Abstract
In New Zealand the number of campylobacteriosis notifications increased markedly between 2000 and 2007. Notably, this country's poultry supply is different than that of many developed countries as the fresh and frozen poultry available at retail are exclusively of domestic origin. To examine the possible link between human cases and poultry, a sentinel surveillance site was established to study the molecular epidemiology of Campylobacter jejuni over a 3-year period from 2005 to 2008 using multilocus sequence typing. Studies showed that 60.1 to 81.4% of retail poultry carcasses from the major suppliers were contaminated with C. jejuni. Differences were detected in the probability and level of contamination and the relative frequency of genotypes for individual poultry suppliers and humans. Some carcasses were contaminated with isolates belonging to more than one sequence type (ST), and there was evidence of both ubiquitous and supplier-associated strains, an epidemiological pattern not recognized yet in other countries. The common poultry STs were also common in human clinical cases, providing evidence that poultry is a major contributor to human infection. Both internationally rare genotypes, such as ST-3069 and ST-474, and common genotypes, such as ST-45 and ST-48, were identified in this study. The dominant human sequence type in New Zealand, ST-474, was found almost exclusively in isolates from one poultry supplier, which provided evidence that C. jejuni has a distinctive molecular epidemiology in this country. These results may be due in part to New Zealand's geographical isolation and its uniquely structured poultry industry.
Campylobacteriosis is a leading enteric zoonosis in the developed world, and the majority of cases are caused by Campylobacter jejuni (22). Poultry sources are suspected to be the major source of human infection with C. jejuni (22, 43, 49), and this conclusion is supported by high levels of contamination of poultry and the detection of identical C. jejuni genotypes in human cases and poultry samples (25). However, C. jejuni can be isolated from a variety of sources, including ruminants (17) and environmental water (41). Due to the complex epidemiology of this pathogen, there is still uncertainty about the relative contributions of individual pathways to the human disease burden (8). In New Zealand the number of notified campylobacteriosis cases increased markedly in the last decade, peaking in 2006 at a total of 15,873 notified cases (422.4 cases per 100,000 population) (1) and costing the economy an estimated US$32 million annually (42). Although there are a number of factors that may have contributed to this increase, it has been noted that the rise in campylobacteriosis cases coincided with a marked increase in the sale and consumption of fresh poultry between 1992 and 2005, while the sale of frozen poultry remained relatively static (1).
New Zealand has one of the highest enteric infectious disease rates in industrialized countries (26), and the high ratio of domestic production animals to humans and the frequent use of rural water supplies in New Zealand have been postulated to be underlying causes (9). In addition, this country's poultry industry is uniquely structured; it is almost entirely focused on the domestic market, and no raw, fresh or frozen, poultry products are imported because of biosecurity threats. Due to its geographical isolation and tight border controls, New Zealand has remained free of poultry diseases endemic in other countries, such as diseases caused by Salmonella enterica serovar Enteritidis PT4 and S. enterica serovar Typhimurium DT 104, Newcastle disease, and infectious bursal disease.
The production of poultry meat in New Zealand is highly integrated; only three companies supply 90% of the chicken meat, which represents 95% of the poultry meat consumed. The remaining 5% of poultry meat consumed includes species such as turkey and duck. The chicken processors own or control most stages of production, processing, and distribution. One of the three dominant companies has one processing plant that distributes nationwide, and one company has multiple plants that tend to be more localized in their distribution, except when they make specialty products, which are distributed nationally. The other companies distribute primarily locally. The broilers are commonly barn raised, not free range, and animal welfare standards require a maximum stocking density of 38 kg (live weight) of broiler chickens per m2. There are approximately 160 broiler farms in a number of specific areas of New Zealand. These farms are usually located near the slaughterhouses that they supply.
