Abstract
Beauveria bassiana is an important insect-pathogenic fungus that invades insects by direct penetration of the host cuticle. To delineate the molecular mechanisms involved in fungal infection, a mitogen-activated protein kinase (MAPK) gene, Bbmpk1, which encodes a YERK1 family MAPK was isolated and characterized. Targeted gene disruption of Bbmpk1 resulted in a complete loss of virulence when applied topically to host insects but did not affect growth of the fungus when conidia were injected directly into the hemocoel. Hyphae of the mutant strain growing in the insect hemocoel were unable to penetrate the cuticle growing outwards and consequently failed to sporulate on the cadaver surface. These data suggest that BbMPK1 is essential for penetration of the insect cuticle both from the outside and from the inside-out in order to escape and disperse from the host. Inactivation of BbMPK1 also caused a significant decrease in fungal adhesion to insect cuticles and eliminated their ability to form appressoria. In order to identify downstream genes regulated by BbMPK1, a suppressive subtractive hybridization (SSH) library was generated comparing mutant and wild-type transcripts isolated during appressorium formation. Thirty-one genes screened from the SSH library were determined to be expressed in the wild-type strain but either significantly reduced or not expressed in the mutant. Ten genes showed high or medium similarity to known protein encoding genes, including proteins involved in cell surface hydrophobicity, lipid metabolism, microtubule dynamics, mitochondrial electron transport, chromatin remodeling, transcription, rRNA processing, small nucleolar RNA accumulation, oxidation of aldehydes, translation, and likely other cellular processes.
Entomopathogenic fungi are important natural regulators of insect populations and have attracted much attention as biological control agents for insect pests (4). However, their use has been limited by low efficacy (36), and understanding the mechanisms of fungal pathogenesis in insects can aid in the development of more efficient mycoinsecticides. Beauveria bassiana is a well-known fungal pathogen infecting a variety of insect pests, including agricultural pests and vectors of human pathogens, and has been intensively investigated as an important environmentally friendly alternative to chemical pesticides (8, 15, 22). Similar to the entomopathogenic fungus Metarhizium anisopliae, as well as many plant-pathogenic fungi, B. bassiana invades its host by direct penetration of the host cuticle. Typically, the fungal conidia adhere to the target insect cuticle, germinate across the surface, and begin the process of penetrating the cuticle. Under certain conditions, germlings can differentiate, producing infection structures known as appressoria that subsequently produce penetration pegs. Penetration of the insect cuticle occurs via a combination of mechanical pressure and cuticle-degrading enzymes, principally proteases and chitinases (4, 9). Once inside the insect hemocoel, the penetrating hyphae differentiate into free-floating single cell propagules known as in vivo hyphal bodies that display distinct morphological and biochemical properties (22, 41). Thus far, only a few genes involved in the infection processes of B. bassiana have been identified. Most notably, a number of cuticle degrading enzymes and their genes have been characterized (1, 8, 52). In addition, two nonribosomal peptide synthetase (NRPS) encoding genes bbBeas and bbBsls, responsible for the synthesis of the insecticidal virulence factors beauvericin and bassianolide, respectively, have been recently identified (49, 50).
Mitogen-activated protein kinase (MAPK) signaling pathways play a pivotal role in sensing extracellular signals and relaying the signals to control gene expression (13, 31). The yeast and fungal extracellular signal-regulated kinase subfamily (YERK1) represented by Fus3/Kss1 in Saccharomyces cerevisiae (19) is among a MAPK superfamily that is essential for pathogenicity of many fungal phytopathogens (47). MAPKs have been directly implicated in regulating infection-related development in diverse plant pathogenic fungi, including Magnaporthe grisea (48), Colletotrichum lagenarium (38), Fusarium oxysporum (6), Botrytris cinerea (54), Cochliobolus heterostrophus (20), Pyrenophora teres (35), and Alternaria brassicicole (3). M. grisea, C. lagenarium, and P. teres produce melanized appressoria that generate strong turgor pressure. In these fungi, MAPK (Fus3/Kss1-homolog) mutants are defective in appressorium formation and are nonpathogenic as a result of their inability to penetrate the plant epidermis and thus colonize host plant tissues (35, 38, 48). Disruption of the MAPK gene CHK1 results in mutants that do not form the small appressoria characteristic of C. heterostrophus. These mutants are able to penetrate into the maize leaf, but the development of disease symptoms is significantly decreased (20). Although disruption of a Fus3/Kss1 homolog-encoding gene, amk1, in A. brassicicole did not influence appressoria formation, the amk1 mutants were nonpathogenic when applied to intact plants (3). Some fungal phytopathogens, such as F. oxysporum and B. cinerea, do not make appressoria but directly penetrate the host tissue. Disruption of MAPK genes in these fungal pathogens also results in defects in pathogenicity due to their inability to penetrate the host cuticle (6, 54).
MAPK genes play a role in signal pathways that regulate cell development via a relay of proteins that ultimately affect gene expression. Thus, the gene expression profiles of mutants blocked in a particular signal transduction pathway, compared to the wild type, can provide a powerful means to identify targets of the pathway. Knowledge of these targets should provide clues to the mechanism by which the pathway affects development. In the rice blast fungus, suppression subtractive hybridization (SSH) was used to isolate genes underexpressed in a mutant lacking the MAPK pmk1 at the appressorium formation stage (51). A similar strategy was used to isolate target genes regulated by the CHK1 MAPK in the southern maize leaf blight pathogen C. heterostrophus during infection on the host plant (21).
Little is known about the role of MAPK signal transduction in insect fungal pathogens. Recently, we characterized a Hog1 MAPK encoding gene in B. bassiana. The Hog1 MAPK was found to regulate stress responses and to be involved in virulence and other characteristics associated with pathogenicity (53). In the present study, we identified Bbmpk1, a gene encoding a YERK1 family MAPK in B. bassiana. Targeted gene disruption of BbMPK1 revealed that it is required for penetration and appressorium formation. In order to identify downstream genes regulated by BbMPK1 and involved in infection, an SSH library was constructed enriched for genes regulated by BbMPK1 at the stage of appressorium formation. Nineteen genes were screened from the SSH library and were found to be expressed in the wild-type strain but not in the mutant. The transcript levels of 12 additional genes were found to be significantly reduced during appressorium formation in the mutants compared to the wild-type parent.
MATERIALS AND METHODS
Fungal and bacterial strains.
B. bassiana Bb0062 single spore isolate was previously described (10). Cultures were grown on Sabouraud dextrose agar (Difco Laboratories, Detroit, MI) supplemented with 1% (wt/vol) yeast extract (SDAY) for 14 days at 26°C with a 15-h light/9-h dark cycle. Escherichia coli DH5α was used for DNA manipulations and transformations. Agrobacterium tumefaciens AGL-1 was used for fungal transformations.
