized that exposure to extreme conditions resulted in the selection of entirely novel extremophilic organisms. Extremophilic cyanobacteria have use in future space applications, such as oxygen, fuel, and biomass production; nutrient acquisition; biomining; and feedstock provision, which are crucial for long-term space flights and extraterrestrial settlements (23, 29).
The rocks were exposed to LEO as part of the ESA-funded Biopan VI experiment. Previous exposure experiments aboard the Biopan facility have shown that some cyanobacteria can survive space conditions. Mancinelli et al. demonstrated that a Synechococcus species, isolated from gypsum-halite crystals collected from the marine intertidal area along the coast of Baja California, Mexico, was able to survive exposure to LEO for 15 days (31). However, nitrogen fixation decreased from approximately 1.5 × 10−3 in controls to 4 × 10−4 μmol ethylene/ng protein in flight samples, as measured by the acetylene reduction assay.
The exposure of the endolithic and epilithic community from Beer to LEO resulted in the isolation of a single extremophilic cyanobacterium, designated OU_20. Further ground-based exposure experiments were conducted to select for cyanobacteria using the individual effects of vacuum, desiccation, and UV radiation. The isolate OU_20 subsequently was isolated after exposure to desiccation and vacuum. However, OU_20 was not detected after exposure to UV radiation (325 to 400 nm).
These data must be reconciled with the data acquired for OU_13, which did not survive exposure to space but survived all of the ground-based experiments, including UV exposure. The survival of exposure to UV radiation may be due to the filamentous nature of OU_13, which enables the cyanobacterium to bore into the rock or grow endolithically into rock cavities, which would protect the cyanobacterium from UV radiation (9). This would be consistent with the depth at which photosynthetically active radiation would be reduced to the minimum required for photosynthesis, which was 450 μm. The failure of OU_13 to survive in space may be caused by differences in the effects of the combined stresses associated with LEO compared to those of OU_20 or differences in the abundance of the organisms in the rocks. If OU_13 was less abundant than OU_20, it may have been killed completely during the exposure to space, whereas small numbers of OU_20 could have survived. Another explanation may be inhomogeneity in the distribution of organisms in different samples, although our culturing experiments suggest that the organisms were distributed quite uniformly on the rock surface.
The morphological features of OU_20 resembled those attributed to the genus Gloeocapsa described by Komárek et al. (www.CyanoDB.cz). Furthermore, TEM images of Gloeocapsa-like cyanobacteria isolated from biofilms colonizing granite rock in Antarctica were similar to those of OU_20 (12, 13). Previous work with Gloeocapsa species has shown that in the natural environment an extracellular sheath holds many cells together, and in such growth forms the effective cellular path lengths for radiation are greatly increased, as is the overall protection against UV radiation (18). The thick mucilaginous sheath observed in the TEM images of OU_20 may have protected the organism from the UV radiation (>290 nm), vacuum, and fluctuations in temperature associated with LEO.
OU_20 was selected, by exposure to LEO, from a diverse community of cyanobacteria. The phylogenetic analysis demonstrated that the community consisted of 14 phylogenetic groups that belonged to the orders Oscillatoriales, Pleurocapsales, and Chroococcales. The phylogenetic similarity of these sequences to those in the GenBank database demonstrates that members of the community are similar to cyanobacteria found in previously studied environments. For example, cyanobacterial sequences comparable to those found in Antarctica, the Taff River, and Lake Ladoga were discovered in the community at Beer (7, 22, 33, 44). The rarefaction analysis of the clone library showed that 92 clones were sufficient to reach the saturation of the community diversity; however, three of the isolates were not identified in the clone library. This might be explained by a very low abundance of these cyanobacteria in the community, such that the rarefaction curve, with 93 clones, reached apparent saturation, but rare phylotypes are still to be discovered.
In this work, we have isolated an extremophilic cyanobacterium from a cliff in Beer, Devon, United Kingdom, which is capable of surviving a variety of extreme conditions, including 10 days in space. We have demonstrated that extreme conditions, including space, can be used to select for novel microorganisms from natural rock-dwelling communities, which could advance our knowledge of environmental tolerances of rock-dwelling cyanobacteria and have implications for future space applications.
Supplementary Material
Acknowledgments
This work was supported by an STFC grant (PP/E001408/1) to C.S.C.
We thank the European Space Agency for the flight opportunity and the Russian Space Agency for access to the launch vehicle and facilities. We also thank Manish Patel and Martin Towner for their help with the xenon light and the Mars simulation chamber. Finally, we thank Heather Davies for the TEM analysis.
