Abstract
During anaerobic growth of Escherichia coli, pyruvate formate-lyase (PFL) and lactate dehydrogenase (LDH) channel pyruvate toward a mixture of fermentation products. We have introduced a third branch at the pyruvate node in a mutant of E. coli with a mutation in pyruvate dehydrogenase (PDH*) that renders the enzyme less sensitive to inhibition by NADH. The key starting enzymes of the three branches at the pyruvate node in such a mutant, PDH*, PFL, and LDH, have different metabolic potentials and kinetic properties. In such a mutant (strain QZ2), pyruvate flux through LDH was about 30%, with the remainder of the flux occurring through PFL, indicating that LDH is a preferred route of pyruvate conversion over PDH*. In a pfl mutant (strain YK167) with both PDH* and LDH activities, flux through PDH* was about 33% of the total, confirming the ability of LDH to outcompete the PDH pathway for pyruvate in vivo. Only in the absence of LDH (strain QZ3) was pyruvate carbon equally distributed between the PDH* and PFL pathways. A pfl mutant with LDH and PDH* activities, as well as a pfl ldh double mutant with PDH* activity, had a surprisingly low cell yield per mole of ATP (YATP) (about 7.0 g of cells per mol of ATP) compared to 10.9 g of cells per mol of ATP for the wild type. The lower YATP suggests the operation of a futile energy cycle in the absence of PFL in this strain. An understanding of the controls at the pyruvate node during anaerobic growth is expected to provide unique insights into rational metabolic engineering of E. coli and related bacteria for the production of various biobased products at high rates and yields.
In Escherichia coli as well as in other aerobic organisms, sugars such as glucose are metabolized in two separate steps: glycolysis, which converts glucose to pyruvate, and tricarboxylic acid (TCA) cycle enzymes, which oxidize acetyl coenzyme A (acetyl-CoA) to CO2 (5, 9). The pyruvate dehydrogenase complex (PDH) connects the glycolytic reactions to TCA cycle enzymes by catalyzing the production of acetyl-CoA from pyruvate. Because of its unique central role in metabolism, PDH is regulated at both the genetic and the biochemical level (7, 12, 27, 33, 34). The NADH generated during the complete oxidation of sugar is reoxidized to NAD+ by O2 through the respiratory electron transport pathway with accompanying energy production (11). Optimum coupling of these enzyme reactions helps to maintain the internal ratios of [NADH] to [NAD+] (redox balance) and of [ATP] to [ADP] plus [AMP] in order to support growth at the highest rate.
The absence of O2 or another external electron acceptor during the growth of E. coli (anaerobic conditions) forces the bacterium to minimize the contribution of the TCA cycle enzymes to biosynthesis from catabolism (4, 14). Under these conditions, pyruvate or acetyl-CoA derived from pyruvate serves as the electron acceptor (reduced to lactate or ethanol, respectively) to maintain the redox balance. The enzymes responsible for redox balance in anaerobic E. coli are pyruvate formate-lyase (PFL), lactate dehydrogenase (LDH), and alcohol/aldehyde dehydrogenase (adhE; ADH-E). The main products of the fermentation of E. coli are a mixture of organic acids, such as acetate, lactate, and formate, in addition to ethanol (2, 4). Succinate, derived from phosphoenolpyruvate (PEP), is a minor product of fermentation and normally accounts for less than 5% of the total products produced from glucose by the culture.
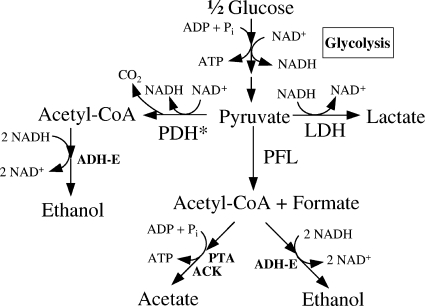
Anaerobic growth of E. coli, compared to aerobic growth, is also limited by energy, leading to an increase in glycolytic flux (19). The conversion of pyruvate to acetate and ethanol yields an additional ATP per glucose, suggesting that this would be the preferred route for pyruvate oxidation during anaerobic growth. This is accomplished by the PFL-dependent production of acetyl-CoA and further conversion to acetate (Fig. 1). This preference for PFL has been demonstrated `with several bacteria under carbon limitation conditions imposed either in a chemostat or in the presence of a poor carbon source (10, 20, 23). This additional ATP also elevates the ATP yield per glucose to 3, with an increase in the growth rate, and has been shown to be essential for the anaerobic growth of E. coli in xylose-mineral salts medium (13). The absence of this third ATP in a pfl mutant has been reported to increase glycolytic flux to lactate to compensate for this decrease in ATP yield per glucose (39). However, the flow of pyruvate carbon to acetate is tempered by the need to maintain redox balance, and this is achieved by the conversion of a second acetyl-CoA to ethanol by ADH-E. Under conditions of energy excess due to a declining growth rate, lactate production is expected to support redox balance maintenance without the additional ATP from the PFL-ADH-E pathway (Fig. 1). The production of this mixture of products in an appropriate ratio helps to maintain the redox balance under anaerobic conditions while also maximizing the ATP yield per glucose to support high growth rates and cell yields.
FIG. 1.
Anaerobic metabolic pathways of E. coli carrying the lpd101 mutation (PDH*).
No PDH-based fermentation reaction to ethanol that can also help maintain cellular redox balance in an anaerobic cell has evolved in E. coli or other closely related bacteria. PDH activity is inhibited by NADH, normally found at higher levels in anaerobically growing cultures than in aerobic cultures (12, 18, 34, 35). Based on genome sequences available in GenBank, the genes encoding the components of PDH are not found in strictly anaerobic bacteria.
We have recently described a mutation (lpd101) in the dihydrolipoamide dehydrogenase (LPD) of the PDH that allowed the enzyme to function in anaerobic cells (designated PDH* here) (17, 18). With this altered PDH*, an anaerobic cell can have three different pathways for pyruvate metabolism (Fig. 1). The three main enzymes that utilize pyruvate as a substrate, PDH*, PFL, and LDH, have different apparent Km values for pyruvate (0.4, 2.0, and 7.2 mM, respectively) (1, 18, 37, 41). PDH requires NAD+ for activity (apparent Km, 0.07 mM), while LDH is dependent on NADH (apparent Km, 0.2 mM) as the second substrate (18, 37).