To enable regulators to implement food safety programs to reduce human campylobacteriosis, there has been great interest in understanding the importance and epidemiology of C. jejuni in the New Zealand poultry production system. A sentinel surveillance site was therefore established in the Manawatu region to quantify the contributions of different sources, including poultry suppliers, to the human disease burden and to study the molecular epidemiology of C. jejuni (33, 34). Isolates were typed using multilocus sequence typing (MLST), a method that has major advantages over other methods of genotyping when the long-term epidemiology of a disease is studied. However, other methods may be more appropriate in other settings, such as outbreak investigations (21), where a higher degree of discrimination may be required. MLST offers a large web-based archive of isolates from many sources and countries: the Campylobacter PubMLST database (10). Sequence typing by MLST is now internationally recognized as a valuable approach for national and international epidemiological characterization and source tracking of major pathogenic microorganisms, such as C. jejuni (46, 48).
The use of integrated surveillance across human, domestic animal, and wildlife populations has been identified as a key component of strategies aimed at prevention and control of emerging pathogens, particularly when the population dynamics of multihost pathogens are poorly understood (52). At the Manawatu surveillance site, samples from human clinical cases, animal-derived food products, and the environment were gathered in a defined geographical area of New Zealand over a 3-year period (19, 34) and genotyped using MLST (11). The resulting data set contained a total of 969 typed samples, 502 of which were from human cases. The temporal and spatial scale of the data allowed us to obtain a more in-depth understanding of local transmission dynamics compared with the results of previous research (43, 50). The application of novel risk attribution approaches to these data previously identified poultry as the major contributor to the human disease burden, with widely varying contributions from different suppliers (33).
In this study we extended the findings of risk attribution and epidemiological studies of human cases (33, 34). Data from the sentinel surveillance site were used to study the epidemiology of C. jejuni for the individual producers that comprise the poultry sector in New Zealand and to better understand the contributions of the producers to the human campylobacteriosis burden. Our study included an investigation of both the probability of contamination and the level of contamination of poultry carcasses and a study of human and poultry MLST sequence types (STs). The resulting data were compared to make inferences about the epidemiology of C. jejuni in the New Zealand poultry industry and to identify determinants for the high number of human cases attributed to this food source.
MATERIALS AND METHODS
Campylobacter isolates.
The isolation and subtyping of human isolates by MLST and the epidemiology of human cases in the study region were described in detail previously (33, 34). Human specimens submitted to MedLab Central, Palmerston North, New Zealand, that were positive for Campylobacter as determined by an enzyme-linked immunosorbent assay (ELISA) (ProSpecT R; Remel, United States) were sent to the Hopkirk Molecular Epidemiology Laboratory over a 3-year period from 1 March 2005 to 29 February 2008, and they included a total of 774 samples that fulfilled the inclusion criteria. Fecal swabs were obtained using Amies charcoal transport swabs (Copan, Italy). Over the same period 12 to 18 fresh whole poultry carcasses, representing the different poultry suppliers in the region, were sampled each month from retail outlets in Palmerston North, with the number of samples collected per supplier reflecting the market share. A total of 500 poultry samples were included in this study (Table 1). The Manawatu region is supplied predominantly by three companies, which for confidentiality reasons are referred to as supplier A, supplier B, and supplier C in this paper (supplier identification letters were assigned arbitrarily).
TABLE 1.
Proportions of poultry samples positive for Campylobacter spp. in the Manawatu region from March 2005 to February 2008
| Supplier | No. of samples | No. of culture-positive samples | Prevalence of presumptive Campylobacter-positive samples (%) | No. of samples with confirmed C. jejuni | Estimated prevalence of C. jejuni (%)a |
|---|---|---|---|---|---|
| A | 239 | 203 | 84.9 | 189 | 80.7 |
| B | 196 | 136 | 69.4 | 117 | 60.1 |
| C | 65 | 55 | 84.6 | 50 | 81.4 |
| All | 500 | 394 | 78.8 | 356 | 72.7 |
Estimates were based on the proportions of samples in which C. jejuni was identified.
Bacterial culture and identification.