Identification of the Fus3/Kss1 MAPK-encoding gene.
Fungal genomic DNA was prepared as described previously (33), and total RNA was isolated from B. bassiana by using an RNeasy plant minikit (Qiagen Sciences). RQ1 RNase-free DNase (Promega) was used to remove DNA contamination in RNA samples. The B. bassiana Fus3/Kss1 mitogen-activated protein kinase (MAPK) gene was cloned by PCR using the degenerate primers Bm-1 and Bm-2 (Table 1), designed to the conserved amino acid sequences (EQYDIQD and FDKHKDN) based on alignments of M. grisea PMK1 (48), C. lagenarium CMK1 (38), F. oxysporum FMK1 (6), and B. cinerea BMP1 (54). PCR was performed using ExTaq DNA polymerase (TaKaRa). Sequencing of the resulting PCR product revealed it to be a fragment of a Fus3 (Kss1)-related MAPK gene. The complete gene was subsequently cloned by PCR walking using a Y-shaped adaptor-dependent extension (YADE) method as described previously (8) with the primers given in Table 1. All PCR products were cloned into the A-T cloning vector pGEM-T Easy (Promega) according to the manufacturer's instructions and subsequently sequenced. The nucleotide sequence of the complete gene, designated Bbmpk1, was assembled using the PCR and YADE products based on overlapping regions. Primers (Table 1) were designed to clone the cDNA sequence of this gene using the 3′ full rapid amplification of cDNA ends (RACE) core set (TaKaRa).
TABLE 1.
Oligonucleotides used in this study
| Purpose and oligonucleotide | Sequence (5′-3′)a | Remarks |
|---|---|---|
| PCR-based cloning of a fragment of Bbmpk1 | ||
| Bm-1 | GAGCAGTACGAYATYCARGA | |
| Bm-2 | TTRTCYTTRTGYTTRTCRAA | |
| YADE-based PCR walking | ||
| Bm5X | ACTCACCAGACAACGCCGTA | Used for cloning 5′ end of Bbmpk1 |
| Bm5Z | ACGACGTCCTGTATATCGTA | Used for cloning 5′ end of Bbmpk1 |
| Bm3X | TCCGATTCCCGAGGAATTCT | Used for cloning 3′ end of Bbmpk1 |
| Bm3Z | TCTTCGACTTCGACAAGCAT | Used for cloning 3′ end of Bbmpk1 |
| 3′ RACE | ||
| BbMs1 | ATCTTGACCGATTGCGCTAC | |
| Bbmpk1 disruption vector construction | ||
| L1 | ACGTACAAGGCAAACGTTGA | Used for amplification of 5′ end of Bbmpk1 |
| L2 | AAGCCTCTGGGCTTCTGAAT | Used for amplification of 5′ end of Bbmpk1 |
| R1 | ACTAGTTCATTCGTACCCAGGTACTT | Used for amplification of 3′ end of Bbmpk1; underlining indicates the introduced SpeI site |
| R2 | TCGAAGAATTCCTCGGGAAT | Used for amplification of 3′ end of Bbmpk1 |
| Reverse complementation of ΔBbmpk1 mutant | ||
| ben1 | ACTGATATCGTCGACAGGGGGCCTTCCA | Used for amplification the benomyl resistance gene ben; underlining indicates the introduced EcoRV site |
| ben2 | CCCAAGCTTGGCCAGCAGTAGACACTT | Used for amplification the benomyl resistance gene ben; underlining indicates the introduced HindIII site |
| Bbmpk1 F | ACTGATATCACGCGGGTTTCTATCTCCT | Used for amplified Bbmpk1; underlining indicates the introduced EcoRV site |
| Bbmpk1 R | TGCTCTAGAATAGCGAGCTGATGACCCCA | Used for amplified Bbmpk1; underlining indicates the introduced XbaI site |
| Screening of vector pBGΔMPK1 and BbMPK1 disruption mutant | ||
| G1 | TTAGATCTCGGTGACGGGCA | 5′ end of egfp |
| B2 | TTAGATCTCGGTGACGGGCA | 3′ end of bar |
| R2′ | TTTTACCGCATGATCTCTTGG | Flanking sequence of 3′ end of primer R2 |
| RT-PCR | ||
| Bbmpk1-1 | AGGCTCTTAAGCACCCGTAT | Used for RT-PCR analysis of Bbmpk1 |
| Bbmpk1-2 | TCGCGTGTGGCATTCTGTAT | Used for RT-PCR analysis of Bbmpk1 |
| β-Tubulin 1 | TACTCTACGATTCGTCAAGT | Used for RT-PCR analysis of β-tubulin |
| β-Tubulin 1 | TGCTGGAACAGAGCCGTCTT | Used for RT-PCR analysis of β-tubulin |
| Bgpd-1 | TCGACCTGACTGCTCGTCTT | Used for RT-PCR analysis of Bbgpd |
| Bgpd-2 | TAGGAGATAAGGTCCAGGA | Used for RT-PCR analysis of Bbgpd |
R = A or G; Y = C or T.
Bbmpk1 disruption and reverse complementation.
Plasmid pK2-BarGFP containing the herbicide resistance gene bar and the enhanced green fluorescent protein gene egfp was generated by inserting a bar::gfp fusion gene (16) into the EcoRI and HindIII sites of the binary vector pPK2 (27) from which an hph cassette was deleted. The 3′ end of Bbmpk1 (3′ Bbmpk1) was cloned by PCR with the primers R1 and R2 (Table 1) using ExTaq DNA polymerase (TaKaRa). The resulting PCR product was cloned into pMD18-T (TaKaRa). The correct insertion of the end of Bbmpk1 in the vector was confirmed by digestion with KpnI to yield a 0.5-kb fragment. The 3′ end of Bbmpk1 was excised from the resulting vector by EcoRI and SpeI, blunt ended using T4 DNA polymerase at EcoRI site, and then inserted into XbaI and blunt-ended HindIII sites of pk2-BarGFP, yielding pK2-BarGFP:3′Bbmpk1. The 5′ end of Bbmpk1 (5′ Bbmpk1) was amplified with the primers L1 and L2 with ExTaq DNA polymerase (TaKaRa). The PCR product was cloned into pGEM-T Easy (Promega) to form pGEM-5′Bbmpk1. The 5′ end of Bbmpk1 was then excised from pGEM-5′mpk1 by partial digestion with EcoRI due to the presence of an internal EcoRI site existing in the fragment and inserted into the unique EcoRI site of pK2-BarGFP:3′Bbmpk1. The resulting gene replacement vector was confirmed by PCR using the primers L1 and B2 (5′ end sequence of bar) (Table 1). The clone was ∼3.7 kb, containing 0.97 kb of the 5′ end of MPK1 and 2.68 kb corresponding to the bar resistance gene cassette. The resultant final plasmid construct, designed as pBGΔmpk1, was mobilized into A. tumefaciens AGL-1 for fungal transformation (10).