Footnotes
Published ahead of print on 12 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostidis, K., and J. Komarek. 1990. Modern approach to the classification-system of Cyanophytes 5. Stigonematales. Archiv. Hydrobiol. 86:1-73. [Google Scholar]
- 3.Bell, R. A. 1993. Cryptoendolithic algae of hot semiarid lands and deserts. J. Phycol. 29:133-139. [Google Scholar]
- 4.Bruce, K. D., W. D. Hiorns, J. L. Hobman, A. M. Osborn, P. Strike, and D. A. Ritchie. 1992. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3413-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruno, L., D. Billi, and P. Albertano. 2006. Genetic characterization of epilithic cyanobacteria and their associated bacteria. Geomicrobiol. J. 23:293-299. [Google Scholar]
- 6.Budel, B., and D. C. J. Wessles. 1991. Rock inhabiting blue green algae/cyanobacteria from hot arid regions. Algol. Stud. 64:385-398. [Google Scholar]
- 7.Camacho, A., C. Rochera, J. J. Silvestre, E. Vicente, and M. W. Hahn. 2005. Spatial dominance and inorganic carbon assimilation by conspicuous autotrophic biofilms in a physical and chemical gradient of a cold sulfurous spring: the role of differential ecological strategies. Microb. Ecol. 50:172-184. [DOI] [PubMed] [Google Scholar]
- 8.Cockell, C., K. Olsson, F. Knowles, L. Kelly, A. Herrera, T. Thorsteinsson, and V. Marteinsson. 2009. Bacteria in weathered basaltic glass, Iceland. Geomicrobiol. J. 26:491-507. [Google Scholar]
- 9.Cockell, C. S., and A. Herrera. 2008. Why are some microorganisms boring? Trends Microbiol. 16:101-106. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Torre, J. R., B. M. Goebel, E. I. Friedmann, and N. R. Pace. 2003. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 69:3858-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de los Ríos, A., M. Grube, L. G. Sancho, and C. Ascaso. 2007. Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks. FEMS Microbiol. Ecol. 59:386-395. [DOI] [PubMed] [Google Scholar]
- 13.de los Ríos, A., J. Wierzchos, L. G. Sancho, and C. Ascaso. 2003. Acid microenvironments in microbial biofilms of antarctic endolithic microecosystems. Environ. Microbiol. 5:231-237. [DOI] [PubMed] [Google Scholar]
- 14.Díez, B., K. Bauer, and B. Bergman. 2007. Epilithic cyanobacterial communities of a marine tropical beach rock (Heron Island, Great Barrier Reef): diversity and diazotrophy. Appl. Environ. Microbiol. 73:3656-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedmann, E. I. 1972. Ecology of lithophytic algal habitats in middle eastern and north American deserts, p. 182-185. In L. E. Rodin (ed.), Ecophysiological foundation of ecosystem productivity in arid zones. Nauka, Moscow, former Soviet Union.
- 16.Friedmann, E. I. 1980. Endolithic microbial life in hot and cold deserts. Orig. Life. 10:223-235. [DOI] [PubMed] [Google Scholar]
- 17.Friedmann, E. I., and R. Ocampo Paus. 1984. Endolithic microorganisms in extreme dry environments: analysis of a lithobiontic microbial habitat. In M. J. Klug and C. A. Reddy (ed.), Current perspectives in microbial ecology. Proceedings of the Third International Symposium on Microbial Ecology. American Society for Microbiology, Washington, DC.
- 18.Garcia-Pichel, F., C. E. Wingard, and R. W. Castenholz. 1993. Evidence regarding the UV sunscreen role of a mycosporine-like compound in the cyanobacterium Gloeocapsa sp. Appl. Environ. Microbiol. 59:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerrath, J. F., J. A. Gerrath, and D. W. Larson. 1995. A preliminary account of the endolithic algae of limestone cliffs of the Niagara Escarpment. Can. J. Bot. 73:788-793. [Google Scholar]
- 20.Gerrath, J. F., J. A. Gerrath, U. Matthes, and D. W. Larson. 2000. Endolithic algae and cyanobacteria from cliffs of the Niagara Escarpment, Ontario, Canada. Can. J. Bot. 78:807-815. [Google Scholar]
- 21.Griffiths, J. 2000. Engineering geomorphological mapping of the Upper Greensand escarpment near Honiton, Devon. Geosci. South-West England 10:64-67. [Google Scholar]
- 22.Gromov, B. V., A. A. Vepritsky, K. A. Mamkaeva, and L. N. Voloshko. 1996. A survey of toxicity of cyanobacterial blooms in Lake Ladoga and adjacent water bodies. Hydrobiologia 322:149-151. [Google Scholar]
- 23.Hendrickx, L., and M. Mergeay. 2007. From the deep sea to the stars: human life support through minimal communities. Curr. Opin. Microbiol. 10:231-237. [DOI] [PubMed] [Google Scholar]
- 24.Herrera, A., and C. S. Cockell. 2007. Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J. Microbiol. Methods 70:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, K. A., and B. Lawley. 2003. A novel Antarctic microbial endolithic community within gypsum crusts. Environ. Microbiol. 5:555-565. [DOI] [PubMed] [Google Scholar]
- 26.Iteman, I., R. Rippka, N. Tandeau De Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiol. 146(Pt 6):1275-1286. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, H., M. Sugihara, K. Kato, and S. Hanada. 2006. Selective phylogenetic analysis targeted at 16S rRNA genes of thermophiles and hyperthermophiles in deep-subsurface geothermal environments. Appl. Environ. Microbiol. 72:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komárek, J., and K. Anagnostidis. 1989. Modern approach to the classification-system of Cyanophytes 4—Nostocales. Archiv. Hydrobiol. 82:247-345. [Google Scholar]
- 29.Lehto, K., E. Kanervo, K. Stahle, H. Lehto, M. Tammi, and J. Virtanen. 2007. Photosynthetic life support systems in the Martian conditions. In C. Cockell and G. Horneck (ed.), ROME: response of organisms to the Martian environment, ESA AP-1299. European Space Agency, Paris, France.