Human fecal swabs were cultured on modified cefoperazone-charcoal-deoxycholate agar (mCCDA) plates (Fort Richard, Auckland, New Zealand) and in Bolton broth (Lab M, Bury, England) and incubated at 42°C in a microaerobic atmosphere (85% N2, 10% CO2, 5% O2) for 2 days. For each swab a single colony resembling Campylobacter species was subcultured on blood agar (BA) (Fort Richard, Auckland, New Zealand) and incubated microaerobically at 42°C for 2 days before DNA preparations were made. In addition, in a small exploratory study multiple colonies (two to five colonies per sample) from individual samples were subcultured. Cultures were frozen at −80°C in glycerol broth (Difco, United States).
Whole chicken carcasses were washed and manually massaged in 200 ml of buffered peptone water (BPW) (Difco, United States). Each chicken wash was centrifuged (16,264 × g, 6°C, 35 min, Sorvall RC5B), and the resulting pellet was resuspended in 5 ml of BPW. Approximately 3 ml of the resuspended pellet was added to 90 ml of Bolton broth, which was incubated at 42°C microaerobically for 2 days. After incubation, the broth was subcultured on mCCDA and incubated microaerobically at 42°C for 2 days. At least two single colonies resembling Campylobacter species were subcultured on BA and incubated microaerobically at 42°C for 2 days before DNA preparations were made. If colonies with different morphologies were present, colonies representing each morphology were selected; otherwise, colonies were selected at random. Cultures were frozen at −80°C.
Isolates belonging to the genus Campylobacter were identified by targeting the 16S rRNA by the method outlined by Linton et al. (27). The species C. jejuni was identified by using the mapA gene, which has been found only in C. jejuni (45). Primers MapA-F (5′-CTTGGCTTGAAATTTGCTTG-3′) and MapA-R (5′-GCTTGGTGCGGATTGTAAA-3′) were designed for this study using the online resource Primer3 (http://frodo.wi.mit.edu/primer3/). No other matches for these primers were found when the GenBank database was searched. These new primers were designed to overcome problems associated with mispriming of the original primers described by Stucki et al. (45), which contained single-base segments that were four or five nucleotides long in both primers. The new primers were validated and shown to be both sensitive and specific by sequencing amplicons and by testing them with a panel of Campylobacter spp. comprising C. jejuni, C. coli, C. upsaliensis, C. lari, C. fetus subsp. fetus, and C. hyointestinalis. The amplification protocols used for pan-Campylobacter and the mapA genes were based on the methods outlined by Linton et al. (27) and Stucki et al. (45), respectively, but they were modified slightly for optimal use in different thermocycler machines and a reverse transcription (RT)-PCR. The PCR products were visualized by electrophoresis in a 1% agarose gel in Tris-borate-EDTA (TBE) buffer, which was then stained with ethidium bromide and exposed to UV light. The presence of a 603-bp product indicated that C. jejuni was present. Species identity was also confirmed by MLST.
Sequence typing.
After species identification, MLST of C. jejuni isolates was performed using seven housekeeping genes, aspA (aspartase A), glnA (glutamine synthase), gltA (citrate synthase), glyA (serine hydroxymethyltransferase), pgm (phosphoglucomutase), tkt (transketolase), and uncA (ATP synthase alpha subunit), based on the method outlined by Dingle et al.(12). Chromosomal DNA was prepared from freshly grown cultures by boiling the cultures for 10 min and then centrifuging the disrupted cells. The supernatant was decanted to a fresh tube and used for amplification. Amplification was performed in a 25-μl reaction mixture using Applied Biosystems AmpliTaq Gold master mixture (Applied Biosystems, Auckland, New Zealand) and 5 pmol of each primer. Products were sequenced with an ABI 3130XL automated DNA sequencer using ABI BigDye v3.1 (Applied Biosystems) and following the manufacturer's instructions. Sequence data were collated, and alleles were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/). Novel alleles and sequence types were submitted for allele and ST designation. Alleles for which there were not clear results were reamplified and sequenced using primers sets described by Miller et al. (31) and the protocol described above.