Bbmpk1 disruption mutants were initially selected on the basis of herbicide (glufosinate-ammonium) resistance and expression of green fluorescent protein (GFP). PCR performed with the primer pairs G1/R2 and G1/R2′ (Table 1), Southern blotting, and reverse transcription-PCR (RT-PCR) were used to confirm disruption of Bbmpk1 in B. bassiana transformants, where G1 was designed to the 5′ end of egfp and R2′ corresponded to the 3′ flanking sequence of R2.
To complement the ΔBbmpk1 mutant, a master vector pK2ben containing the benomyl-resistant tubulin gene ben from Neurospora crassa as a selectable marker was constructed (32). The ben cassette was cloned from pMYX2 by PCR using primers Ben1 and Ben2 (Table 1) and the PrimeSTARTM HS DNA polymerase (TaKaRa). The resultant fragment was then inserted in HindIII and blunted-EcoRI sites of the binary vector pPK2 (27), replacing the hygromycin marker in the original vector. The Bbmpk1 gene containing its upstream and downstream sequences was amplified with the primers Bbmpk1 F and Bbmpk1 R (Table 1) using PrimeSTARTM HS DNA polymerase (TaKaRa). After being confirmed by sequencing, the resultant PCR product was digested with XbaI and EcoRV and inserted into XbaI and blunt-ended HindIII sites of pK2ben to form pK2ben-Bbmpk1. This plasmid was mobilized into A. tumefaciens AGL-1 for fungal transformation (10). Reverse complementation transformants were initially selected for benomyl resistance (1.5 μg of benomyl/ml). PCR performed with the primers Bbmpk1 F and Bbmpk1 R (Table 1) was then used to confirm the insertion of the wild-type Bbmpk1. The expression of Bbmpk1 was then confirmed by RT-PCR with the primers Bbmpk1-1 and Bbmpk1-2 (Table 1).
Blotting analysis.
Approximately 25 μg of genomic DNA digested with appropriate restriction enzymes for each sample was used in Southern blot analyses. The DNA probes were labeled with [α-32P]dCTP using a Ready-To-Go DNA labeling kit (Amersham Biosciences). For dot blot analysis, the cDNA probes were labeled with digoxigenin according to the instructions provided in the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche).
Protein extracts were prepared from mycelium inoculated in cicada (Graptopsaltria nigrofuscata) cuticle broth (53) and cultured at 26°C with aeration. Ten-milliliter aliquots of the growing culture were sampled at 12, 24, 36, and 48 h postinoculation and cleared of hyphal debris by filtration through a Whatman grade 4 filter paper. The proteins in the filtrate were precipitated by using acetone and dissolved in 8 M urea. For Western blot analyses, protein samples (5 μg) were analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-10% PAGE) and subsequently transferred to a polyvinylidene difluoride membrane (Roche). Immunoblotting and blot development were performed according to the instructions provided with an Opti-4CN Western blot kit (Bio-Rad). Cuticle-degrading enzymes Cdep1 (Pr1 protease) and Bbchit1 (chitinase) were detected by using antibodies raised in rabbit (7, 8).
Quantification of conidia yield and insect bioassays.
Conidial production on agar plates was determined according to a previously described method (53).
Third-instar larvae of Pieris brassicae reared on cabbage in the greenhouse were used for insect bioassays. Conidia of the wild-type and Bbmpk1 disruption strains were applied either topically by immersion of larvae in an aqueous suspension containing 107 conidia/ml for 20 s or by injecting the second proleg with 3 μl of an aqueous suspension containing 107 spores per ml (42). Each treatment had three replicates with 10 insects each, and the experiments were repeated twice. Mortality was recorded every 12 h. Additional insects were injected and bled 48 h later to observe the extent of hyphal body differentiation within the insect hemocoel.
Determination of adherence and appressorium formation.
Conidial adherence to cicada hind wings was investigated as described previously (43, 53). The wings were immersed in a conidial suspension in water at a concentration of 2 × 107 spores/ml for 20 s, and placed on 0.7% water agar. After incubation for 8 h, the conidia were counted under a light microscope before and after washing out the less-adherent conidia in 0.05% Tween 20 for 30 s. Appressoria were induced on cicada hind wings according to a previously described method (44). Portions (50 μl) of 2 × 107 conidia/ml were spotted onto the wing sterilized using 37% H2O2. After incubation up to 22 h at 26°C, the appressoria were examined microscopically.
RT-PCR analysis.
Two micrograms of total RNA was reverse transcribed using an oligo(dT)-primed cDNA synthesis kit (MBI Fermentas), and the first-strand cDNA was used as the template for PCR amplification. The expression of Bbmpk1, Bbgpd, and β-tubulin was investigated during growth in Czapek-Dox broth containing 0.5% (wt/vol) peptone for 24 h. Primer sequences for the three genes used are shown in Table 1. The expression of genes from SSH library was investigated at the stage of appressorium formation. Wild-type (WT) and ΔBbmpk1 conidia were inoculated on cicada hind wings (to induce appressorium formation) and incubated for 22 h at 26°C. Total RNA was extracted and subjected to RT-PCR analysis using Bbgpd and β-tubulin as references. Primer sequences for the SSH clones are shown in Table S2 in the supplemental material. All RT-PCR analyses were performed for 25 cycles.
Construction and verification of the subtracted library.
Fungal samples for cDNA library construction were prepared as follow: 50 μl of WT and ΔBbmpk1 conidial suspension at a concentration of 2 × 107 spore/ml was inoculated onto a cicada hind wing (to induce appressorium formation) and incubated for 22 h at 26°C. RNAs were extracted from 10 inoculated wings for each strain. A suppression subtractive library was prepared according to the manufacturer's instructions (PCR-Select cDNA subtraction kit; Clontech, Palo Alto, CA), using RNA isolated from the WT strain and the ΔBbmpk1 mutant as the tester and driver transcript populations, respectively. Clones representing transcripts expressed differentially in WT and mutant strains were ligated into vector pGEM-T Easy (Promega), transformed to E. coli DH5α. Colonies containing inserts (white) were inoculated onto Hybond N+ nylon membranes (Amersham) placed on LB agar plates, and a reference set of these colonies was prepared by replica plating. The colonies formed on the membrane were lysed, and the resultant DNA was denatured and fixed onto the membranes in situ as described previously (12). The membranes were hybridized with cDNA pools representing tester populations (WT mRNA) and driver populations (mutant mRNA) according the instructions of the PCR-Select differential screening kit (Clontech). Dot blots were probed with DIG-labeled first-strand cDNA according the instructions provided with the DIG High Prime DNA Labeling and Detection Starter Kit (Roche). Clones uniquely expressed in the wild-type strain were sequenced (Invitrogen).
