Abstract
Zearalenone (ZON) is a potent estrogenic mycotoxin produced by several Fusarium species most frequently on maize and therefore can be found in food and animal feed. Since animal production performance is negatively affected by the presence of ZON, its detoxification in contaminated plant material or by-products of bioethanol production would be advantageous. Microbial biotransformation into nontoxic metabolites is one promising approach. In this study the main transformation product of ZON formed by the yeast Trichosporon mycotoxinivorans was identified and characterized by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and LC-diode array detector (DAD) analysis. The metabolite, named ZOM-1, was purified, and its molecular formula, C18H24O7, was established by time of flight MS (TOF MS) from the ions observed at m/z 351.1445 [M-H]− and at m/z 375.1416 [M+Na]+. Employing nuclear magnetic resonance (NMR) spectroscopy, the novel ZON metabolite was finally identified as (5S)-5-({2,4-dihydroxy-6-[(1E)-5-hydroxypent-1-en-1-yl]benzoyl}oxy)hexanoic acid. The structure of ZOM-1 is characterized by an opening of the macrocyclic ring of ZON at the ketone group at C6′. ZOM-1 did not show estrogenic activity in a sensitive yeast bioassay, even at a concentration 1,000-fold higher than that of ZON and did not interact with the human estrogen receptor in an in vitro competitive binding assay.
Zearalenone (ZON) is the main member of a growing family of biologically important “resorcylic acid lactones” (RALs), which have been found in nature. ZON is produced by several Fusarium species, which colonize maize, barley, oat, wheat, and sorghum and tend to develop ZON during prolonged cool, wet growing and harvest seasons (38). Maize is the most frequently contaminated crop plant, and therefore, ZON can be found frequently in animal feeding stuff. Occurrence, toxicity, and metabolism data of ZON were summarized by the European Food Safety Authority (EFSA) (5) and in recent reviews (12, 38).
The potent xenohormone ZON leads to hyperestrogenism symptoms and in extreme cases to infertility problems, especially in pigs (15). Ovarian changes in pigs have been noted with toxin levels as low as of 50 μg/kg in the diet (1). Ruminants are more tolerant to ZON ingestion; however, hyperestrogenic syndrome, including restlessness, diarrhea, infertility, decreased milk yields, and abortion, have been well documented with cattle and sheep (4, 29).
Because widespread ZON contamination in feed can occur in problematic years, efficient ways to detoxify are desirable. The transformation of mycotoxins to nontoxic metabolites by pure cultures of microorganisms or by cell-free enzyme preparations (3) is an attractive possibility. Microbial metabolization of ZON to alpha-ZOL and beta-ZOL cannot be regarded as detoxification, because both ZOL products are still estrogenic (14). Also, formation of ZON-glucosides and -diglucosides (8, 17) and ZON-sulfate (7) cannot be considered true detoxification but rather formation of masked mycotoxins, because the conjugates may be hydrolyzed during digestion (11, 23), releasing ZON again (2).
As the estrogenic activity of ZON and its derivates can be explained by its chemical structure, which resembles natural estrogens (20), it can be expected that cleavage of the lactone undecyl ring system of ZON results in permanent detoxification.
El-Sharkawy and Abul-Hajj (9) were the first to report inactivation of ZON after opening of the lactone ring by Gliocladium roseum. This filamentous fungus was capable of metabolizing ZON in yields of 80 to 90%. Also Takahashi-Ando et al. (31) described the degradation reaction of ZON with Clonostachys rosea (synonym of G. roseum). A hydrolase (encoded by a gene designated ZHD101) cleaves the lactone ring, and as recently proved (37; unpublished data) by subsequent decarboxylation of the intermediate acid, the compound 1-(3,5-dihydroxyphenyl)-10′-hydroxy-1′E-undecene-6′-one is formed. In contrast to ZON and 17β-estradiol, which showed potent estrogenic activity, this cleavage product did not show any estrogenic activity in the human breast cancer MCF-7 cell proliferation assay (16). Further details, e.g., on the conditions of the maximum activity of ZHD101 and its exploitation in genetically modified grains, can be found in later published work of this research group (32, 33).
Only a few authors reported the loss of estrogenicity in microbial metabolites of ZON, which are based on reactions other than cleavage of the lactone undecyl ring system. El-Sharkawy and Abul-Hajj demonstrated (10) that binding to rat uterine estrogen receptors requires a free 4-OH phenolic group (devoid of methylation or glycosylation). Loss of estrogenicity was, for instance, observed with 2,4-dimethoxy-ZON, one of the metabolites produced by Cunninghamella bainieri ATCC 9244B. Nevertheless, this rule cannot be generalized, as 8′-hydroxyzearalenone formed by Streptomyces rimosus NRRL 2234, despite having a free 4-phenolic hydroxyl group, did not bind to the estrogen receptor. Also, other authors reported that 8′-hydroxyzearalenone and 8′-epi-hydroxyzearalenone are nonestrogenic (13). However, so far, no practical application in feed or food detoxification has been found for the microorganisms producing these compounds.
It has been shown previously that the yeast Trichosporon mycotoxinivorans has a very high capability to degrade both ochratoxin A (OTA) and ZON (22, 26, 27). When T. mycotoxinivorans is used as a feed additive preparation, microbial degradation of the mycotoxins is assumed to take place in the gastrointestinal tract of the animal after consumption of contaminated feed. The protective effect of T. mycotoxinivorans against OTA toxicity has already been shown with broiler chicken (24).