The PDH* serves as the first enzyme in a pathway that oxidatively decarboxylates pyruvate to acetyl-CoA and NADH, followed by reduction of the acetyl-CoA by alcohol dehydrogenase to ethanol in a two-step process using 2 NADHs (Fig. 1). The NADH produced during the conversion of  glucose to acetyl-CoA dictates that the acetyl-CoA generated by PDH be used for redox balance (ethanol) and not for ATP generation (acetate), unless some of the NADH is used for biosynthesis by the growing cell (17). PDH* and LDH serve essentially the same physiological role in the cell, oxidizing NADH to support continued operation of glycolysis, although it is not readily apparent with PDH*. Although PDH* contributes to an increase in NADH pool, the redox balance is still maintained by coupling PDH* to NADH-dependent reduction of acetyl-CoA to ethanol by ADH-E (Fig. 1). This potential competition between LDH and PDH has been eliminated in the wild type through inhibition of the activity of PDH by NADH (12, 18, 32). However, the in vivo role of PDH* in a mutant that has all three pathways has not been investigated, since the flow of pyruvate through any of the three reactions during growth and postgrowth fermentation of sugars to products is expected to be dependent on the redox state, the ATP requirement, and other physiological conditions of the anaerobic cell. Using a combination of metabolic flux analysis and mutations in one or more of the genes encoding these enzymes, we have evaluated the flow of pyruvate carbon among the three potential pathways. The results are presented in this communication.
glucose to acetyl-CoA dictates that the acetyl-CoA generated by PDH be used for redox balance (ethanol) and not for ATP generation (acetate), unless some of the NADH is used for biosynthesis by the growing cell (17). PDH* and LDH serve essentially the same physiological role in the cell, oxidizing NADH to support continued operation of glycolysis, although it is not readily apparent with PDH*. Although PDH* contributes to an increase in NADH pool, the redox balance is still maintained by coupling PDH* to NADH-dependent reduction of acetyl-CoA to ethanol by ADH-E (Fig. 1). This potential competition between LDH and PDH has been eliminated in the wild type through inhibition of the activity of PDH by NADH (12, 18, 32). However, the in vivo role of PDH* in a mutant that has all three pathways has not been investigated, since the flow of pyruvate through any of the three reactions during growth and postgrowth fermentation of sugars to products is expected to be dependent on the redox state, the ATP requirement, and other physiological conditions of the anaerobic cell. Using a combination of metabolic flux analysis and mutations in one or more of the genes encoding these enzymes, we have evaluated the flow of pyruvate carbon among the three potential pathways. The results are presented in this communication.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are listed in Table 1. All strains are derived from E. coli K-12 strain W3110 (ATCC 27325). Strain YK1 is a kanamycin-sensitive derivative of strain SE2378 that carries the lpd101 mutation described previously (17). Gene deletions were constructed using the method described by Datsenko and Wanner (6). Mutations were transduced using bacteriophage P1 as described previously (29).
TABLE 1.
Bacterial strains used in this study
| Straina | Relevant genotypeb | Source or reference |
|---|---|---|
| W3110 | F− IN(rrnD-rrnE)1 λ− | ATCC 27325 |
| AH218 | BW25113 Δ(focA-pflB)-FRT-Km-FRT | 13 |
| AH240 | Δ(focA-pflB)-FRT-Km-FRT | W3110 × P1 (AH218) |
| AH241 | ΔldhA | 17 |
| BW25113 | lacIqrrnBT14 ΔlacZW116hsdR514 araBADAH33 ΔrhaBADLD78 | 6 |
| PMD23 | ΔhypF-FRT-Km-FRT | 15 |
| SE2378 | lpd101 ΔldhA Δ(focA-pflB)-FRT-Km-FRT pdhR | 17 |
| SE2382 | lpd101 ΔldhA Δ(focA-pflB)-FRT-Km-FRT | 18 |
| YK1 | lpd101 ΔldhA Δ(focA-pflB)-FRT pdhR Kms | SE2378; Kms |
| YK98 | BW25113 Δlpd-FRT-Km-FRT | This study |
| YK134 | Δlpd-FRT-Km-FRT | W3110 × P1 (YK98) |
| YK142 | lpd101 | YK134 × P1 (SE2382) |
| YK143 | ΔldhA Δlpd-FRT-Km-FRT | AH241 × P1 (YK98) |
| YK167 | lpd101 Δ(focA-pflB)-FRT-Km-FRT | YK142 × P1 (AH240) |
| QZ1 | ΔldhA lpd101 | YK143 × P1 (YK142) |
| QZ2 | lpd101 ΔhypF-FRT-Km-FRT | YK142 × P1 (PMD23) |
| QZ3 | ΔldhA lpd101 ΔhypF-FRT-Km-FRT | QZ1 × P1 (PMD23) |
All strains are derivatives of E. coli K-12 strain W3110, a nearly wild type strain deposited by J. Lederberg, unless noted otherwise (BW25113 and YK98).
FRT, FLP recombination target.
Growth medium.
L broth (LB) (29) supplemented with 20 g/liter of glucose was used for all chemostat cultivations. The glucose concentration was increased to 50 g/liter in batch fermentation experiments.
Bioreactor conditions.
For metabolic flux analysis, fermentations were conducted under anaerobic chemostat conditions in a 2.5-liter Bioflo III fermenter (New Brunswick Scientific, New Brunswick, NJ) with a 1.2-liter culture volume. Anaerobic conditions were established by sparging the culture with N2 at a flow rate of 0.2 liter min−1. The pH, temperature, and agitation were maintained at 7.0, 37°C, and 250 rpm, respectively. The culture pH was maintained by the addition of KOH. A dilution rate of 0.1 h−1 was maintained throughout the experiment by controlling the medium feed rate. The continuous culture reached steady state after 5 to 6 residence times. Samples were taken for analysis every 5 h. Three samples were taken from the steady-state culture for determination of fermentation products, and the average of the three was used for the calculation. Deviations from this mean were less than 5%, and the results from one complete representative experiment are presented. For batch fermentations, cells were cultivated with 50 g/liter glucose, starting with a 16-h-old aerobic culture as the inoculum (1%, vol/vol), as previously described (17). All fermentation experiments were repeated at least three times, and the variation among experimental results was less than 15%. Results from one representative experiment are presented in this communication.