Data analysis.
Statistical analyses of the data were performed using one-way analysis of variance, and the significance between treatments was tested using the lease significant difference or the Dunnett T3 method. All statistical analyses were carried out with the SPSS 8.0 program.
Nucleotide sequence accession numbers.
Data reported here have been deposited in the GenBank database under the following accession numbers: AY348317 for Bbmpk1 genomic DNA, AY333430 for Bbmpk1 mRNA, and AY679162 for Bbgpd genomic DNA. The GenBank/EMBL/DDBJ accession numbers for the SSH sequences are shown in Table S1 in the supplemental material.
RESULTS
Gene cloning of Bbmpk1.
A Fus3/Kss1-related MAPK gene, designated Bbmpk1, was cloned from B. bassiana by PCR amplification using genomic DNA as the template and degenerate primers, with additional flanking sequences obtained via YADE as described in Materials and Methods. Based on the putative coding sequence identified using BLASTX, additional primers were designed to clone the Bbmpk1 cDNA sequence using 3′ RACE (rapid amplification of cDNA ends). The cDNA fragment contained a 1,071-bp open reading frame that encoded a 356-amino-acid protein with a predicted molecular mass of 41.2 kDa and a deduced pI of 6.6. Three introns were identified in the DNA sequence by comparison of the genomic sequence to the cDNA sequence. The predicted BbMPK1 protein had typical features of MAPKs, including 11 protein kinase domains and double TEY phosphorylation sites. The amino sequence of BbMPK1 showed 94 to 96% identity to F. oxysporum FMK1 (6), Trichoderma virens TMKA (30), M. grisea PMK1 (48), C. lagenarium CMK1 (38), and B. cinerea BMP1 (54). Southern blot analysis with genomic DNA digested with XbaI, BamHI, SmaI, and SpeI indicated that Bbmpk1 is a single-copy gene in B. bassiana (data not shown).
Bbmpk1 influences conidiation of B. bassiana.
To investigate the function of Bbmpk1 in B. bassiana, a gene disruption strategy was used. The Bbmpk1 gene replacement vector pBGΔmpk1 was constructed by replacing a 142-bp sequence in the Bbmpk1 coding region with the bar::gfp fusion gene cassette (see Fig. S1A in the supplemental material). The disruption mutants of Bbmpk1 were screened by PCR using two different primer pairs, G1/R2 and G1/R2′ (Table 1) (data not shown) and then verified by Southern blotting and RT-PCR (see Fig. S1B and C in the supplemental material). Three Bbmpk1 disruption mutants, ΔBbmpk1 2112, ΔBbmpk1 2167 and ΔBbmpk1 2179, were randomly selected for further study.
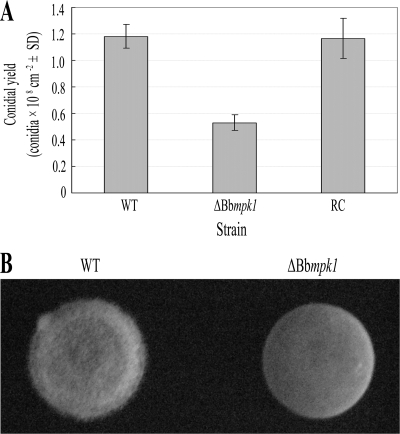
The ΔBbmpk1 mutants formed typical white colonies on SDAY plate and produced morphologically normal conidia. No obvious reduction in colony growth rate and conidial germination was observed in ΔBbmpk1 mutants (data not shown). However, disruption of Bbmpk1 resulted in a reduction in conidiation, with a 55.4% decrease in conidia/day/cm2 compared to the WT on SDAY (F2,14 = 60.58; least significant difference for the post hoc test, P < 0.001), while the complemented ΔBbmpk1 was similar to WT in conidial yield (Fig. 1A), suggesting that Bbmpk1 is involved in conidiation.
FIG. 1.
(A) Conidial yield of WT, ΔBbmpk1, or reverse complementation transformant (RC) strains produced on Sabouraud dextrose agar (Difco Laboratories) supplemented with 1% (wt/vol) yeast extract. Conidial production was measured as described earlier (53). The error bars indicate the standard deviations from five repeats of the experiment. (B) Hyphal growth on the surface of a drop of liquid. Drops of Sabouraud dextrose broth (Difco Laboratories) supplemented with 1% (wt/vol) yeast extract containing conidia of the WT strain (left) or the Bbmpk1 disruption mutant (right) were placed in a petri dish and incubated for 96 h at 100% humidity.
To determine the ability of hyphae to breach the liquid-air interface, 150-μl drops of Sabouraud dextrose broth supplemented with 1% (wt/vol) yeast extract containing conidia at a concentration of 105 spores/ml were placed in a plastic petri dish and inoculated at 100% humidity and 26°C. After 96 h, the mutant formed a smooth velum in the air-liquid interface, while the WT grew a fluffy velum (Fig. 1B) similar to that of F. oxysporum (6), indicating that disruption of Bbmpk1 results in alterations in the surface properties of B. bassiana.
Disruption of Bbmpk1 leads to loss of virulence in B. bassiana.
Insect bioassays showed that the pathogenicity of B. bassiana to Pieris brassicae larva was completely lost when the gene was disrupted. Whereas the median lethal time (LT50) for the wild-type strain was 3.2 days, no significant mortality due to application of the Bbmpk1 mutant conidia to the host larvae was noted even after 6 days. These data indicate that Bbmpk1 is essential for the pathogenicity of B. bassiana. Application of WT conidia resulted in the appearance of a cuticle melanization response within 2 days postinoculation, whereas no such response was noted for insects treated with conidia derived from the Bbmpk1 mutant strain. Application of WT conidia resulted in insect death within 4 days after inoculation, and hyphae were observed penetrating the cuticle outward from within the insect body, with new conidia produced on the cadaver surface ∼6 days after inoculation. In contrast, the mutant did not cause any infection symptoms and most of the treated insects underwent normal pupation ∼6 days after inoculation (Fig. 2A). Similar results were obtained in insect bioassays using aphids, Myzus persicae (data not shown). The full virulence was restored by reintroducing Bbmpk1 into the mutant (Fig. 2A).
FIG. 2.
(A) Symptoms of Pieris brassicae larva after topical application of 107 conidia/ml suspensions of WT, ΔBbmpk1, or reverse complementation transformant (RC) strains (control insects were dipped in water). (B) Symptoms of insects following injection into the second proleg with 3 μl of 107 conidia/ml suspensions (control insects were injected with 3 μl of water).