In the present study we report the isolation, analytical characterization, and structure elucidation, as well as the evaluation, of the estrogenic activity of the main degradation product of ZON produced by T. mycotoxinivorans.
MATERIALS AND METHODS
Microbial cultivation of T. mycotoxinivorans and degradation of ZON.
Erlenmeyer flasks with 30 ml yeast medium (10 g/liter glucose, 20 g/liter malt extract, 10 g/liter yeast extract, 5 g/liter peptone of casein) were inoculated with T. mycotoxinivorans (22) directly with aliquots from a culture stock stored at −80°C and incubated at 37°C and 200 rpm on an orbital shaker. After 48 h, biomass was harvested by centrifugation, resuspended in the same volume of sterile 0.9% NaCl containing 10 mg/liter ZON (Biopure Referenzsubstanzen GmbH, Tulln, Austria), and incubated under the same conditions. As controls, flasks containing solutions without ZON (matrix control) or without biomass (substrate control) were incubated in parallel. Samples (1.0 ml) were taken at given time points for up to 6 days and heat inactivated in glass vials for 5 min in a boiling water bath. Samples were stored frozen (−20°C) until analysis.
For preparative-scale production of the ZON metabolite ZOM-1, a total culture volume of 900 ml (150 ml per flask) was incubated under the conditions described above. Biomass was harvested, washed with 0.9% NaCl solution, dissolved in 900 ml minimal medium (MM) (27) containing 50 mg/liter of ZON, portioned in 50-ml volumes, placed in a 300-ml Erlenmeyer flask, and incubated at 35°C for 192 h. Heat inactivation in an autoclave followed for 10 min. Samples were stored frozen (−20°C) until analysis.
LC-MS/MS and LC-UV analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses were performed with a QTrap-LC-MS/MS system (Applied Biosystems, Foster City, CA), equipped with an electrospray ionization (ESI) source and an 1100 series high-pressure liquid chromatography (HPLC) system (Agilent, Waldbronn, Germany), including an 1100 series diode array detector (DAD).
Enhanced mass spectra (EMS) and enhanced product ion (EPI) scans, as well as other MS operation modes, e.g., precursor ion scan (data not shown), were used to gain structural information about the molecules. For analysis, samples were clarified by centrifugation (Beckman GS-6; 10 min at 3,500 rpm), and the supernatant was transferred to HPLC vials for direct injection.
Chromatographic separation was achieved at 25°C using a gradient on an 250- by 3.00-mm (length by inner diameter [i.d.]), 5-μm-particle-size Phenomenex Luna C18(2) column (Phenomenex Inc., Torrance, CA), including an identical matrix guard column. The injection volume was 10 μl, while the flow rate was 0.5 ml/min. Based on the mobile phases A, H2O/HCOOH (50 μl/liter), and B, MeOH/HCOOH (50 μl/liter), the elution started at 15% solvent B, with a linear gradient to 100% solvent B from 0 to 20 min. After holding for 7 min, the initial conditions returned within 1 min and were held for reequilibration until the end of the run at 38 min.
The ESI source was operated at 400°C in negative ionization mode. EMS parameters in the negative ionization mode were as follows: curtain gas (CUR), 20 lb/in2; nebulizer gas (GS1), 25 lb/in2; auxiliary gas (GS2), 65 lb/in2; ion spray voltage (IS), −4,200 V; declustering potential (DP), −30 V; entrance potential (EP), −10 V; collision energy (CE), −30 V; mass range, 100 to 850 amu; scan rate, 1,000 amu/s; and linear iontrap (LIT) fill time, 40 ms, where 1 lb/in2 is 6.895 kPa. MS/MS spectra were recorded in the EPI mode with the following parameters: LIT fill time, 40 ms; scan rate, 1,000 amu/s; Q1 resolution, unit; Q0 trapping, yes; and MR pause, 5.0 ms. The collision energy and the declustering potential for m/z 351.1 were DP, −51 V; and CE, −38 eV.
Data acquired by the DAD were recorded at 220 and 270 nm.
ZOM-1 isolation and purification.
Ethyl acetate was added at a ratio of 0.5:1 (vol/vol) to the clarified culture fluid, and extraction was repeated three times. The combined organic fractions were dried over anhydrous sodium sulfate (Na2SO4) and concentrated to dryness in a rotary evaporator, and the residue was reconstituted in ethyl acetate and transferred to a 4-ml screw vial in which it was evaporated again in a steam of nitrogen gas to yield 42.5 mg of crude residue. The crude residue was reconstituted in 2 ml of a mixture of MeOH/H2O (70:30) and cleared by centrifugation, and the supernatant was transferred to an HPLC vial.
ZOM-1 isolation was done at 25°C on a semipreparative Phenomenex Luna C18(2) column (Phenomenex Inc., Torrance, CA), 250- by 10.0-mm (length by i.d.), 5-μm particle size, including a C18 security guard column (10.0 by 10.0 mm; 5 μm). The injection volume was 80 μl, while the flow rate was 3.0 ml/min. The mobile phases consisted of A (H2O/HCOOH [50 μl/liter]) and B (MeOH/HCOOH [50 μl/liter]). Elution started isocratically at 65% B for 14 min, increased to 95% to wash the column, and returned to 65% B for column equilibration. Fractions belonging to ZOM-1 were collected after peak detection at 270 nm. After solvent evaporation using nitrogen gas flow and overnight lyophilization, the yield of isolated ZOM-1 was 16.7 mg.