Enzyme activity.
A 250-ml culture from a batch fermentation in LB with glucose (30 g/liter) was harvested at the mid-exponential phase of growth, and the cells were collected after centrifugation (10,000 × g, 20 min, 4°C). Cells were washed twice with 25 ml of 50 mM potassium phosphate buffer (pH 7.5) and were then resuspended in 5 ml of the same buffer. All operations were conducted at 4°C. Cells were passed through a French pressure cell at 20,000 lb/in2. The crude extract was centrifuged first at 10,000 × g for 30 min, and the supernatant was further clarified by centrifugation at 30,000 × g for 60 min. The supernatant was used for enzyme assays.
PDH activity was determined by monitoring pyruvate-dependent reduction of NAD+ at 340 nm (ɛ, 6,220 M−1 cm−1) at room temperature, as described previously (18). LDH activity was determined by measuring pyruvate-linked NADH oxidation at 340 nm (43). ADH-E activity was assayed by measuring the increase in absorbance at 340 nm resulting from the reduction of NAD+ in the presence of ethanol (ADH assay; Worthington Biochemical Corporation). One unit of enzyme activity is defined as one micromole of product produced per minute.
Total RNA was extracted from the same cultures used for enzyme activity determination, and the specific mRNA level in the RNA sample was determined as described by Kim et al. (18).
Analytical methods.
Sugar and fermentation products were determined by high-performance liquid chromatography using an HP1090 chromatograph (Agilent Technologies, Santa Clara, CA) fitted with an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA) and UV (210 nm) and refractive index detectors, in series (38). Cell density was monitored at 420 nm (DU640 spectrophotometer; Beckman Coulter, Fullerton, CA) and reported as the dry weight of cells. The dry weight of cells of a culture at 1 optical density (OD) unit at 420 nm equals 0.22 g/liter. The protein concentration was determined by the Bradford method with bovine serum albumin as the standard (3).
Metabolic flux analysis.
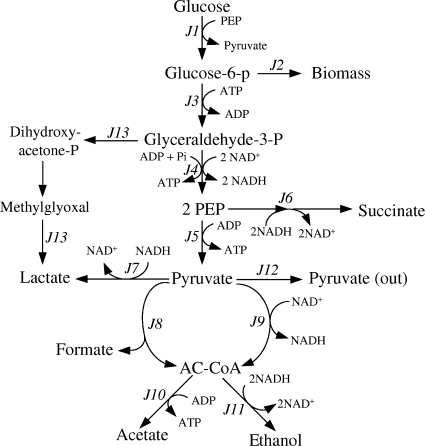
Metabolic flux analysis was used for the calculation of in vivo fluxes (36). The methodology relies on the metabolite balances, biochemical constraints, and pseudo-steady-state assumption for intracellular metabolites (40). The stoichiometric reactions presented in Fig. 2 were used for the calculation of flux distributions based on experimental data. The resulting metabolic network consists of 13 reactions, 8 of which (J1, J6, J7, J8, J10, J11, J12, and J13) can be measured. All the lactate produced by the ldhA-plus strains was assumed to be produced by LDH (reaction J7). In ldhA mutants, the lactate was assumed to be from the melthylglyoxal pathway (reaction J13). In order to distinguish between the contributions of PDH* and PFL, further metabolism of formate was eliminated by introducing an hypF mutation to remove formate hydrogen-lyase (FHL) activity. In such an hypF mutant, the level of formate in the medium is a measure of PFL activity.
FIG. 2.
Central metabolic pathways of E. coli used for flux analysis. Although PDH* also produces acetyl-CoA (reaction J9), due to NADH production most of this acetyl-CoA was assumed to flow only through reaction J11 to maintain redox balance.
Materials.
Inorganic salts, organic chemicals, and medium components were purchased either from Fisher Scientific (Pittsburgh, PA) or from Sigma Chemical Co. (St. Louis, MO).
RESULTS AND DISCUSSION
Preferred pathway at the pyruvate node.
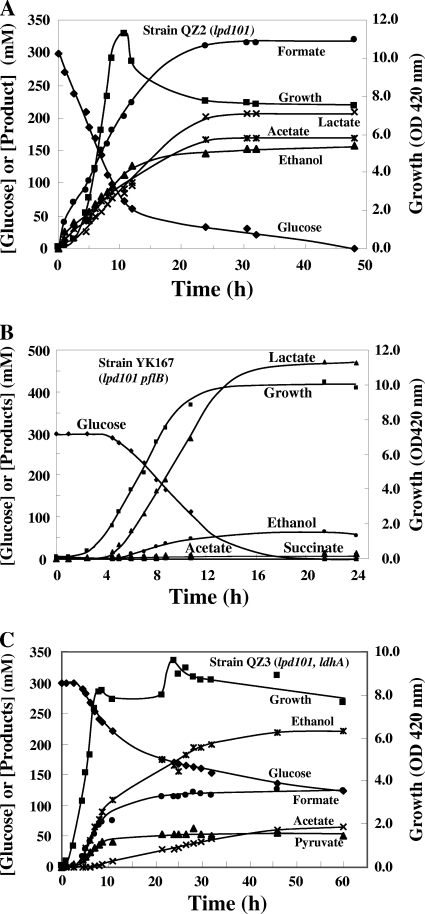
In strain QZ2, which carries the lpd101 mutation, rendering the PDH* functional during anaerobic growth, pyruvate flux through PDH* in batch cultures was minimal during anaerobic growth (Fig. 3A). Strain QZ2 produced acetate, ethanol, formate, and lactate in pH-controlled batch fermentation. The specific rates of production of acetate, ethanol, and formate during the exponential phase of growth were 29.1, 27.2, and 51.5 mmol h−1 (g of cells [dry weight])−1, respectively. The presence of equimolar amounts of acetate and ethanol in the medium suggests that PFL-produced acetyl-CoA is the precursor for these two products. This is also confirmed by the specific rate of production of formate, that is, the combined rates of acetate and ethanol production, a measure of the PFL pathway. The specific rate of lactate production (26.1 mmol h−1 [g of cells {dry weight}]−1) during this growth phase was about 50% of the formate production rate. The presence of lactate in the fermentation broth suggests that a significant fraction of the reductant generated during glycolysis was channeled to reduce pyruvate to lactate in order to maintain redox balance. An interesting observation is the absence of PDH*-based acetyl-CoA production in this culture, since this acetyl-CoA can be converted to ethanol while also supporting optimum redox balance (17, 18). Although PDH* requires NAD+ for activity, in contrast to LDH, a coupled PDH* and ADH pathway (pyruvate to ethanol) does oxidize 1 net NADH, as does LDH (Fig. 1). However, in vivo, LDH appears to be the preferred enzyme for NADH oxidation in the cell. In support of this pyruvate flux distribution, the PDH activity of strain QZ2 (0.14 U mg protein−1) was found to be only about 30% of that of an isogenic “wild-type” strain, PMD23 (0.46 U mg protein−1) (Table 2).