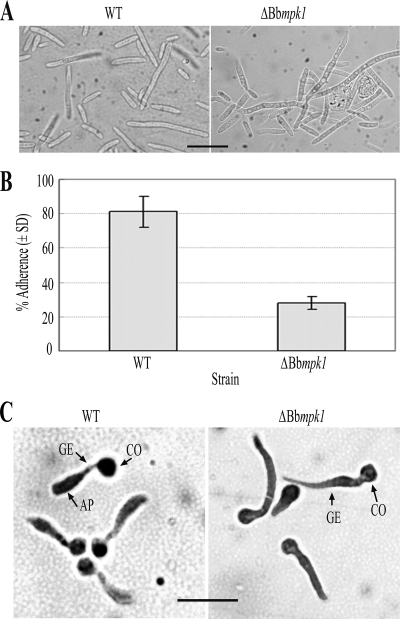
When fungal conidia were injected directly into P. brassicae larvae through the intersegmental membrane into the hemocoel, similar mortality values were observed between WT and Bbmpk1 mutant strains and, 4 days after injection, all of the insects in both treatment groups were moribund (Fig. 2B). Some morphological differences were noted between WT and ΔBbmpk1 hemolymph derived (in vivo) hyphal bodies (Fig. 3A). However, in contrast to the WT strain and the complemented ΔBbmpk1, the Bbmpk1 mutant failed to sporulate on the insect cadaver, suggesting an inability of the hyphae produced in the insect hemocoel to penetrate the cuticle outwards (Fig. 2B).
FIG. 3.
(A) Hyphal body differentiation at 3 days after injection of conidia into P. brassicae larva. Bar, 20 μm. (B) Conidial adherence to cicada hind wings. The assay was performed as described previously (43). The experiment was repeated three times with three replicates for each repeat. (C) Appressorial differentiation. Appressorium formation was induced on cicada hind wings as described previously (44). AP, appressorium; GE, germ tube; CO, conidium. Bar, 20 μm.
Bbmpk1 influences adherence of conidia and is essential for appressorium formation.
To investigate the effect of the Bbmpk1 disruption on virulence-related phenotypic parameters, spore adherence and appressorium formation was examined. Conidial adherence was assayed using cicada hind wings as described previously (43, 53). Conidial suspensions were applied in the cicada substrata and, 8 h after inoculation, only 27.8% ± 3.8% of the mutant conidia remained on the wings after a wash step using 0.05% (vol/vol) Tween 20, compared to 81.1% ± 10.5% of the WT strain conidia (df = 4; t = 9.56; P < 0.01) (Fig. 3B), indicating that conidial adherence to the insect cuticle was severely impaired by the gene disruption.
Appressorium formation was induced by application of the fungal conidia on cicada hind wings and allowing for fungal germination and growth (44). These experiments revealed that most of the WT B. bassiana conidia germinated and formed appressoria on the wings ∼22 h after inoculation. However, no appressorium formation was observed under identical condition in the ΔBbmpk1 mutant, although the conidia did germinate and grow (Fig. 3C).
BbMPK1 does not influence expression of cuticle-degrading enzymes Cdep1 and Bbchit1.
B. bassiana expresses two well-characterized cuticle-degrading enzymes, Cdep1, a Pr1-like protease implicated in the degradation of cuticular proteins and peptides, and Bbchit1, a chitinase involved in the degradation of the chitin that comprises the insect cuticle. The effect of disruption of Bbmpk1 on the expression of these two cuticle-degrading enzymes was investigated by Western blots. The results showed that the expression patterns of Cdep1 and Bbchit1 were similar in the WT and ΔBbmpk1 strains when induced by insect cuticle (see Fig. S2 in the supplemental material).
Construction and differential screening of an appressorium-stage SSH cDNA library.
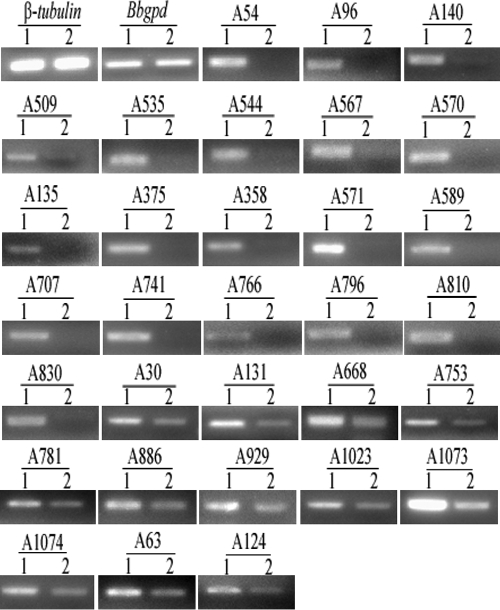
In order to identify potential BbMPK1-regulated target genes involved in appressoria formation, a SSH library was constructed using WT mRNA isolated from cells at the stage of appressorium formation (grown on cicada hind wings at 26°C for 22 h) as the tester and Bbmpk1 mutant mRNA isolated from cells grown under the same condition as the driver. From the resultant subtracted library, 1243 bacterial (transformant) colonies were preserved individually as the subtraction library. The library was then screened by dot blotting according the instructions of a PCR-select differential screening kit (Clontech). A total of 195 cDNA clones were found to hybridize exclusively to the WT tester mRNA pool (appressorium cDNA probe) and were selected for sequencing. Sequence analysis indicated that the insert sizes for the isolated clones ranged between 201 and 781 bp and corresponded to 31 unique genes. RT-PCR analysis showed that 19 genes were expressed in the WT strain but not in the mutant, and 12 additional genes showed markedly reduced transcript levels in the mutant (Fig. 4).
FIG. 4.
RT-PCR analysis of 31 selected SSH library genes. RNAs were extracted from conidia of the WT and ΔBbmpk1 mutant strains that were inoculated on cicada hind wings to induce appressorium formation (22 h after inoculation) and used for RT-PCR. All RT-PCR analysis was performed for 25 cycles using Bbgpd and β-tubulin as references. Lane 1, WT; lane 2, ΔBbmpk1 mutant.
Bioinformatic analysis revealed that 5 clones (A30, A781, A886, A929, and A1023) showed high similarity to known genes (identities ≥ 89%), specifically to histone H3, a putative aldehyde dehydrogenase, translation elongation factor 1α, ADP-ribosylation factor, and ATP-dependent RNA helicase. Five clones (A131, A140, A509, A668, and A1073) displayed medium similarity to known genes (identities ≥ 64%) that included an rRNA intron-encoded homing endonuclease, hemagglutinin esterase (infectious salmon anemia virus), cytochrome c oxidase polypeptide Via, the CHK1 checkpoint-like protein, and a hydrophobin (Hyd2). One clone (A544) displayed weak similarity to a hydrophobin 3 precursor (Fusarium culmorum) (identities = 44%). Eight clones had weak or high similarity to hypothetical protein encoding genes, and the remainder had no similarity to known genes (Table 2).