Data generated by this sampling strategy have been used in studies that examined the epidemiology of human clinical cases arising from multiple animal reservoirs (33, 34) in addition to the study described in this paper.
Enumeration of Campylobacter on poultry carcasses.
Beginning in October 2006, Campylobacter isolates on all chicken carcasses were enumerated using a Wasp spiral plater (Don Whitley, England) and a manual spread plate. Duplicate mCCDA plates were inoculated with 50-μl (spiral plater) or 1-ml (spread plate) aliquots of chicken washes and 100-μl (spiral plater) aliquots of resuspended chicken wash pellets. The plates were incubated microaerobically at 42°C for 2 days. Colonies were counted manually or by using a plate reader (aCOLyte; Synbiosis, England).
Statistical analysis of Campylobacter count data.
The aim of this study was to estimate both the proportion of carcasses that were positive and the levels of Campylobacter present on positive carcasses. The method used for analysis employed a novel application of recently developed statistical tools for analysis of count data where there is a large proportion of zeros and there are several replicates at the sample level. Bacterial count data have two components: (i) whether bacteria are present or absent and (ii) how many bacteria are present. In order to capture these two components in our model, we used a Bayesian zero-inflated Poisson model (37). This model includes two linear predictors, one informing the probability that the bacteria are present and the other informing the number of bacteria that are obtained assuming that they are present.
For the zero-inflation part of the model we assumed that sample s was positive for Campylobacter with probability ps. Since ps is a probability, the logit link function was used when the linear predictor was formed. We wished to model the change in the probability of contamination quarter by quarter for each company, and so we included a random effect for each quarter-company pair, αc(s), q(s). Thus, logit(ps) = α0 + αc(s), q(s), where c(s) is the company that produced sample s and q(s) is the quarter in which sample s was produced. This is a Bayesian model, and so priors must be chosen. We assumed the following weakly informative priors: α0 ∼ N(0,10), αc,q ∼ N(0,κα−1), and κα ∼ Gamma(2,2).
The second part of the model describes the amount of Campylobacter, assuming that it is present. We assumed that the number of Campylobacter bacteria in each replicate i for a contaminated sample had a Poisson distribution, with the volume of rinse that was plated as an offset; i.e., Yi ∼ Pois(Viμi), where Yi is the number counted for replicate i, Vi is the volume of rinse that is plated, and μi is the concentration of Campylobacter in replicate i. To make a linear predictor for μi, we used the log link function and included random effects for each company-quarter pair as described above [βc(s),q(s)]. To attempt to separate the laboratory variation from the sample variation, we also included random effects at the replicate level (δi) and the sample level [γs(i)]. Thus, log(μi) = β0 + βc(i), q(i) + γs(i) + δi. Again we assumed weakly informative priors: β0 ∼ N (0,10), βc,q ∼ N (0,κβ−1), γs ∼ N (0,κγ−1), δi ∼ N (0,κδ−1), κβ ∼ Gamma (2,2), κγ ∼ Gamma (2,2), and κδ ∼ Gamma (2,2).
The output from these models is presented below as a series of graphs describing the probability that a carcass contained Campylobacter by supplier and by quarter and the estimated number of viable Campylobacter CFU on positive carcasses, again by supplier and by quarter. This method ensured that all of the individual replicate counts for each sample were analyzed appropriately.
Statistical analysis of ST distributions.
The proportional similarity index (PSI) or Czekanowski index is an objective and simple measure of the area of intersection between two frequency distributions (38). It estimates the similarity between the frequency distributions of, for instance, bacterial subtypes from different sources. The PSI is calculated as follows: PSI = 1 − 0.5∑i|pi − qi| = ∑i min(pi,qi), where pi and qi are the proportions of strains belonging to type i of all strains typed from sources P and Q, respectively (16, 38). The value for the PSI ranges from 1 for identical frequency distributions to 0 for distributions with no common types. Bootstrap confidence intervals for this measure were estimated based on the approach developed by Garrett et al. (20).