FIG. 3.
Growth and fermentation characteristics of E. coli strains QZ2, YK167, and QZ3 with different enzyme compositions at the pyruvate node in pH-controlled batch cultures. See Materials and Methods for details. The concentrations of lactate and succinate in the strain QZ3 fermentation broth at 60 h were 19 mM and 20 mM, respectively. Formate was not detected in the broth of strain YK167.
TABLE 2.
Enzyme activities of select mutants with alterations in the pyruvate node
| Strain | Relevant genotype | Enzymes presenta | Enzyme activity (μmol min−1 mg of protein−1)b |
||
|---|---|---|---|---|---|
| LDH | PDH | ADH | |||
| PMD23 | Wild type | PFL, LDH | 1.52 | 0.46 | 0.04 |
| QZ2 | lpd101 | PFL, LDH, PDH* | 0.41 | 0.14 | 0.05 |
| QZ3 | lpd101 ldhA | PFL, PDH* | 0.01 | 0.44 | 0.04 |
| YK167 | lpd101 pflB | LDH, PDH* | 4.17 | 0.32 | 0.06 |
PFL, pyruvate formate-lyase; LDH, fermentative lactate dehydrogenase; PDH*, pyruvate dehydrogenase complex with a mutation in dihydrolipoamide dehydrogenase.
Determined in extracts of cells harvested from fermenter cultures in the mid-exponential phase of growth. ADH, alcohol dehydrogenase. See Materials and Methods for assay conditions and other details.
Pyruvate flux in strain YK167, which produces only LDH and PDH*, and not PFL activity, was also directed toward LDH and not PDH* (Fig. 3B), and the PDH activity of this culture was only slightly lower than the value obtained with strain PMD23 (Table 2). However, strain YK167 extracts contained a level of LDH activity about 2.7 times higher than that of strain PMD23, and this higher LDH activity could account for the flux through LDH over PDH* at the pyruvate node. In agreement with this higher LDH activity, the ldhA mRNA level of strain YK167 was found to be 2.5-fold higher than that of strain PMD23. These results indicate that in the presence of both LDH and PDH*, LDH is the preferred physiological pathway of pyruvate metabolism in anaerobic E. coli strains for redox maintenance.
The rate of ethanol production (20.5 mmol h−1 [g of cells {dry weight}]−1) by strain QZ3, lacking LDH activity, was the same as the formate production rate (20.6 mmol h−1 [g of cells {dry weight}]−1) during the growth phase (Fig. 3C). Acetate production during growth was only 1.2 mmol h−1 (g of cells [dry weight])−1. Strain QZ3 also produced a significant amount of pyruvate (10.6 mmol h−1 [g of cells {dry weight}]−1). The reducing equivalents (NADH) generated during pyruvate production and secretion were apparently used to reduce the acetyl-CoA produced by PFL to ethanol, accounting for the equimolar amounts of ethanol and formate in the medium. These results suggest that PDH* did not contribute to pyruvate flux in strain QZ3 during this initial phase of growth. Only when the culture reached the stationary phase (after about 10 h) could a contribution of PDH to pyruvate flux be detected based on the ratios of the acetate, ethanol, and formate production rates; rates of specific productivity for acetate, ethanol, and formate were 1.1, 3.6, and 2.1 mmol h−1 (g of cells [dry weight])−1.
Although the initial growth rates of all three strains were comparable, a significant amount of glucose remained in the medium of strain QZ3 even after 116 h of incubation, in contrast to levels in the other two strains with LDH activity (strains QZ2 and YK167) (Fig. 3A and B). This lower rate of glucose consumption by strain QZ3 compared to strain QZ2 suggests that PDH* and PFL are rate-limiting at the pyruvate node. This is in agreement with the observation that pyruvate was detected in the broth of strain QZ3 alone among the three strains tested (Fig. 3). However, the PDH* activity of strain QZ3 was comparable (about 0.4 U mg protein−1 in crude extracts) to that of the wild-type strain PMD23 (Table 2), a characteristic that may be related to a higher accumulation of pyruvate, a known activator of the pdh operon and of pyruvate secretion by strain QZ3.
Pyruvate distribution in a steady-state culture.
Anaerobic batch cultures of E. coli in which the growth rate is constantly changing may not be sensitive enough to measure flux through PDH*. To overcome this limitation, various mutants lacking one or more of the enzymes at the pyruvate node were cultured in a chemostat operating at a dilution rate of 0.1 h−1. In these experiments, a rich medium was used to minimize the amount of carbon that was diverted to biosynthesis.
Strains with active LDH and PFL.
The biochemistry framework depicted in Fig. 2 was used to determine the flux values using a steady-state glucose-limited chemostat culture. The detailed flux distributions of these cultures are presented in Table 3. The cell mass of strain PMD23, which is wild type for fermentation except for the absence of FHL activity (hypF), stabilized at about 1.8 g/liter, and the specific glucose consumption rate was about 5.8 mmol h−1 (g of cells [dry weight])−1. The major fermentation products of strain PMD23 were acetate, lactate, and ethanol, with the carbon flux ratio to these products at about 1:1:1 (J7, J10, and J11) (Tables 3 and 4). Carbon flux to formate was the sum of the fluxes to acetate and ethanol, as expected. The production of lactate indicates that this culture is apparently ATP limited and is not limited for reductant, since the rich medium provided the needed precursors for the biosynthesis of macromolecules. The additional reductant generated due to an increase in carbon flux for the production of ATP is apparently channeled to lactate to maintain redox balance. This is about 30% of the carbon flux at the pyruvate node (Table 4). The higher-than-expected flux to lactate in strain W3110 derivatives compared to the flux in a strain MC4100 derivative reported by Yang et al. (42) could reflect the significant differences between the genotypes of the two strains (25).