TABLE 2.
mRNA size and DNA sequence analysis of SSH cDNA clones expressed in the WT strain but not expressed or with markedly decreased expression in the Bbmpk1 mutant during appressorium formation
| Clonea | Size (bp) | E value | Identity (%) | Accession no. | Homologous sequence in GenBank |
|---|---|---|---|---|---|
| A30 | 472 | 9.00E−42 | 89 | XP_956003 | Neurospora crassa histone H3 |
| A54* | 584 | 8.00E−04 | 33 | XP_001586699 | Sclerotinia sclerotiorum predicted protein |
| A96* | 275 | 5.00E−40 | 96 | YP_173374 | Nicotiana tabacum hypothetical protein NitaMp027 |
| A131 | 245 | 2.00E−08 | 83 | XP_002294430 | Thalassiosira pseudonana rRNA intron-encoded homing endonuclease |
| A140* | 451 | 6.00E−04 | 82 | CAR56929 | Infectious salmon anemia virus hemagglutinin esterase |
| A509* | 564 | 5.00E−43 | 64 | EEY15879 | Verticillium albo-atrum cytochrome c oxidase polypeptide VIa |
| A535* | 544 | 6.00E−32 | 72 | XP_380967 | Gibberella zeae hypothetical protein FG00791.1 |
| A544* | 561 | 0.038 | 44 | ABE27987 | Fusarium culmorum hydrophobin 3 precursor |
| A567* | 418 | 2.00E−24 | 50 | XP_001389285 | Aspergillus niger hypothetical protein An01g08630 |
| A570* | 358 | 1.00E−43 | 89 | XP_001407297 | Magnaporthe grisea hypothetical protein MGG_12068 |
| A668 | 450 | 2.00E−09 | 79 | CAJ83813 | Xenopus tropicalis CHK1 checkpoint homolog |
| A753 | 392 | 2.00E−05 | 33 | XP_001595237 | Sclerotinia sclerotiorum hypothetical protein SS1G_03326 |
| A781 | 516 | 3.00E−82 | 100 | ABG77527 | Beauveria bassiana putative aldehyde dehydrogenase |
| A886 | 513 | 6.00E−95 | 99 | AAX86765 | Beauveria bassiana translation elongation factor 1 alpha |
| A929 | 712 | 3.00E−99 | 97 | XP_381190 | Gibberella zeae ADP-ribosylation factor |
| A1023 | 351 | 6.00E−44 | 92 | XP_964893 | Neurospora crassa ATP-dependent RNA helicase |
| A1073 | 620 | 1.00E−28 | 68 | ABP58683 | Beauveria bassiana hydrophobin Hyd2 |
| A1074 | 559 | 6.00E−26 | 36 | XP_001215278 | Aspergillus terreus conserved hypothetical protein |
| A63 | 201 | Unknown | |||
| A124 | 330 | Unknown | |||
| A135* | 305 | Unknown | |||
| A358* | 242 | Unknown | |||
| A375* | 347 | Unknown | |||
| A571* | 476 | Unknown | |||
| A589* | 616 | Unknown | |||
| A707* | 781 | Unknown | |||
| A741* | 484 | Unknown | |||
| A766* | 201 | Unknown | |||
| A796* | 501 | Unknown | |||
| A810* | 379 | Unknown | |||
| A830* | 324 | Unknown |
*, clones expressed in the WT strain but not in the Bbmpk1 mutant.
DISCUSSION
In several phytopathogenic fungi, Fus3/Kss1 related MAPKs are essential for appressorium formation and virulence. Mutants of Fus3/Kss1 homolog-encoding genes in M. grisea, C. lagenarium, C. gloeosporioides, and P. teres failed to produce appressoria and were avirulent (35, 38, 48). The B. bassiana MPK1 described in this report shares some conserved characteristic features of MAPKs related to Fus3/Kss1, and our data indicated that it is required for pathogenicity and differentiation of infection structures. Overall, these results suggest convergent roles for the phytopathogenic and entomopathogenic YERK1 family Fus3/Kss1 MAPKs in appressorium formation and virulence. In fungal phytopathogens, a general feature of YERK1 family MAPK mutants is that, in addition to being deficient in infection-related morphogenesis and penetration, they are also unable to grow invasively on living plant tissue (6, 38, 48, 54). However, hyphae of the ΔBbmpk1 mutant injected into the insect bodies were able to proliferate in the insect coelom, indicating some divergent regulatory pathways in entomopathogenic fungi. Cuticle penetration is thought to require both enzymatic degradation and mechanical pressure exerted by the penetrating hyphae. However, the hyphal bodies of the ΔBbmpk1 mutant grown in the insect coelom appeared to be unable to penetrate the cuticle outward and consequently failed to sporulate on the host cadaver surface. These results indicate a deficiency in penetration phenotype for ΔBbmpk1 and suggest possible defects in establishing the required mechanical pressure to penetrate through the cuticle particularly since no effect was observed on the expression of the protease and chitinases enzymes that participate in enzymatic degradation of the cuticle during penetration.
The formation of appressoria is a pivotal process for many plant and insect fungal pathogens that utilize host cuticle penetration leading to mycosis (4). In M. grisea, appressoria generate hydrostatic turgor by accumulating molar concentrations of glycerol (5, 29). Glycogen and lipid mobilization in M. grisea did not occur in a Δpmk1 mutant, which lacked the MAPK required for appressorium differentiation (40). In B. bassiana, we found that transcript levels of an ADP-ribosylation factor homolog was significantly decreased in ΔBbmpk1 mutants at the stage of appressorium formation, suggesting that BbMPK1 is possibly involved in the regulation of vesicular traffic, lipid metabolism, microtubule dynamics, and development (23).
Hydrophobins are generally thought to be ubiquitous proteins of fungal walls. These proteins form an outer layer coat on conidia and in certain cases on hyphae and play a role in a broad range of processes during fungal growth and development. They are involved in the formation of aerial structures and in attachment to hydrophobic surfaces (2, 15, 46). For instance, the hydrophobin MPG1 of M. grisea directs formation of a rodlet layer on conidia, which contributes to the hydrophobicity of the surface, and a Δmpg1 mutant showed reduced infectivity coupled to an inability to form appressoria (39). In our previous study, disruption of a HOG1 kinase gene, Bbhog1, in B. bassiana caused a dramatic decrease in transcript levels of two hydrophobin-encoding genes hyd1 and hyd2 (53). ΔBbhog1 mutants also showed greatly reduced pathogenicity, decreased spore viability, reduced ability to attach to insect cuticles, and a reduction in appressorium formation. In the present study, disruption of Bbmpk1 in B. bassiana caused a change in the expression pattern of two hydrophobin genes. First, expression of a hydrophobin-encoding gene, A1073 (similar to the B. bassiana hydrophobin Hyd2), was dramatically reduced in the ΔBbmpk1 mutant. Second, another gene, A544, which showed similarity to the F. culmorum hydrophobin 3 precursor, was found to be expressed in the wild-type strain but not in the ΔBbmpk1 mutant. Moreover, ΔBbmpk1 mutants showed a defect in surface hydrophobicity and a reduction in attachment to insect cuticles. These data are consistent with the idea that hydrophobins mediate surface hydrophobicity which partially controls attachment and appressorium formation (14).