- 30.Lepere, C., A. Wilmotte, and B. Meyer. 2000. Molecular diversity of Microcystis strains (Cyanophyceae, Chroococcales) based on 16S rDNA sequences. Syst. Geogr. Plants 70:275-283. [Google Scholar]
- 31.Mancinelli, R. L., M. R. White, and L. J. Rothschild. 1998. Biopan-survivial I: exposure of the osmophiles Synechococcus sp. (Nageli) and Haloarcela sp. to the space environment. Adv. Space Res. 22:327-334. [Google Scholar]
- 32.McKay, C. P., and E. I. Friedmann. 1985. The cryptoendolithic microbial environment in the Antarctic cold desert: temperature variations in nature. Polar Biol. 4:19-25. [DOI] [PubMed] [Google Scholar]
- 33.Nelissen, B., R. De Baere, A. Wilmotte, and R. De Wachter. 1996. Phylogenetic relationships of nonaxenic filamentous cyanobacterial strains based on 16S rRNA sequence analysis. J. Mol. Evol. 42:194-200. [DOI] [PubMed] [Google Scholar]
- 34.Norris, T. B., and R. W. Castenholz. 2006. Endolithic photosynthetic communities within ancient and recent travertine deposits in Yellowstone National Park. FEMS Microbiol. Ecol. 57:470-483. [DOI] [PubMed] [Google Scholar]
- 35.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson-Francis, K., R. de la Torre, M. C. Towner, and C. S. Cockell. 2009. Survival of akinetes (resting-state cells of cyanobacteria) in low Earth orbit and simulated extraterrestrial conditions. Orig. Life Evol. Biosph. 39:565-579. [DOI] [PubMed] [Google Scholar]
- 37.Ortega-Morales, B. O., C. C. Gaylarde, G. E. Englert, and P. M. Gaylarde. 2005. Analysis of salt-containing biofilms on limestone buildings of the Mayan culture at Edzna, Mexico. Geomicrobiol. J. 22:261-268. [Google Scholar]
- 38.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 39.Raven, J. A., J. E. Kubler, and J. Beardall. 2000. Put out the light, and then put out the light. J. Marine Biolog. Assoc. United Kingdom 80:1-25. [Google Scholar]
- 40.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 41.Sigler, W. V., R. Bachofen, and J. Zeyer. 2003. Molecular characterization of endolithic cyanobacteria inhabiting exposed dolomite in central Switzerland. Environ. Microbiol. 5:618-627. [DOI] [PubMed] [Google Scholar]
- 42.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 43.Taton, A., S. Grubisic, E. Brambilla, R. De Wit, and A. Wilmotte. 2003. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 69:5157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taton, A., S. Grubisic, D. Ertz, D. A. Hodgson, R. Piccardi, N. Biondi, M. R. Tredici, M. Mainini, D. Losi, F. Marinelli, and A. Wilmotte. 2006. Polyphasic study of Antarctic cyanobacterial strains. J. Phycol. 42:1257-1270. [Google Scholar]
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Thielen, N., and D. J. Garbary. 1999. Life in the rocks: endolithic algae, p. 243-253. In J. Seckbach (ed.), Enigmatic microorganisms and life in extreme environments. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 47.Vinogradova, O. N., O. V. Kovalenko, S. P. Wasser, E. Nevo, and M. Weinstein-Evron. 1998. Species diversity gradient to darkness stress in blue-green algae/cyanobacteria: a microscale test in a prehistoric cave, Mount Carmel, Israel. Isr. J. Plant Sci. 46:229-238. [Google Scholar]
- 48.Walker, J. J., and N. R. Pace. 2007. Phylogenetic composition of Rocky Mountain endolithic microbial ecosystems. Appl. Environ. Microbiol. 73:3497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterbury, J. B., and R. Y. Stanier. 1978. Patterns of growth and development in Pleurocapsalean cyanobacteria. Microbiol. Rev. 42:2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitton, B. A., and M. Potts. 2000. The ecology of cyanobacteria—their diversity in time and space. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 51.Yu, Y. N., M. Breitbart, P. McNairnie, and F. Rohwer. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.