Structure 2.2 (15, 36) was used as a frequency-based clustering model to investigate the population structure of C. jejuni in the New Zealand poultry supply. This Bayesian MCMC method attempts to assign individuals to clusters on the basis of their genotypes while simultaneously estimating allele frequencies for each cluster. This approach assumes a model with K populations, each of which is characterized by a set of allele frequencies at each locus, and assumes that loci are unlinked and at equilibrium with one another within populations (36). Using the approach taken by Sheppard et al. (43), we further assumed the NO ADMIXTURE model and that the allele frequencies were uncorrelated. We are interested in exploring the structure of the distribution of STs for the three poultry companies. Here we considered only a case in which there are three clusters (K = 3), and we used the source of the isolates as prior information for the model. Larger values of K were investigated, but these values resulted in little change in the mean log-likelihood and the program was run without population information to ensure that the predetermined populations were in broad agreement with the genetic information. We set the parameter GENSBACK to 1; this parameter represents the number of generations back that Structure considers that a sample may have had an ancestor belonging to a different cluster. The model was run with a burn-in period of 100,000 iterations, and thereafter sampling of the chain occurred over a further 100,000 iterations. The posterior probabilities of the final model were visualized as confluent, stacked bar plots using the Distruct program. Subpopulations were represented by different shades, and individuals were indicated by bars partitioned into shaded segments that corresponded to the posterior probabilities of assignment (39).
RESULTS
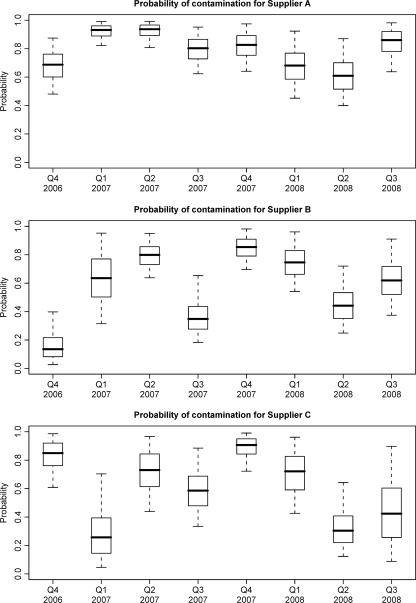
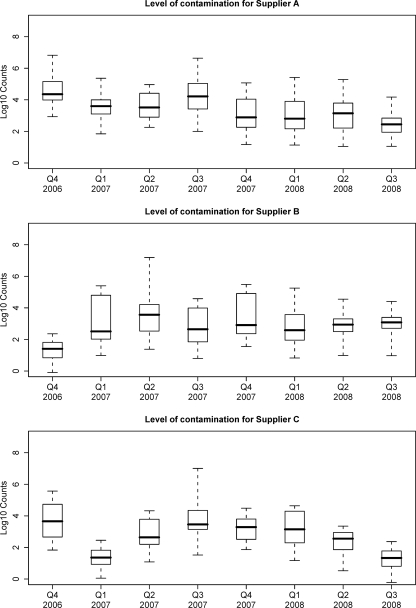
Retail poultry samples had a high prevalence of contamination with Campylobacter spp. throughout the 3-year period. Overall, 78.8% of the carcasses were positive for Campylobacter, 72.7% of the carcasses were positive for C. jejuni, and the estimates of the prevalence of C. jejuni for the individual suppliers ranged from 60.1 to 81.4% (Table 1). Figures 1 and 2 show the estimated proportions of positive carcasses and the counts conditional on being positive for suppliers A, B, and C and for each quarter from October 2006 to February 2008, respectively. There was a moderately high probability of contamination for all suppliers throughout the study period; over 80% of the carcasses from supplier A were positive throughout 2007, and over 60% of the carcasses were positive for most suppliers in most quarters (Fig. 1). The average number of Campylobacter CFU on positive carcasses was initially estimated to be high (over 104 CFU/carcass) for supplier A but declined steadily over the study period. The levels on positive carcasses for supplier B remained relatively constant but lower throughout the study period and the levels for supplier C declined beginning in mid-2007 from a medium level of contamination to a low level of contamination (Fig. 2).