In addition, our data show that BbMPK1 may also contribute to the regulation of the mitochondrial electron transport chain (37) since a cytochrome c oxidase homolog was identified to be expressed in the WT parent but not in the mutant during appressoria formation. Transcript levels of six genes (A30, A131, A668, A1023, A886 and A781), which had similarity to histone H3, rRNA intron-encoded homing endonuclease, CHK1 checkpoint-like protein, ATP-dependent RNA helicase, translation elongation factor 1 alpha and a putative aldehyde dehydrogenase, respectively, were also markedly reduced in ΔBbmpk1 mutants during appressorium formation. These results suggest that BbMPK1 probably influences DNA repair (11, 18), transcription (11, 17), chromatin remodeling (11, 25), rRNA processing, small nucleolar RNA accumulation (11), translation and posttranslational modification (34, 55), oxidation of aldehydes (24, 45), and likely other cellular processes. In addition, a virus hemagglutinin esterase homolog (A140) was found to be exclusively expressed in WT, but its origin and role in eukaryotic cells remains unknown. Further experiments are needed to confirm and elucidate the relationship between BbMPK1 signal pathway and the putative target genes involved in the corresponding biological processes.
As noted above, regulatory genes affect a broad set of targets, and their disruption often results in pleiotropic losses of function. In several plant pathogenic fungi, expression of some cell wall-degrading enzymes are regulated by Fus3/Kss1 homologs MAPK. One of the targets of Kss1 is PGU1, a gene that encodes a secreted endopolygalacturonase that may aid in invasion of the natural plant substrate of S. cerevisiae (26). PL1, which encodes a pectate lyase, is underexpressed in a mutant of the vascular wilt fungus F. oxysporum lacking the MAPK Fmk1 (6). Expression of two cellulolytic enzymes encoding genes, CBH7 and EG6, was strictly regulated by Chk1 MAPK signal pathway in C. heterostrophus (21). In A. brassicicole, the amk1 disruption mutants expressed extremely low amounts of several hydrolytic enzyme genes that were induced over 10-fold in the WT during infection (3). Similarly, a MAPK suppresses the expression of cuticle-degrading enzyme encoding genes related to mycoparasitism in Trichoderma virens (28). However, disruption of Bbmpk1 in B. bassiana did not influence expression of two insect cuticle-degrading enzymes, a Pr1-like protease, Cdep1, and an secreted chitnase, Bbchit1, both of which are important virulence factors (8, 52). The results suggest that expression of cuticle-degrading enzymes in B. bassiana is under the control of a different regulatory mechanism.
Supplementary Material
Acknowledgments
This study was supported by grants from the Natural Science Foundation of China (grant 30871668), the Ministry of Science and Technology of China (grants 2009CB118904 and 2006AA10A212), and the Program for the First Excellent Talents of University in Chongqing.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bidochka, M., and G. G. Khachatourians. 1990. Identification of Beauveria bassiana extracellular protease as a virulence factor in pathogenicity toward the migratory grasshopper, Melanoplus sanguinipes. J. Invertebr. Pathol. 56:362-370. [Google Scholar]
- 2.Cho, E. M., B. H. Kirkland, D. J. Holder, and N. O. Keyhani. 2007. Phage display cDNA cloning and expression analysis of hydrophobins from the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology 153:3438-3447. [DOI] [PubMed] [Google Scholar]
- 3.Cho, Y., R. A. Cramer, K. H. Kim, J. Davis, T. K. Mitchell, P. Figuli, B. M. Pryor, E. Lemasters, and C. B. Lawrence. 2007. The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of genes encoding hydrolytic enzymes in Alternaria brassicicola. Fungal Genet. Biol. 44:543-553. [DOI] [PubMed] [Google Scholar]
- 4.Clarkson, J. M., and A. K. Charnley. 1996. New insights into the mechanisms of fungal pathogenesis in insect. Trends Microbiol. 4:197-203. [DOI] [PubMed] [Google Scholar]
- 5.de Jong, J. C., B. J. McCormack, N. Smirnoff, and N. J. Talbot. 1997. Glycerol generates turgor in rice blast. Nature 389:244-245. [Google Scholar]
- 6.Di Pietro, A., F. I. García-Maceira, E. Méglecz, and M. I. G. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 7.Fan, Y., Y. Zhang, Z. Luo, J. Feng, W. Fang, and Y. Pei. 2006. Preparation of a serine protease of Beauveria bassiana by the Pichia pastoris expression system. Mycosystema 25:56-62. [Google Scholar]
- 8.Fang, W., B. Leng, Y. Xiao, K. Jin, J. Ma, Y. Fan, J. Feng, X. Yang, Y. Zhang, and Y. Pei. 2005. Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl. Environ. Microbiol. 71:363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang, W., M. Pava-ripoll, S. Wang, and R. J. St. Leger. 2009. Protein kinase A regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fungal Genet. Biol. 46:277-285. [DOI] [PubMed] [Google Scholar]
- 10.Fang, W., Y. Zhang, X. Yang, X. Zheng, H. Duan, Y. Li, and Y. Pei. 2004. Agrobacterium tumefaciens-mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J. Invertebr. Pathol. 85:18-24. [DOI] [PubMed] [Google Scholar]
- 11.Gribun, A., K. L. Y. Cheung, J. Huen, J. Ortega, and W. A. Houry. 2008. Yeast Rvb1 and Rvb2 are ATP-Dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 376:1320-1333. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein, M., and D. S. Hogness. 1975. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc. Natl. Acad. Sci. U. S. A. 72:3961-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herskowitz, I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80:187-197. [DOI] [PubMed] [Google Scholar]
- 14.Holder, D. J., B. H. Kirkland, M. W. Lewis, and N. O. Keyhani. 2007. Surface characteristics of the entomopathogenic fungus Beauveria (Cordyceps) bassiana. Microbiology 153:3448-3457. [DOI] [PubMed] [Google Scholar]
- 15.Holder, D. J., and N. O. Keyhani. 2005. Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl. Environ. Microbiol. 71:5260-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, K., Y. Zhang, Z. Luo, Y. Xiao, Y. Fan, D. Wu, and Y. Pei. 2008. An improved method for Beauveria bassiana transformation using phosphinothricin acetlytransferase and green fluorescent protein fusion gene as a selectable and visible marker. Biotechnol. Lett. 30:1379-1383. [DOI] [PubMed] [Google Scholar]
- 17.Jo, Y. K., G. L. Wang, and M. J. Boehm. 2007. Expression analysis of rice defense-related genes in turgrass in response to Magnaporthe grisea. Phytopathology 97:170-178. [DOI] [PubMed] [Google Scholar]