ZOM-1 characterization via time of flight (TOF) MS.
Accurate mass measurements of the purified ZOM-1 were performed on an ESI micrOTOF mass spectrometer (Bruker Daltonik, Bremen, Germany) with flow injection.
Data acquisition was controlled by Bruker Daltonics micrOTOF control version 1.1. Data evaluation was performed using the Generate Molecular Formula (GMF) software suite within Bruker Daltonics micrOTOF DataAnalysis version 3.3, evaluating both accurate mass position and true isotopic patterns for the calculation of molecular formulae. The mass accuracy tolerance window of generated masses was set to 30 ppm in order to check how many hits are possible in the mass range of interest. Only molecular formulae of ions which yielded a “sigma value” of <0.02 (sigma is an indicator value for the isotopic pattern fit) were considered confident.
Calibration was done internally using sodium formate clusters [Na(NaCOOH)x]+ in positive ionization mode and formate adducts of sodium formate clusters in the form of [HCOO(NaCOOH)y]− in negative ionization.
Samples containing approximately 1 μg/liter ZOM-1 in MeOH/H2O (1:4 [vol/vol]) and the calibration standard were introduced into the mass spectrometer via a Hamilton syringe at a flow rate of 180 μl/h, and measurements were carried out in the positive as well as negative ion mode using a scan range of m/z 50 to 1,000 and the following settings for both polarities: end plate offset, 500 V; capillary voltage, 4,500 V; nebulizer pressure (N2 gas), 0.5 bar; dry gas flow (N2), 5 liter/min; dry temperature, 200°C; flight tube voltage value, 9,000 V; and reflector voltage, 1,300 V. The following transfer parameters were applied in positive ion mode: capillary exit at 120 V, skimmer 1 at 50 V, hexapole 1 at 24 V, skimmer 2 at 23 V, hexapole 2 at 21 V, hexapole RF at 150 voltage per pole (Vpp), transfer time at 45 μs, and pre puls storage at 5 μs. In the negative ion mode, the transfer parameters were capillary exit at −120 V, skimmer 1 at −40 V, hexapole 1 at −24 V, skimmer 2 at −26 V, hexapole 2 at −21 V, hexapole RF at 120 Vpp, transfer time at 45 μs, and pre puls storage at 5 μs.
Nuclear magnetic resonance (NMR) spectroscopy of ZOM-1.
For structure elucidation of ZOM-1, 1H, 13C-APT, 1H1H correlation spectroscopy (COSY), 1H13C heteronuclear single quantum correlation (HSQC), and 1H13C heteronuclear multiple bond correlation (HMBC) spectra were recorded. Approximately 15 mg ZOM-1 was dissolved in about 0.5 ml of CD3OD and filtered, and the solution was transferred into a 5-mm NMR tube. NMR spectra were obtained on a Bruker Avance DRX-400 FT-NMR spectrometer, and the chemical shifts were established on the basis of the residual CD3OD resonances (3.30 ppm for 1H NMR, 48.0 ppm for 13C NMR). All pulse programs were taken from the Bruker software library. The NMR data were evaluated using WIN-NMR 6.0 (Bruker-Franzen Analytik GmbH).
Biological activity.
Comparison of estrogenic activities of ZON and ZOM-1 was carried out using the estrogen bioindicator yeast strain YZRM7 as previously described (21). Briefly, YZRM7, which requires for growth the activation of the pyrimidine biosynthetic gene URA3 by the expressed human estrogen receptor in the presence of an exogenous estrogenic substance, was grown, diluted to an optical density at 600 nm (OD600) of 0.05, and inoculated in 5-μl spots on defined solid yeast medium SC-His-Ura containing 0 to 400 nM ZON (Sigma Z-2125; 5 mg/ml stock solution in 70% ethanol) or 0 to 3,200 nM ZOM-1 (0.12 mg/ml stock solution in methanol). The plates were incubated for 3 days at 30°C and photographed. Yeast strain YZGA376, which is capable of pyrimidine biosynthesis (21), was used as a positive control.
The ability of ZON and ZOM-1 to bind to the human estrogen receptor was assayed in vitro using the HitHunter EFC estrogen chemiluminescence assay kit (DiscoveRx/Amersham Biosciences) and the human estrogen receptor-α (Sigma) as previously described for zearalenone-4-O-glucoside (25). Luminescence of the β-galactosidase product was measured after 1.5 h of incubation with a luminometer (Victor 2; Wallac/Perkin Elmer, Monza, Italy).
RESULTS
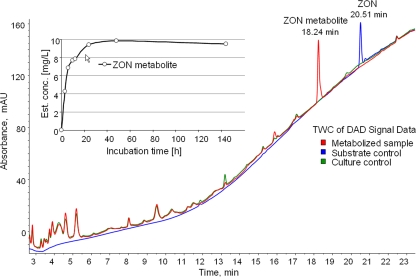
T. mycotoxinivorans was incubated for 0 to 144 h in saline medium with 10 mg/liter ZON, and culture medium was analyzed by LC-MS/MS and LC-DAD. Saline was used in order to keep matrix effects in the subsequent analysis to facilitate the identification of newly formed compounds as easy as possible. Due to the substrate controls (ZON incubated in culture medium without T. mycotoxinivorans), loss of ZON due to adsorption or decomposition during incubation could be excluded. Comparison between DAD chromatograms after cultivation of T. mycotoxinivorans alone (culture controls) and cocultivation of T. mycotoxinivorans and ZON, both treated in the same manner, resulted in the identification of one main ZON-derived metabolite (ZOM-1) eluting at a retention time of 18.2 min. (Fig. 1). The increase of ZOM-1 concentration over incubation time is illustrated in Fig. 1 (inset). ZOM-1 concentration was estimated by assuming a UV response equal to that for ZON at the same concentration. This seemed to be feasible since no other major metabolites were detected in the samples, and the UV spectra of both ZON and ZOM-1 were similar (data not shown). After 48 h of incubation, 95% of ZON was metabolized. It was converted to ZOM-1, which appeared to be stable for several days, indicating that ZOM-1 constitutes a stable metabolization end product under the conditions applied.