FIG. 1.
Probabilities of Campylobacter contamination on poultry carcasses for each supplier for different quarters.
FIG. 2.
Levels of Campylobacter contamination on positive poultry carcasses for each supplier for different quarters.
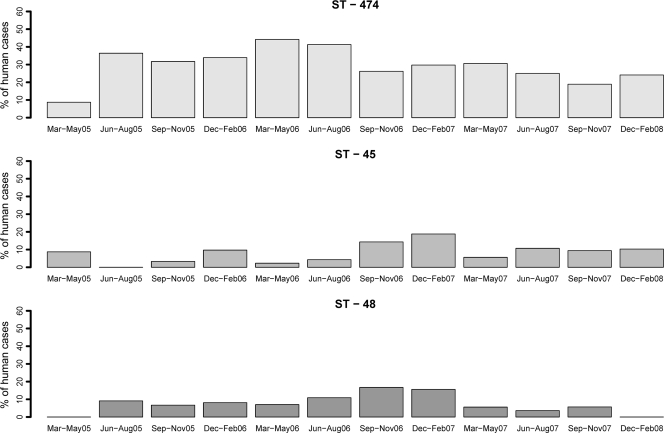
A total of 62 different STs were detected in human and poultry samples. The four major human STs were ST-474, ST-48, ST-45, and ST-53, which accounted for 30.7%, 8.4%, 8.2%, and 5.4% of the human cases (Table 2). The three most frequently isolated human STs, ST-474, ST-45, and ST-48, were prevalent in human cases throughout the study period (Fig. 3) and were also consistently detected in poultry samples over the same time period. The dominant human ST, ST-474, was very prevalent in samples from supplier A but was rarely detected in samples from other poultry suppliers. Other STs, such as ST-45 and ST-48, occurred frequently in human samples and poultry samples from all three suppliers. Of the 62 STs detected, 51 were found in human samples, 18 were found in samples from supplier A, 17 were found in samples from supplier B, and 11 were found in samples from supplier C. Twenty-nine human STs, representing 14.5% of all human cases, were not found in any of the poultry samples. The samples from supplier A contained 7 unique STs, the samples from supplier B contained 10 unique STs, and the samples from supplier C contained 5 unique STs (Table 2). ST-3609, which was unique to supplier B, was isolated from 12.6% of this supplier's positive samples but was not isolated from human cases. This highly prevalent ST is closely related to ST-48, a common human pathogen ST, differing by only one point mutation in the gltA gene. To date ST-3609 has been found only in New Zealand.
TABLE 2.
Relative frequencies of sequence types in samples from human cases and poultry suppliers in the Manawatu regiona
| ST | % of isolates fromb: |
|||
|---|---|---|---|---|
| Human casesc | Supplier A | Supplier B | Supplier C | |
| 21 | 1.4 | 0.0 | 0.8d | 0.0 |
| 25 | 0.2 | 1.5 | 0.0 | 5.7 |
| 38 | 2.6 | 0.0 | 0.0 | 0.0 |
| 42 | 3.8 | 3.1d | 0.0 | 0.0 |
| 45 | 8.2 | 27.5 | 12.6 | 25.7 |
| 48 | 8.4 | 0.8 | 22.7 | 20.0 |
| 50 | 4.6 | 3.8 | 17.6 | 0.0 |
| 52 | 3.4 | 4.6d | 0.0 | 0.0 |
| 53 | 5.4 | 7.6 | 3.4 | 8.6 |
| 61 | 2.8 | 0.0 | 0.0 | 0.0 |
| 81 | 0.2 | 0.0 | 0.0 | 0.0 |
| 137 | 0.2 | 0.0 | 0.0 | 0.0 |
| 190 | 4.2 | 6.1d | 0.0 | 0.0 |
| 219 | 0.2 | 0.0 | 0.0 | 0.0 |
| 227 | 0.0 | 0.8d | 0.0 | 0.0 |
| 257 | 2.4 | 7.6 | 3.4 | 0.0 |
| 354 | 4.6 | 2.3 | 0.0 | 0.0 |
| 403 | 0.2 | 0.0 | 0.0 | 0.0 |
| 422 | 0.6 | 0.0 | 0.0 | 0.0 |