- 18.Kosoy, A., and M. J. O' Connell. 2008. Regulation of Chk1 by its C-terminal domain. Mol. Biol. Cell 19:4546-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kültz, D. 1998. Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 46:571-588. [DOI] [PubMed] [Google Scholar]
- 20.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. U. S. A. 96:13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lev, S., and B. A. Horwitz. 2003. A mitogen-activated protein kinase pathway modulates the expression of two cellulase genes in Cochliobolus heterostrophus during plant infection. Plant Cell 15:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis, M. W., I. V. Robalino, and N. O. Keyhani. 2009. Uptake of the fluorescent probe FM4-64 by hyphae and hemolymph-derived in vivo hyphal bodies of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3110-3120. [DOI] [PubMed] [Google Scholar]
- 23.Li, Y., W. G. Kelly, J. M. Logsdon, Jr., A. M. Schurko, B. D. Harfe, K. L. Hill-Harfe, and R. A. Kahn. 2004. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in Caenorhabditis elegans. FASEB J. 18:1834-1850. [DOI] [PubMed] [Google Scholar]
- 24.Lindahl, R. 1992. Aldehyde dehydrogenases and their role in carcinogenesis. Crit. Rev. Biochem. Mol. Biol. 27:283-335. [DOI] [PubMed] [Google Scholar]
- 25.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 26.Madhani, H. D., T. Galitski, E. S. Lander, and G. R. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. U. S. A. 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCluskey, K. 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52:245-262. [DOI] [PubMed] [Google Scholar]
- 28.Mendoza-Mendoza, A., M. J. Pozo, D. Grzegorski, P. Martínez, J. M. García, V. Olmedo-Monfil, C. Cortés, C. Kenerley, and A. Herrera-Estrella. 2003. Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 100:15965-15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Money, N. P. 1997. Mechanism linking cellular pigmentation and pathogenicity in rice blast disease. Fungal Genet. Biol. 22:151-152. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishida, E., and Y. Gotoh. 1993. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem. Sci. 18:128-131. [DOI] [PubMed] [Google Scholar]
- 32.Orbach, M. J., E. B. Porra, and C. Yanofsky. 1986. Cloning and characterization of the gene for β-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol. Cell. Biol. 6:2452-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 34.Roger, A. J., O. Sandblom, W. F. Doolittle, and H. Philippe. 1999. An evaluation of elongation factor 1α as a phylogenetic marker for eukaryotes. Mol. Biol. Evol. 16:218-233. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Roldan, M. C., F. J. Maier, and W. Schafer. 2001. PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant-Microbe Interact. 14:116-125. [DOI] [PubMed] [Google Scholar]
- 36.St Leger, R. J., L. Joshi, M. J. Bidochka, and D. W. Roberts. 1996. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. U. S. A. 93:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Susan, S., and R. Brambl. 1981. Mitochondrial biogenesis during fungal spore germination: respiration and cytochrome c oxidase in Neurospora crassa. J. Bacteriol. 147:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13:374-383. [DOI] [PubMed] [Google Scholar]
- 39.Talbot, N. J., M. J. Kershaw, G. E. Wakley, O. M. H. De Vries, J. G. H. Wessels, and J. E. Hamer. 1996. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8:985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thines, E., R. W. S. Weber, and N. J. Talbot. 2000. MAP kinase and protein kinase A-dependent mobilization of triacylglycerol and glycogen during appressorium turgor generation by Magnaporthe grisea. Plant Cell 12:1703-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanchoo, A., M. W. Lewis, and N. O. Keyhani. 2009. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and hemolymph derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 155:3121-3133. [DOI] [PubMed] [Google Scholar]
- 42.Wang, C., and R. J. St. Leger. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 282:21110-21115. [DOI] [PubMed] [Google Scholar]
- 43.Wang, C., and R. J. St. Leger. 2007. The MAD1 adhesion of Metarhizium anisopliae links adhesion with blastospore production and virulence to insects, and the MAD2 adhesion enables attachment to plants. Eukaryot. Cell 6:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, C., and R. J. St. Leger. 2005. Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot. Cell 4:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiner, H. 1989. Role of alcohol and aldehyde dehydrogenase in vivo: speculations on their natural substrates, p. 147-160. In K. E. Crow and R. D. Batt (ed.), Human metabolism of alcohol. vol. 2, CRC Press, Boca Raton, FL. [Google Scholar]
- 46.Whiteford, J. R., and P. D. Spanu. 2002. Hydrophobins and the interactions between fungi and plants. Mol. Plant Pathol. 3:391-400. [DOI] [PubMed] [Google Scholar]
- 47.Xu, J. R. 2000. Map kinases in fungal pathogens. Fungal Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 48.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696-2706. [DOI] [PubMed] [Google Scholar]
- 49.Xu, Y., R. Orozco, E. M. K. Wijeratne, A. A. L. Gunatilaka, S. P. Stock, and I. Molnár. 2008. Biosynthesis of the cyclooligomer depsipeptide beauvericin, a virulence factor of the entomopathogenic fungus Beauveria bassiana. Chem. Biol. 15:898-907. [DOI] [PubMed] [Google Scholar]
- 50.Xu, Y., R. Orozco, E. M. K. Wijeratne, P. Espinosa-Artiles, A. A. L. Gunatilaka, S. P. Stock, and I. Molnár. 2009. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 46:353-364. [DOI] [PubMed] [Google Scholar]
- 51.Xue, C., G. Park, W. Choi, L. Zheng, R. A. Dean, and J. R. Xu. 2002. Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14:2107-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., M. Feng, Y. Fan, Z. Luo, X. Yang, D. Wu, and Y. Pei. 2008. A cuticle-degrading protease (CDEP-1) of Beauveria bassiana enhances virulence. Biocontrol Sci. Technol. 18:543-555. [Google Scholar]
- 53.Zhang, Y., J. Zhao, W. Fang, J. Zhang, Z. Luo, M. Zhang, Y. Fan, and Y. Pei. 2009. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl. Environ. Microbiol. 75:3787-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, L., M. Campbell, J. Murray, S. Lam, and J. R. Xu. 2000. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13:724-732. [DOI] [PubMed] [Google Scholar]
- 55.Zobel-Thropp, P., M. C. Yang, L. Machado, and S. Clarke. 2000. A novel posttranslational modification of yeast elongation factor 1A. J. Biol. Chem. 275:37150-37158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.