FIG. 1.
Comparison (large graph) of the total wavelength chromatogram (TWC) of the metabolized sample (red curve) and the culture (green curve) and the substrate (blue curve) control samples after 144 h of incubation. In the metabolized sample the substance causing the most intense peak (at 18.2 min) was identified as the ZON metabolite (ZOM-1). Depiction (inset) of estimated concentration of the ZON metabolite ZOM-1 during an incubation experiment of 10 mg/liter ZON with Trichosporon mycotoxinivorans.
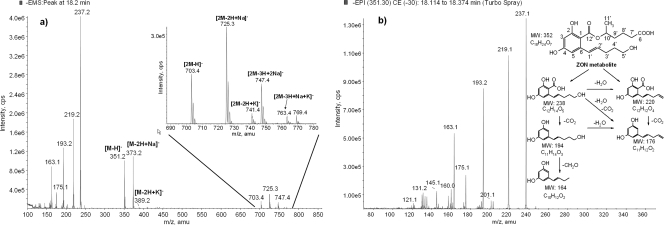
The data of the LC-MS total ion chromatogram (TIC) obtained in the negative enhanced full scan mode (EMS) confirmed the formation of one main ZON metabolite at 18.2 min. After subtraction of the culture control TICs, the resulting full-scan mass spectrum of the putative ZOM-1 (Fig. 2a) was evaluated in detail. Initially this spectrum was difficult to interpret; however, after these data were combined with additional measurements in the positive ionization mode (showing [M+Na]+ at m/z 375 as the most abundant ion [data not shown]), a molar mass (M) of 352 g/mol was assumed for ZOM-1. The mass shift of 34 amu might indicate the addition of 2 hydrogen and 2 oxygen atoms to ZON. While the signal at m/z 351 was assigned to the deprotonated molecular ion [M-H]−, m/z 373 was assumed to originate from its sodium adduct [M-2H+Na]−. Since sodium adduct formation in negative ESI is rare but has been reported for aromatic acids (28), the signal at m/z 373 might correspond to the presence of a free carboxylic group in the ZON metabolite.
FIG. 2.
(a) Full-scan spectrum (ESI negative mode) of the ZON metabolite at 18.2 min (144 h of incubation time) after background subtraction. Highlighted are the deprotonated molecular ion [M-H]− at m/z 351 and further signals of its sodium and potassium adducts (m/z 373, m/z 389) as well as the molecular dimer ion at m/z 703 and further alkali-bridged dimer ions at m/z 725, 741, 747, and 763 (zoomed). (b) Structure of the ZON metabolite (ZOM-1) of T. mycotoxinivorans and consecutive numbering of the carbon atoms. Enhanced product ion data of its deprotonated molecule ion [M-H]− at m/z 351 and scheme of proposed major fragmentation pathways. MW indicates molecular weight.
As demonstrated by the EPI spectrum in negative ionization of the deprotonated molecular ion [M-H]− at m/z 351 (Fig. 2b), the signals at m/z 237, 219, 193, 175, and 163 in the full-scan mass spectrum originated from fragmentation of m/z 351 in the ion source. The product ion spectrum showed similarities to the MS/MS spectrum of ZON, e.g., the loss of 114 amu and formation of the ion at m/z 175 (30), indicating that the substance produced during the incubation experiment is in fact related to ZON. Proposed fragmentation pathways of ZOM-1 in the negative ionization mode are presented in Fig. 2b. The fragments are displayed in the neutral state. However, full-structure elucidation of the new metabolite required the use of additional analytical techniques, such as high-resolution MS and NMR.
For further characterization of ZOM-1, a large-scale incubation experiment with ZON and T. mycotoxinivorans was performed. After extraction and preparative HPLC purification and isolation, a putative molecular formula of C18H24O7 was assigned to the purified ZOM-1 by TOF MS measurements. In detail, measurement in the negative ionization mode showed the formation of [M-H]− at m/z 351.1445. Evaluation with the GMF software revealed the molecular formula C18H23O7. The relative deviation between the measured mass and the theoretical mass of this signal was 1.1 ppm, and the sigma factor was calculated to be 0.0082, indicating very good agreement between measured spectra and calculated molecular mass as well as isotope pattern. The spectrum also included the main fragments of [M-H]− at m/z 237 and 219 as well as an adduct formation at m/z 373. GMF evaluation suggested the molecular formula of C12H13O5, C12H11O4, and C18H22NaO7 for these signals, respectively. This confirmed our early interpretation of m/z 373 as the sodium adduct [M+Na-H]− and supported our structural interpretation of the fragments at m/z 237 and 219, as shown in Fig. 2b. Measurement in the positive ionization mode (data not presented) showed only the formation of [M+Na]+ at m/z 375.1416 and not of [M+H]+. GMF evaluation suggested the molecular formula C18H24NaO7 with an accuracy of −0.554 ppm and a calculated sigma factor of 0.0043. The spectrum also included an [M-H + 2Na]+ adduct formed at m/z 397.1231.