TABLE 3.
Metabolic flux distributions of different mutant strains of E. coli affected at the pyruvate nodea
| Strain (relevant genotype) | Metabolic flux (mmol h−1 [g of cells {dry wt}]−1) for the following reactionb: |
Dry wt of cells (g/liter) | YATP (g of cells [mol of ATP]−1) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| J1 (G-6-P) | J3 (G-3-P) | J4 (PEP) | J5 (PYR) | J6 (SUCC) | J7c (lactate) | J8 (formate) | J9 (PDH) | J10 (acetate) | J11 (ethanol) | J12 (pyruvate) | J13c (lactate) | NADH formation | NADH consumption | |||
| PMD23 (hypF) | 5.85 | 4.87 | 9.74 | 3.58 | 0.30 | 2.82 | 6.60 | 0.02 | 3.58 | 3.04 | 0.00 | 0.00 | 9.76 | 9.50 | 1.84 | 10.9 |
| QZ2 (lpd101 ΔhypF) | 5.00 | 4.70 | 9.40 | 4.10 | 0.30 | 2.93 | 6.00 | 0.16 | 3.37 | 2.79 | 0.01 | 0.00 | 9.56 | 9.11 | 2.18 | 12.9 |
| QZ3 (ΔldhA lpd101 ΔhypF) | 4.45 | 4.29 | 8.44 | 3.79 | 0.20 | 0.00 | 3.81 | 3.91 | 1.93 | 5.78 | 0.52 | 0.15 | 12.34 | 12.12 | 1.93 | 13.6 |
| YK167 [lpd101 Δ(focA-pflB)] | 7.30 | 7.26 | 14.53 | 6.31 | 0.91 | 8.88 | 0.00 | 4.30 | 1.01 | 3.30 | 0.43 | 0.00 | 18.83 | 17.30 | 1.50 | 6.9 |
| SE2382 [lpd101 ΔldhA Δ(focA-pflB)] | 3.62 | 3.38 | 6.20 | 2.20 | 0.38 | 0.00 | 0.00 | 4.23 | 0.92 | 3.31 | 1.59 | 0.56 | 10.44 | 7.94 | 0.69 | 7.4 |
| YK1 [lpd101 ΔldhA Δ(focA-pflB) pdhR] | 4.06 | 3.86 | 7.72 | 3.33 | 0.33 | 0.00 | 0.00 | 7.39 | 1.13 | 6.26 | 0.01 | 0.00 | 15.11 | 13.19 | 0.60 | 4.9 |
All cultures were grown anaerobically in a chemostat with glucose limitation at a dilution rate of 0.1 h−1 as described in Materials and Methods. Reactions J1, J6, J7, J8, J10, J11, J12 and J13 were measured directly, while the other reaction rates were computed.
Products of reactions are given in parentheses. G-6-P, glucose-6-phosphate; G-3-P, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; SUCC, succinate.
The lactate produced by the ldhA deletion mutants (strains QZ3 and SE2382) is assumed to be derived from the methylglyoxal (J13) pathway; in strains with LDH activity, the amount of lactate produced by the methylglyoxal pathway (J13) was considered to be negligible.
TABLE 4.
Anaerobic flux ratios at the pyruvate and acetyl-CoA branch points in select mutants with mutations in PDH, LDH, and/or PFL
| Strain | Enzyme(s)a | Anaerobic flux ratio at the following branch point: |
||||
|---|---|---|---|---|---|---|
| Pyruvate |
Acetyl-CoA |
|||||
| PFL | PDH | LDH | Acetate | Ethanol | ||
| PMD23 | PFL, LDH | 0.70 | 0.002 | 0.30 | 0.54 | 0.46 |
| QZ2 | PFL, LDH, PDH* | 0.66 | 0.01 | 0.33 | 0.55 | 0.45 |
| QZ3 | PFL, PDH* | 0.48 | 0.50 | 0.00 | 0.25 | 0.75 |
| YK167 | LDH, PDH* | 0.00 | 0.33 | 0.67 | 0.23 | 0.77 |
| SE2382 | PDH* | 0.00 | 1.00 | 0.00 | 0.22 | 0.78 |
| YK1 | PDH* | 0.00 | 1.00 | 0.00 | 0.15 | 0.85 |
Enzyme(s) expected to be functional at the pyruvate node in the indicated strain based on its genotype and phenotype and on the enzyme activities in the extract.
The introduction of an active PDH into strain PMD23 through the lpd101 mutation (PDH*; strain QZ2) did not significantly alter the glucose flux through the three enzymes (Tables 3 and 4). This lack of carbon flux through PDH* in a strain with active LDH is in agreement with the results of the batch culture experiments (Fig. 3A) and suggests that LDH is the preferred enzyme in E. coli to support redox balance.
Strain QZ3, lacking LDH activity (PFL+ PDH*).
The deletion of the ldhA gene (strain QZ3) forced about 50% of the pyruvate carbon through PDH* (Tables 3 and 4). This can be seen by the higher ethanol production rate relative to that of acetate. Since the PFL-based reactions yield an acetate-to-ethanol ratio of 1.0, the additional ethanol produced by strain QZ3 is expected to come from the PDH*/ADH pathway (Fig. 1). In strain QZ3, the amount of carbon diverted to PDH* was slightly higher than the amount of carbon flux through LDH to lactate in strain QZ2 (3.9 versus 2.9 mmol h−1 [g of cells {dry weight}]−1, respectively). Surprisingly, the pyruvate flux through PFL was about 35% lower in strain QZ3 than in strain QZ2, although the steady-state cell masses of the two cultures in the chemostat were comparable (about 2.0 g of cells [dry weight] liter−1). The reason for this decline in pyruvate flux through PFL in a strain lacking LDH is not clear, although the lower apparent pyruvate Km for PDH* (0.4 mM versus 2.0 mM for PFL) may be a contributing factor.