| 436 | 0.8 | 0.0 | 0.0 | 0.0 |
| 451 | 1.6 | 0.0 | 0.0 | 5.7d |
| 459 | 0.2 | 0.0 | 0.0 | 0.0 |
| 474 | 30.7 | 20.6 | 1.7 | 0.0 |
| 520 | 1.6 | 3.8 | 0.0 | 5.7 |
| 578 | 0.2 | 0.0 | 0.0 | 0.0 |
| 583 | 1.8 | 6.1 | 0.0 | 11.4 |
| 658 | 0.4 | 0.0 | 0.0 | 0.0 |
| 677 | 1.0 | 0.0 | 0.8d | 0.0 |
| 829 | 0.2 | 0.0 | 0.0 | 0.0 |
| 1457 | 0.2 | 0.0 | 0.0 | 0.0 |
| 1517 | 0.8 | 0.0 | 5.0d | 0.0 |
| 1581 | 0.2 | 0.0 | 4.2d | 0.0 |
| 1707 | 0.2 | 0.0 | 0.0 | 0.0 |
| 1818 | 0.0 | 0.0 | 0.8d | 0.0 |
| 1911 | 0.0 | 0.0 | 0.8d | 0.0 |
| 2026 | 2.2 | 0.0 | 0.0 | 0.0 |
| 2219 | 0.2 | 0.0 | 0.0 | 0.0 |
| 2343 | 0.2 | 0.0 | 0.0 | 0.0 |
| 2345 | 0.6 | 0.8 | 9.2 | 0.0 |
| 2350 | 0.4 | 0.0 | 0.0 | 0.0 |
| 2391 | 0.2 | 0.8d | 0.0 | 0.0 |
| 2397 | 0.0 | 0.0 | 2.5d | 0.0 |
| 2535 | 0.0 | 0.0 | 0.0 | 2.9d |
| 3072 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3222 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3230 | 0.0 | 0.0 | 0.0 | 2.9d |
| 3538 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3609 | 0.0 | 0.0 | 12.6d | 0.0 |
| 3676 | 0.4 | 0.0 | 0.0 | 0.0 |
| 3711 | 0.4 | 0.0 | 0.0 | 2.9d |
| 3712 | 0.4 | 0.0 | 0.0 | 0.0 |
| 3715 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3717 | 0.2 | 0.0 | 0.0 | 8.6d |
| 3718 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3719 | 0.0 | 0.0 | 0.8d | 0.0 |
| 3720 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3721 | 0.0 | 0.8d | 0.0 | 0.0 |
| 3725 | 0.0 | 0.0 | 0.8d | 0.0 |
| 3726 | 0.0 | 1.5d | 0.0 | 0.0 |
| 3727 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3784 | 0.2 | 0.0 | 0.0 | 0.0 |
| 3792 | 0.2 | 0.0 | 0.0 | 0.0 |
Some of the data have been used in other studies of the epidemiology of human cases (34).
The total numbers of isolates obtained from the difference sources were as follows: 502 isolates from human samples, 131 isolates from supplier A samples, 119 isolates from supplier B samples, and 35 isolates from supplier C samples.
STs of human isolates have been described previously (33).
ST that was unique to this supplier in this study.
FIG. 3.
Relative frequencies of the three major human C. jejuni MLST genotypes in clinical cases from the MidCentral District surveillance site, New Zealand, in each quarter between March 2005 and February 2008.
One of the 10 humans from which two isolates were obtained was infected with two different STs. Between two and five isolates were obtained from 58 poultry samples, and up to three different STs were found in 22 individual samples. Different STs were isolated from 6 of 16 samples from supplier A, from 24 of 38 samples from supplier B, and from 2 of 4 samples from supplier C (Table 3).