The complete molecular structure of ZOM-1 was finally determined by NMR analysis. From 1H and 13C-APT spectra (Table 1), it was evident by comparison that a substantial part of the ZON structure is preserved in ZOM-1, namely, the tetra-substituted aromatic ring, including the benzoic ester moiety and the olefinic double bond. This was proven by two-dimensional spectra (1H1H COSY, 1H13C HSQC, and 1H13C HMBC), which also allowed the elucidation of the remaining part of the structure.
TABLE 1.
13C NMR and 1H NMR chemical shifts of the ZON metabolitec
| Position | ZON metabolite |
||
|---|---|---|---|
| 13C (ppm)a,b | 1H (ppm)a | J (Hz) | |
| 1 | 104.1 (s) | ||
| 2 | 164.3 (s) | ||
| 3 | 101.6 (d) | 6.22 (1H) | 2.5 (d) |
| 4 | 162.6 (s) | ||
| 5 | 108.2 (d) | 6.38 (1H) | 2.5 (d) |
| 6 | 143.8 (s) | ||
| 1′ | 131.7 (d) | 6.96 (1H) | 15.5 (d), 1.5 (t) |
| 2′ | 131.6 (d) | 5.92 (1H) | 15.4 (d), 6.9 (t) |
| 3′ | 29.4 (t) | 2.29 (2H) | 7.6 (t), 6.9 (d), 1.5 (d) |
| 4′ | 32.3 (t) | 1.73 (2H) | (m) |
| 5′ | 61.5 (t) | 3.64 (2H) | 6.6 (t) |
| 6′ | 176.3 (s) | ||
| 7′ | 33.7 (t) | 2.36 (2H) | 6.5 (t) |
| 8′ | 21.1 (t) | 1.80-1.65 (2H) | (m) |
| 9′ | 35.5 (t) | 1.85-1.70 (2H) | (m) |
| 10′ | 72.3 (d) | 5.21 (1H) | (m) |
| 11′ | 19.4 (q) | 1.38 (3H) | 6.3 (d) |
| 12′ | 171.3 (s) | ||
13C and 1H NMR spectra were recorded at 100 MHz and 400 MHz, respectively. Chemical shifts in ppm are referenced to tetramethylsilane (TMS) as an internal standard. Spectra were recorded in CD3OD.
Multiplicity was determined from APT spectra.
See Fig. 2b for numbering of the carbon atoms. Multiplicities (shown in parentheses) are abbreviated as follows: s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet.
A long-range correlation of the benzoic ester carbon (C12′) to the significant H10′ signal at 5.2 ppm provided an entry point for the analysis of the substituent attached there. From position 10′, the methyl group 11′ and the adjacent CH2 group (9′) were easily identified, although the protons of the latter are part of an overlapped region of six signals between 1.90 and 1.60 ppm. Because of this overlap, the next members in the chain were identified via two- and three-bond C-H correlations, leading to two further CH2 groups 8′ and 7′ (21.1 and 33.7 ppm in the carbon spectrum). The chemical shift and coupling pattern of the 7′ signals indicated a neighboring C=O fragment, which was assigned to the 176.3-ppm carbon signal via the HMBC spectrum. Its characteristic shift and the absence of any long-range correlation beyond showed it to be a carboxylic acid forming the end of the chain.
COSY correlations from the olefinic signals (1′ and 2′) of the substituent in position 6 of the aromatic ring led to the CH2 group 3′ and from there to the aliphatic overlap region. Thus, the next steps were again established by long-range correlations, leading to CH2 groups 4′ and 5′. The shifts of the 5′ signals (61.5 ppm and 3.64 ppm, respectively) and the lack of any further couplings led to the conclusion of a terminal CH2OH group.
Therefore, by means of a complete NMR spectroscopic analysis, we concluded that transformation of ZON by T. mycotoxinivorans into ZOM-1 occurs by cleavage of the lactone undecyl ring system at the ketone group at C6′, leading to formation of 5-({2,4-dihydroxy-6-[(1E)-5-hydroxypent-1-en-1-yl]benzoyl}oxy)hexanoic acid (Fig. 2b) in accordance with the molecular formula elucidated by mass spectrometry.
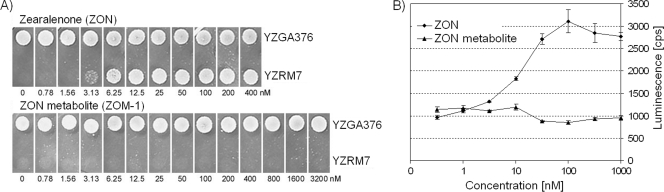
With the purified ZOM-1, an estrogenicity test was conducted. The growth of the indicator yeast strain YZRM7, as well as the positive-control strain YZGA376, on SC-His-Ura plates supplemented with either ZON or ZOM-1 is shown in Fig. 3A. Whereas a concentration of 3 nM ZON (∼1 μg/liter ZON) allowed growth of the estrogen-dependent bioassay strain YZRM7, an even 1,000-fold-higher concentration of ZOM-1 did not. This indicates that either ZOM-1 is not estrogenic in vivo or the ZOM-1 metabolite with its carboxy group is effectively excluded from the cytosol of yeast. To exclude this second possibility we directly tested the interaction of ZOM-1 with the human estrogen receptor protein in vitro. The assay that we used is based on alpha complementation of β-galactosidase by an enzyme acceptor and a steroid hormone-linked donor peptide. Enzyme complementation is prevented if the estrogen receptor protein binds to the conjugated steroid. The addition of a competitor compound that binds to the estrogen receptor and displaces the steroid hormone-linked donor peptide allows reconstitution of active β-galactosidase. The enzymatic activity is then measured by the hydrolysis of a β-galactosidase substrate yielding a luminescent product. As shown in Fig. 3B, addition of ZON caused a dose-dependent increase of β-galactosidase activity. In contrast, ZOM-1 at the same concentrations was inactive.