Strain QZ3 also produced pyruvate as a fermentation product, as seen with batch fermentations (Fig. 3C), suggesting that the conversion of pyruvate to acetyl-CoA by PFL and PDH* is rate-limiting in this strain. Even with this potential limitation, the steady-state cell masses of the cultures (about 2 g/liter) and calculated yields per mole of ATP (YATP) for strains QZ2 and QZ3 (12.9 and 13.6 g of cells, respectively) were comparable (Table 2) and similar to reported YATP for E. coli (16, 22, 24, 42). These results show that PDH* effectively competes with PFL for pyruvate, even though PDH* activity requires NAD+. This is in contrast to the inability of PDH* to compete for pyruvate when all three enzymes are present (strain QZ2) and confirms that the LDH pathway outcompetes PDH* for pyruvate, in support of maintaining the redox balance in E. coli strain W3110.
Strain YK167, lacking PFL activity (LDH+ PDH*).
A mutant that lacks PFL activity (YK167) while still retaining LDH and PDH* activities had an almost 50% increase in glucose consumption and flux over those for strains with active PFL (QZ2 and QZ3) (Table 3). Apparently, the higher glucose flux to lactate is geared to ATP production at the glycolysis step, compensating for the absence of postpyruvate ATP production. This is comparable to a similar increase in glucose flux reported for a pfl or ackA pta mutant with the native PDH over that of its corresponding parent strain (39, 42). Although PDH* was unable to compete with LDH for pyruvate in strain QZ2, in strain YK167 flux through PDH* was about 33% of the total pyruvate (Tables 3 and 4). This flux through PDH* in strain YK167 was comparable to the observed PDH* flux in strain QZ3, which lacks LDH activity (about 4 mmol h−1 [g of cells {dry weight}]−1). In the absence of PFL, PDH* is the only other enzyme that can generate acetyl-CoA, which can be converted to acetate with associated ATP production. However, the NADH generated during pyruvate oxidation by PDH* is expected to limit the amount of ATP that can be produced by the PDH*, phosphotransacetylase (PTA), and acetate kinase (ACK) pathway due to the need for redox balance. The rate of PDH*-dependent acetate production may be dictated by the rate at which the growing culture can utilize NADH. Although the growth rate of strain YK167 was comparable to those of the wild-type strains in batch cultures, the YATP for this strain in the chemostat was significantly lower, 6.9 g of cells per mol of ATP, than the values for strains with active PFL, which were higher than 10.0 g per mol of ATP (Table 3). This is unexpected and is different from the YATP of 15.2 g per mol of ATP reported for an ackA pta mutant of a strain MC4100 derivative (42). This difference in YATP between the pfl mutant, strain YK167, and the ackA pta mutant of Yang et al. (42) could result from the differences between the genotypes of the two strains of E. coli K-12: strain W3110, used in this study (ATCC 27325), is believed to be nearly wild type (ATCC), while strain MC4100 has multiple alterations and deletions (25), with unknown physiological consequences. Strain MC4100 is also reported to have a lower level of the anaerobic control protein FNR than another wild-type strain, MG1655 (31). The potential effect of the lower level of FNR in modulating the YATP of an anaerobic culture is not known and needs investigation.
The YATP calculations were based on the predicted ATP yield by the fermentation products from the chemostat cultures and do not take into account any loss of ATP through a futile cycle (21, 26, 30). It is possible that in the absence of PFL, a futile cycle is draining ATP from glycolytic reactions in strain YK167. One possible mechanism of ATP loss could be an alternate pathway for phosphoenolpyruvate conversion to pyruvate through oxaloacetate and malate catalyzed by PEP-carboxylase, malate dehydrogenase, and NAD+-dependent malate dehydrogenase (decarboxylating). Due to the absence of PFL and the accumulation of pyruvate, the PEP-carboxylase may initiate this alternate pathway, with the loss of ATP. This pathway is redox neutral and is not expected to alter the redox balance. It is also possible that in the absence of PFL, the transport and assimilation of amino acids and other components from the medium may be limiting growth, leading to a condition of ATP excess and thus initiating a potential futile energy cycle(s).
Strain SE2382, lacking both LDH and PFL activities (PDH*).
Glucose flux in strain SE2382, which lacks both PFL and LDH activities, was slightly lower than that in strain QZ3 (Δldh) but about 50% lower than that in strain YK167 (ldh+ ΔpflB) (Table 2). Levels of pyruvate flux through the PDH* in strains YK167 and SE2382 were similar, suggesting that the loss of LDH activity in strain SE2382 due to mutation is the primary reason for the decrease in glucose flux. As expected, the lower rate of glycolysis in strain SE2382 led to a lower cell mass in steady-state cultures. The calculated YATP for strains SE2382 and YK167 were comparable and were about 50% of the value for the PFL-positive strain QZ3. These results show that the absence of PFL is responsible for the lower cell yield of the pfl mutants in steady-state cultures, apparently due to a lower ATP yield, irrespective of the presence or absence of LDH activity.
Strain SE2382 also produced pyruvate as a product, suggesting that the rate-limiting step in glucose fermentation is PDH* activity. In addition, strain SE2382 also produced lactate, probably through the methylglyoxal pathway, indicating an accumulation of glyceraldehyde-3-phosphate and associated dihydroxyacetone-3-phosphate, the substrate for methylglyoxal synthase, which is the starting point for LDH-independent lactate production. No pyruvate or lactate was detected in the broth of strain YK1, which carries additional mutations in PdhR and the intergenic region between pdhR and aceE (Table 2) (18). These additional mutations increased the expression of the pdh operon by about 4-fold over that for strain SE2382 (aceE mRNA levels, 2.2 versus 9.4 pg μl−1 of total RNA for strains SE2382 and YK1, respectively) and increased flux through PDH* by about 1.75-fold. Besides the absence of lactate and pyruvate in the broth and the higher rate of ethanol production by strain YK1, there was no other significant difference between strains SE2382 and YK1 in the chemostat cultures (Tables 3 and 4). The calculated YATP for strain YK1 (4.9 g of cells per mol of ATP) was the lowest of those for all the strains investigated.