FIG. 3.
(A) Growth of strains YZRM7 and YZGA376 observed with SC-His-Ura medium plates after 3 days of incubation at 30°C either with ZON or with the ZON metabolite (ZOM-1) at different concentrations. Whereas a concentration of 3 nM ZON (∼1 μg/liter ZON) allows growth of the bioassay strain YZRM7 due to activation of the estrogen-inducible URA3 gene, a 1,000-fold-higher concentration of the ZON metabolite does not. (B) Detection of the luminescent product of β-galactosidase activity released after 90 min of incubation. Competitive displacement of the estrogen receptor from the estrogen-linked donor peptide by ZON reconstitutes β-galactosidase activity, while the ZON metabolite is inactive.
DISCUSSION
In this work we studied the degradation of ZON by T. mycotoxinivorans, a basidiomycete yeast which is used as a microbial feed additive against mycotoxins. A nonestrogenic ZON metabolite (ZOM-1) was the main product of this ZON degradation.
In order to facilitate metabolite identification and characterization, high concentrations of ZON (10 and 50 mg/liter) were used in a simple cultivation medium lacking some nutrients. Therefore, the incubation period had to be extended since under these conditions the ZON metabolizing activity of T. mycotoxinivorans is reduced. Moreover, additional metabolization experiments using complex culture media were carried out, which demonstrated a fast transformation of ZON to ZOM-1 and further currently uncharacterized metabolites in minor amounts (data not shown).
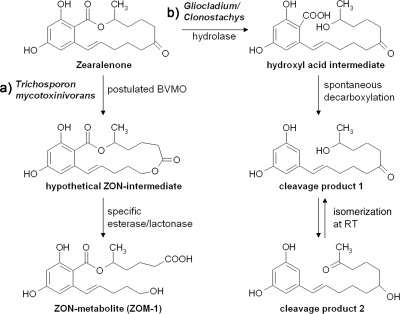
LC-MS/MS and TOF MS measurements demonstrated the structural similarity of ZOM-1 to ZON and revealed its fragment pattern and molecular formula. Subsequent NMR analysis of the purified metabolite confirmed its identity as (5S)-5-({2,4-dihydroxy-6-[(1E)-5-hydroxypent-1-en-1-yl]benzoyl}oxy)hexanoic acid. The opening of the ring at the keto group of the macrocyclic ring of ZON and formation of a carboxy and hydroxy group is different from the hydrolysis of the preexisting lactone in ZON by Gliocladium/Clonostachys (followed by decarboxylation). The product ZOM-1 probably is formed by the proposed two-step mechanism illustrated in Fig. 4. First, the macrocyclic ring is extended by insertion of an oxygen next to the carbonyl group, and the thereby newly formed lactone is then opened in a second step to give ZOM-1. The first Baeyer-Villiger reaction (for a review, see reference 34) typically requires a NAD(P)H-dependent flavoenzyme of the class of Baeyer-Villiger monooxygenases (35). Alternatively, as shown for the ring extension of a plant hormone, brassinolide, a (untypical) cytochrome P450 monooxygenase (19), might be involved as well as other largely uncharacterized metallo-oxidoreductases, such as those encoded by the aflatoxin biosynthetic AflY gene (6). As a caveat, we have to state that we were not able to obtain experimental evidence for the proposed Baeyer-Villiger intermediate. A reason could be that the Baeyer Villiger reaction is slow and rate limiting, while the opening of the newly formed lactone by a specific esterase/lactonase (18) is very rapid. It is interesting to note that this hypothetical lactonase must have high specificity, since the preexisting lactone in ZON, which is targeted by the lactone hydrolase of Gliocladium, remains unchanged in ZOM-1.
FIG. 4.
Description of the proposed lactone ring opening of zearalenone by Trichosporon mycotoxinivorans (a) and by Gliocladium roseum/Clonostachys rosea (b). (a) Hypothetical pathway of ZOM-1 formation. The macrocyclic ring is first extended by insertion of an oxygen next to the carbonyl group (Baeyer-Villiger oxidation), and the thereby newly formed lactone is then opened in a second step to give ZOM-1. (b) The preexisting lactone in ZON is hydrolyzed, and following decarboxylation the cleavage product 1, which can isomerize at room temperature (RT) to the cleavage product 2, is formed.
The most significant finding of our work is that the ZOM-1 metabolite is not estrogenic in vivo and does not interact in vitro with the estrogen receptor protein. This result is in accordance with the result of a previous estrogenicity test with human breast cancer cell line MCF-7, performed with the nonpurified substance and showing the loss of ZOM-1 estrogenicity (27).
The novel detoxification mechanism of ZON by T. mycotoxinivorans is therefore highly attractive for biological detoxification of ZON. Since T. mycotoxinivorans can be fermented, concentrated, freeze-dried, and stabilized without losing its deactivating abilities, its utilization as a feed additive for mycotoxin detoxification seems practicable. With the elucidation of the genetic basis of the detoxification reaction and cloning of the corresponding gene(s), it may also become feasible to develop enzymatic detoxification systems (36) or to engineer this detoxification pathway in other organisms.
Acknowledgments
This work was supported by grants from the Christian Doppler Society and the Austrian Research Promotion Agency (FFG) and was done in cooperation with Biomin GmbH.
We are grateful to Martin Täubel and Alexander Frank for their assistance with the degradation experiments. We also thank Dieter Moll for critically reading the manuscript.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Bauer, J., K. Heinritzi, M. Gareis, and B. Gedek. 1987. Veränderungen am Genitaltrakt des weiblichen Schweines nach Verfütterung praxisrelevanter Zearalenonmengen. Tierärztl. Praxis 15:33-36. [PubMed] [Google Scholar]
- 2.Berthiller, F., R. Schuhmacher, G. Adam, and R. Krska. 2009. Formation, determination and significance of masked mycotoxins. Anal. Bioanal. Chem. 395:1243-1252. [DOI] [PubMed] [Google Scholar]
- 3.Binder, E. M. 2007. Managing the risk of mycotoxins in modern feed production. Anim. Feed Sci. Tech. 133:149-166. [Google Scholar]
- 4.Bloomquist, C., J. N. Davidson, and E. G. Pearson. 1982. Zearalenone toxicosis in prepubertal dairy heifers. J. Am. Vet. Med. Assoc. 180:164-165. [PubMed] [Google Scholar]
- 5.EFSA J. 2004. Opinion of the scientific panel on contaminants in the food chain on a request from the commission related to zearalenone as undesirable substance in animal feed. EFSA J. 89:1-35. [Google Scholar]
- 6.Ehrlich, K. C., B. Montalbano, S. M. Boué, and D. Bhatnagar. 2005. An aflatoxin biosynthesis cluster gene encodes a novel oxidase required for conversion of versicolorin A to sterigmatocystin. Appl. Environ. Microbiol. 71:8963-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Sharkaway, S. H., M. I. Selim, M. S. Afifi, and F. T. Halaweish. 1991. Microbial transformation of zearalenone to a zearalenone sulfate. Appl. Environ. Microbiol. 57:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Sharkawy, S. H. 1989. Microbial transformation of zearalenone. III. Formation of zearalenone-2-4-O-ß-diglucoside. Acta Pharm. Jugosl. 39:303-310. [Google Scholar]
- 9.El-Sharkawy, S. H., and Y. Abul-Hajj. 1988. Microbial cleavage of zearalenone. Xenobiotica 18:365-371. [DOI] [PubMed] [Google Scholar]
- 10.El-Sharkawy, S. H., and Y. Abul-Hajj. 1988. Microbial transformation of zearalenone. II. Reduction, hydroxylation, and methylation products. J. Org. Chem. 53:515-519. [Google Scholar]
- 11.Gareis, M., J. Bauer, J. Thiem, G. Plank, S. Grabley, and B. Gedek. 1990. Cleavage of zearalenone-glycoside, a “masked” mycotoxin, during digestion in swine. Zentralbl. Veterinarmed. B 37:236-240. [DOI] [PubMed] [Google Scholar]
- 12.Gromadzka, K., A. Waśkiewicz, J. Chełkowski, and P. Goliński. 2008. Zearalenone and its metabolites: occurrence, detection, toxicity and guidelines. World Mycotoxin J. 1:209-220. [Google Scholar]
- 13.Hagler, W. M., C. J. Mirocha, S. V. Pathre, and J. C. Behrens. 1979. Identification of the naturally occurring isomer of zearalenol produced by Fusarium roseum ‘Gibbosum' in rice culture. Appl. Environ. Microbiol. 37:849-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurd, R. N. 1977. Structure activity relationships in zearalenones, p. 379-391. In J. V. Rodricks, C. W. Hesseltine, and M. A. Mehlman, (ed.), Mycotoxins in human and animal health. Pathotox Publishers, Park Forest South, IL.