It is known that flux through PFL would lead to ATP production, which is essential for increasing the growth rate of an anaerobic culture (13, 19, 39). Under the redox balance condition, the reactions shown in Fig. 2 can be used to maximize ATP production with linear optimization. The flux distributions in the mutant strain QZ2 were calculated to be almost the same as the calculated ATP-maximized fluxes. There was no flux to PDH. When the flux to PDH was increased, the ATP generated from glucose actually decreased. The genome-level flux balance model also gave the same results (28) when the optimized objective was set as the maximum biomass generation. These results suggest that the cell prefers the PFL pathway over the PDH* pathway, probably due to the ATP yield.
Conclusion.
Flux through LDH or PDH* alone serves to maintain redox balance in the wild type, and the low flux to PDH* (about 2%) in the “wild-type” E. coli strain that favors LDH is unexpected, considering the apparent pyruvate Km values for the three competing enzymes. Although PDH requires NAD+ for activity, the apparent NAD+ Km for this enzyme is 70 μM, compared to an apparent NADH Km of 200 μm for LDH (18, 37). The reported intracellular NAD+ concentration in E. coli grown anaerobically in a glucose-limited chemostat ranges from 1 to 2.5 mM depending on the strain (8), a concentration that is significantly higher than the apparent NAD+ Km for PDH* (18). Considering that the highest reported [NADH]/[NAD+] ratio of an anaerobic E. coli strain is less than 1.0 (8), kinetically, PDH* would be the preferred enzyme at the pyruvate node. However, in the presence of LDH and PFL, flux through PDH* is negligible (strain QZ2 [Table 3]), indicating that some other factor(s) besides enzyme kinetics plays a role in distributing pyruvate among the three enzymes. In strain QZ2, the level of PDH in cell extracts was about 0.14 U mg protein−1, a reduction in activity of about 70% from those of strain QZ3 and “wild-type” strain PMD23 (about 0.4 U mg protein−1). Although strain QZ2 had a lower PDH activity than strain PMD23, the ratios of LDH to PDH* activities in the extracts of these two strains were about 3.0 and may not account for the significant difference in pyruvate flux through PDH*. However, it should be noted that besides the kinetic constants, the intracellular enzyme concentration, the levels of various regulatory metabolites, and substrate and product concentrations, etc., do play significant roles in the flux through a specific enzyme, and a combination of these factors may divert pyruvate carbon to LDH. It is noteworthy that the high glucose flux in strain YK167, lacking PFL activity, was supported by elevating the LDH activity and not by increasing flux through PDH* (Tables 2 and 3), although PDH* can support redox balance. These results suggest a complex physiological interaction among the three pathways at the pyruvate node that includes control at the genetic level.
PDH is highly regulated by various metabolic intermediates, including pyruvate (7, 12, 33, 34), and accumulation of some of the effectors in the cell may have a negative effect on the activity of PDH*. Although this is possible, flux through PDH* in mutant strains lacking either PFL or LDH, or both, argues against complete inhibition of PDH activity by metabolic inhibitors. Apparently, there are other, unknown intricate physiological factor(s) that regulate flux at the pyruvate node toward lactate production for redox balance. One possibility is that ADH is rate-limiting and the accumulation of acetyl-CoA is inhibitory to PDH* activity (33). In strains PMD23, QZ2, YK167, and SE2382, the overall flux through ADH (step J11) (Table 3 and Fig. 2) is an almost constant value of about 3 mmol·h−1·(g of cells [dry weight])−1. Although flux through reaction J11 (Fig. 2) increased to 5.8 mmol·h−1·(g of cells [dry weight])−1 in strain QZ3 (Δldh lpd101), the level of ADH activity in the extract of this strain was only slightly higher (0.05 U mg protein−1) than those for strains PMD23 and QZ2 (0.04 U mg protein−1). These physiological and biochemical results suggest that ADH activity may not be limiting flux through PDH* in strain QZ2. This is further confirmed by the absence of increased flux through PDH* even after an increase in the expression of the adhE gene, from a plasmid, in strain QZ2 (data not presented).
Pyruvate is reported to function as an allosteric activator of LDH, and this may favor pyruvate flux through LDH (37). Pyruvate is also a known activator of pdh operon expression (27), and thus, pyruvate may not provide a unique advantage for flux through one enzyme complex or the other.
The results presented above suggest that the mechanism that splits pyruvate among the three available pathways (PFL, LDH, and PDH*) is apparently determined by energy requirement and redox balance constraints. Under anaerobic conditions, the cell prefers PFL due to the energy supply and LDH for redox balance maintenance, even in the presence of an active PDH. The lack of flux through PDH* in a strain with all three enzymes could result from a reluctance of the cell to elevate the level of NADH and further increase the [NADH]/[NAD+] ratio, although this NADH can be reoxidized in subsequent ADH-dependent reactions. An understanding of the controls at the pyruvate node, as well as the potential function of a futile cycle(s) in the absence of PFL, can provide unique insights toward rational metabolic engineering of E. coli and related bacteria for the production of various biobased products, including ethanol, at high rates and yields.
Acknowledgments
We thank Phi M. Do for providing strain PMD23 with the ΔhypF mutation.
This study was supported in part by a grant from the Department of Energy (DE-FG36-04GO14019) and the State of Florida, University of Florida Agricultural Experiment Station.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Bisswanger, H., and U. Henning. 1971. Regulatory properties of the pyruvate-dehydrogenase complex from Escherichia coli. Positive and negative cooperativity. Eur. J. Biochem. 24:376-384. [DOI] [PubMed] [Google Scholar]
- 2.Bock, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 5:223-234. [DOI] [PubMed] [Google Scholar]
- 5.Cronan, J. E., Jr., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxalate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta, A. 1991. Characterization of the inhibition of Escherichia coli pyruvate dehydrogenase complex by pyruvate. Biochem. Biophys. Res. Commun. 176:517-521. [DOI] [PubMed] [Google Scholar]
- 8.de Graef, M. R., S. Alexeeva, J. L. Snoep, and M. J. Teixeira de Mattos. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J. Bacteriol. 181:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraenkel, D. G. 1996. Glycolysis, p. 189-198. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 10.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 217-261. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 12.Hansen, H. G., and U. Henning. 1966. Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. Biochim. Biophys. Acta 122:355-358. [DOI] [PubMed] [Google Scholar]
- 13.Hasona, A., Y. Kim, F. G. Healy, L. O. Ingram, and K. T. Shanmugam. 2004. Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J. Bacteriol. 186:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iuchi, S., and E. C. Lin. 1993. Adaptation of Escherichia coli to redox environments by gene expression. Mol. Microbiol. 9:9-15. [DOI] [PubMed] [Google Scholar]
- 15.Kajiya, M., K. Sato, M. J. Silva, K. Ouhara, P. M. Do, K. T. Shanmugam, and T. Kawai. 2009. Hydrogen from intestinal bacteria is protective for concanavalin A-induced hepatitis. Biochem. Biophys. Res. Commun. 386:316-321. [DOI] [PubMed] [Google Scholar]
- 16.Kayser, A., J. Weber, V. Hecht, and U. Rinas. 2005. Metabolic flux analysis of Escherichia coli in glucose-limited continuous culture. I. Growth-rate-dependent metabolic efficiency at steady state. Microbiology 151:693-706. [DOI] [PubMed] [Google Scholar]
- 17.Kim, Y., L. O. Ingram, and K. T. Shanmugam. 2007. Construction of an Escherichia coli K-12 mutant for homoethanologenic fermentation of glucose or xylose without foreign genes. Appl. Environ. Microbiol. 73:1766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, Y., L. O. Ingram, and K. T. Shanmugam. 2008. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol. 190:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koebmann, B. J., H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawford, H. G., and J. D. Rousseau. 1995. Comparative energetics of glucose and xylose metabolism in ethanologenic recombinant Escherichia coli B. Appl. Biochem. Biotechnol. 51-52:179-195. [DOI] [PubMed] [Google Scholar]
- 21.Liao, J. C., Y. P. Chao, and R. Patnaik. 1994. Alteration of the biochemical valves in the central metabolism of Escherichia coli. Ann. N. Y. Acad. Sci. 745:21-34. [DOI] [PubMed] [Google Scholar]
- 22.Mainzer, S. E., and W. P. Hempfling. 1976. Effects of growth temperature on yield and maintenance during glucose-limited continuous culture of Escherichia coli. J. Bacteriol. 126:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melchiorsen, C. R., K. V. Jokumsen, J. Villadsen, H. Israelsen, and J. Arnau. 2002. The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis. Appl. Microbiol. Biotechnol. 58:338-344. [DOI] [PubMed] [Google Scholar]
- 24.Paalme, T., R. Elken, A. Kahru, K. Vanatalu, and R. Vilu. 1997. The growth rate control in Escherichia coli at near to maximum growth rates: the A-stat approach. Antonie Van Leeuwenhoek 71:217-230. [DOI] [PubMed] [Google Scholar]
- 25.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian, H., and D. A. Beard. 2006. Metabolic futile cycles and their functions: a systems analysis of energy and control. Syst. Biol. 153:192-200. [DOI] [PubMed] [Google Scholar]
- 27.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 28.Reed, J. L., T. D. Vo, C. H. Schilling, and B. O. Palsson. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosentel, J. K., F. Healy, J. A. Maupin-Furlow, J. H. Lee, and K. T. Shanmugam. 1995. Molybdate and regulation of mod (molybdate transport), fdhF, and hyc (formate hydrogenlyase) operons in Escherichia coli. J. Bacteriol. 177:4857-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell, J. B. 2007. The energy spilling reactions of bacteria and other organisms. J. Mol. Microbiol. Biotechnol. 13:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Sawers, R. G. 2005. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J. Bacteriol. 187:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmincke-Ott, E., and H. Bisswanger. 1981. Dihydrolipoamide dehydrogenase component of the pyruvate dehydrogenase complex from Escherichia coli K12. Comparative characterization of the free and the complex-bound component. Eur. J. Biochem. 114:413-420. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, E. R., and L. J. Reed. 1970. Regulation of the activity of the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 9:1434-1439. [DOI] [PubMed] [Google Scholar]
- 34.Shen, L. C., and D. E. Atkinson. 1970. Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J. Biol. Chem. 245:5974-5978. [PubMed] [Google Scholar]
- 35.Snoep, J. L., M. R. de Graef, A. H. Westphal, A. de Kok, M. J. Teixeira de Mattos, and O. M. Neijssel. 1993. Differences in sensitivity to NADH of purified pyruvate dehydrogenase complexes of Enterococcus faecalis, Lactococcus lactis, Azotobacter vinelandii and Escherichia coli: implications for their activity in vivo. FEMS Microbiol. Lett. 114:279-283. [DOI] [PubMed] [Google Scholar]
- 36.Stephanopoulos, G. N., A. A. Aristidou, and J. Nielsen. 1998. Metabolic engineering: principles and methodologies. Academic Press, Burlington, MA.
- 37.Tarmy, E. M., and N. O. Kaplan. 1968. Kinetics of Escherichia coli B d-lactate dehydrogenase and evidence for pyruvate-controlled change in conformation. J. Biol. Chem. 243:2587-2596. [PubMed] [Google Scholar]
- 38.Underwood, S. A., S. Zhou, T. B. Causey, L. P. Yomano, K. T. Shanmugam, and L. O. Ingram. 2002. Genetic changes to optimize carbon partitioning between ethanol and biosynthesis in ethanologenic Escherichia coli. Appl. Environ. Microbiol. 68:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Utrilla, J., G. Gosset, and A. Martinez. 2009. ATP limitation in a pyruvate formate lyase mutant of Escherichia coli MG1655 increases glycolytic flux to d-lactate. J. Ind. Microbiol. Biotechnol. 36:1057-1062. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Q., X. Chen, Y. Yang, and X. Zhao. 2006. Genome-scale in silico aided metabolic analysis and flux comparisons of Escherichia coli to improve succinate production. Appl. Microbiol. Biotechnol. 73:887-894. [DOI] [PubMed] [Google Scholar]
- 41.Yang, Y. T., G. N. Bennett, and K. Y. San. 2001. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab. Eng. 3:115-123. [DOI] [PubMed] [Google Scholar]
- 42.Yang, Y. T., K. Y. San, and G. N. Bennett. 1999. Redistribution of metabolic fluxes in Escherichia coli with fermentative lactate dehydrogenase overexpression and deletion. Metab. Eng. 1:141-152. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, S., K. T. Shanmugam, and L. O. Ingram. 2003. Functional replacement of the Escherichia coli d-(−)-lactate dehydrogenase gene (ldhA) with the l-(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl. Environ. Microbiol. 69:2237-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]