- 15.Hussein, H. S., and J. M. Brasel. 2001. Toxicity, metabolism and impact of mycotoxins on humans and animals. Toxicology 167:101-134. [DOI] [PubMed] [Google Scholar]
- 16.Kakeya, H., N. Takahashi-Ando, M. Kimura, R. Onose, I. Yamaguchi, and H. Osada. 2002. Biotransformation of the mycotoxin zearalenone to a non-estrogenic compound by a fungal strain Clonostachys sp. Biosci. Biotechnol. Biochem. 66:2723-2726. [DOI] [PubMed] [Google Scholar]
- 17.Kakimura, H. 1986. Conversion of zearalenone to zearalenone glycoside by Rhizopus sp. Appl. Environ. Microbiol. 52:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kataoka, M., K. Honda, K. Sakamoto, and S. Shimizu. 2007. Microbial enzymes involved in lactone compound metabolism and their biotechnological applications. Appl. Microbiol. Biotechnol. 75:257-266. [DOI] [PubMed] [Google Scholar]
- 19.Kim, T. W., J. Y. Hwang, Y. S. Kim, S. H. Joo, S. C. Chang, J. S. Lee, S. Takatsuto, and S. K. Kim. 2005. Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17:2397-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuiper, G. G., J. G. Lemmen, B. Carlsson, J. C. Corton, S. H. Safe, P. T. Van der Saag, B. Van der Burg, and J. A. Gustafsson. 1998. Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology 139:4252-4263. [DOI] [PubMed] [Google Scholar]
- 21.Mitterbauer, R., H. Weindorfer, N. Safaie, R. Krska, M. Lemmens, P. Ruckenbauer, K. Kuchler, and G. Adam. 2003. A sensitive and inexpensive yeast bioassay for the mycotoxin zearalenone and other compounds with estrogenic activity. Appl. Environ. Microbiol. 69:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molnar, O., G. Schatzmayr, E. Fuchs, and H. Prillinger. 2004. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 27:661-671. [DOI] [PubMed] [Google Scholar]
- 23.Plasencia, J., and C. J. Mirocha. 1991. Isolation and characterization of zearalenone sulfate produced by Fusarium spp. Appl. Environ. Microbiol. 57:146-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Politis, I., K. Fegeros, S. Nitsch, G. Schatzmayr, and D. Kanatas. 2005. Use of Trichosporon mycotoxinivorans to suppress the effects of ochratoxicosis on the immune system of broiler chicks. Br. Poultry Sci. 46:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Poppenberger, B., F. Berthiller, H. Bachmann, D. Lucyshyn, C. Peterbauer, R. Mitterbauer, R. Schuhmacher, R. Krska, J. Glössl, and G. Adam. 2006. Heterologous expression of Arabidopsis UDP-glucosyltransferases in Saccharomyces cerevisiae for production of zearalenone-4-O-glucoside. Appl. Environ. Microbiol. 72:4404-4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schatzmayr, G., D. Heidler, E. Fuchs. M. Mohnl, M. Täubel, A. P. Loibner, R. Braun, and E. M. Binder. 2003. Investigation of different yeast strains for the detoxification of ochratoxin A. Mycotoxin Res. 19:124-128. [DOI] [PubMed] [Google Scholar]
- 27.Schatzmayr, G., F. Zehner, M. Täubel, D. Schatzmayr, A. Klimitsch, A. P. Loibner, and E. M. Binder. 2006. Microbiologicals for deactivating mycotoxins. Mol. Nutr. Food Res. 50:543-551. [DOI] [PubMed] [Google Scholar]
- 28.Schug, K. A. 2002. Pseudo-molecular ion formation by aromatic acids in negative ionization mode electrospray ionization mass spectrometry. Ph.D. dissertation. Virginia Polytechnic Institute and State University, Blacksburg, VA.
- 29.Schuh, M., and W. Baumgartner. 1988. Mikrobiologisch und mykotoxikologisch kontaminierte Futtermittel als Krankheitsursache bei Rindern. Wien Tierärztl. Mschr. 75:329-332. [Google Scholar]
- 30.Shin, B. S., S. H. Hong, S. W. Hwang, H. J. Kim, J. B. Lee, H. S. Yoon, D. J. Kim, and S. D. Yoo. 2009. Determination of zearalenone by liquid chromatography/tandem mass spectrometry and application to a pharmacokinetic study. Biomed. Chromatogr. 23:1014-1021. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi-Ando, N., M. Kimura, H. Kakeya, H. Osada, and I. Yamaguchi. 2002. A novel lactonohydrolase responsible for the detoxification of zearalenone: enzyme purification and gene cloning. Biochem. J. 365:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi-Ando, N., S. Ohsato, T. Shibata, H. Hamamoto, I. Yamaguchi, and M. Kimura. 2004. Metabolism of zearalenone by genetically modified organisms expressing the detoxification gene from Clonostachys rosea. Appl. Environ. Microbiol. 70:3239-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi-Ando, N., T. Tokai, H. Hamamoto, I. Yamaguchi, and M. Kimura. 2005. Efficient decontamination of zearalenone, the mycotoxin of cereal pathogen, by transgenic yeasts through the expression of a synthetic lactonohydrolase gene. Appl. Microbiol. Biotechnol. 67:838-844. [DOI] [PubMed] [Google Scholar]
- 34.ten Brink, G. J., I. W. C. E. Arends, and R. A. Sheldon. 2004. The Baeyer-Villiger reaction: new developments toward greener procedures. Chem. Rev. 104:4105-4124. [DOI] [PubMed] [Google Scholar]
- 35.Torres Pazmiño, D. E., and M. W. Fraaije. 2007. Discovery, redesign and applications of Baeyer-Villiger monooxygenases, p. 107-127. In T. Matsuda (ed.), Future directions in biocatalysis. Elsevier, Amsterdam, Netherlands.
- 36.Torres Pazmiño, D. E., R. Snajdrova, B. J. Baas, M. Ghobrial, M. D. Mihovilovic, and M. W. Fraaije. 2008. Self-sufficient Baeyer-Villiger monooxygenases: effective coenzyme regeneration for biooxygenation by fusion engineering. Angew. Chem. Int. Ed. Engl. 47:2275-2278. [DOI] [PubMed] [Google Scholar]
- 37.Vekiru, E. 2009. Towards the development of a novel biological feed additive for mycotoxin degradation: isolation, structural elucidation and quantification of biotransformation-products of the estrogenic zearalenone. Ph.D. thesis. Vienna University of Technology, Vienna, Austria.
- 38.Zinedine, A., J. M. Soriano, J. C. Moltó, and J. Maňes. 2007. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: an oestrogenic mycotoxin. Food Chem. Toxicol. 45:1-18. [DOI] [PubMed] [Google Scholar]