TABLE 3.
Multiple typed samples
| Parameter | Human | Supplier A | Supplier B | Supplier C |
|---|---|---|---|---|
| Samples with multiple STs/total no. | 1/10 | 6/16 | 24/38 | 2/4 |
| Proportion of multiple STs | 0.10 | 0.38 | 0.63 | 0.50 |
| Maximum no. of different STs found in one sample | 2 | 2 | 3 | 2 |
The PSI was calculated to explore formally the similarity of genotypes from human cases and the different poultry suppliers. This analysis revealed a significantly higher degree of similarity between STs identified in human cases and genotypes identified in samples from supplier A (PSI, 0.58; 95% CI, 0.48 to 0.64) than between STs from human cases and the other suppliers (Table 4). Isolates in samples from the other suppliers were less similar to human isolates, and the estimated PSI for suppliers B and C were 0.32 (95% CI, 0.26 to 0.36) and 0.28 (95% CI 0.21 to 0.31), respectively. When the similarity between samples from the poultry suppliers was examined, the greatest similarity was that between samples from suppliers A and C (PSI, 0.46; 95% CI, 0.27 to 0.50) and the lowest similarity was that between samples from suppliers A and B (PSI, 0.26; 95% CI, 0.18 to 0.35).
TABLE 4.
Proportional similarity index for each supplier compared with the other suppliers and with the distribution of human genotypes
| Source | Proportional similarity index (bootstrapped 95% confidence interval)a |
||
|---|---|---|---|
| Supplier A | Supplier B | Supplier C | |
| Supplier A | 1.00 | ||
| Supplier B | 0.26 (0.18, 0.35) | 1.00 | |
| Supplier C | 0.46 (0.27, 0.50) | 0.36 (0.22, 0.44) | 1.00 |
| Human cases | 0.58 (0.48, 0.64) | 0.32 (0.26, 0.36) | 0.28 (0.21, 0.31) |
Higher values indicate higher levels of similarity between the STs identified for the different sources.
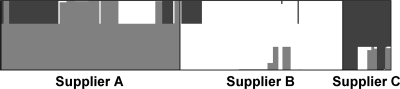
The population structure of C. jejuni in samples from New Zealand poultry suppliers was explored using Structure 2.2. The posterior probabilities for each isolate are shown in a stacked bar plot in Fig. 4, where the stacked bars represent the probability that each individual isolate belongs to each of the three clusters. This figure allows visual assessment of the three clusters against the three poultry suppliers. Isolates from supplier A showed the most diversity, and the isolates had reasonable probabilities of belonging to any of the three clusters (0.611, 0.154, and 0.234 for clusters 1 to 3, respectively). On the other hand, isolates from supplier B had the highest probability of belonging to cluster 2 (0.845), and the contributions of clusters 1 and 3 were minor (0.065 and 0.090, respectively). The diversity of isolates recovered from supplier C's products was similar to that of isolates from supplier A's products, with probabilities of belonging to clusters 1, 2, and 3 of 0.195, 0.149, and 0.657, respectively, but the highest probability was for cluster 3 instead of cluster 1.
FIG. 4.
Population structure of C. jejuni for different poultry suppliers estimated by cluster analysis using Structure 2.2. The posterior probabilities that each isolate belongs to cluster 1 (light gray), cluster 2 (white), and cluster 3 (dark gray) are shown. Isolates are grouped by supplier and ordered by ST.
DISCUSSION
In New Zealand the prevalence of Campylobacter spp. in poultry products is generally higher than that for other meats (51), and in other countries a similar high prevalence in chicken meat has been observed (e.g., 60.9% in a United Kingdom study [28] and 60% in a Belgium study [24]). In this study 78.8% of fresh carcasses were positive, and differences in contamination probability and levels were observed for the different suppliers. A high level of contamination of retail poultry in New Zealand was also reported by a study conducted in 2003 and 2004 by Wong et al. (51). In addition, the prevalence of and mean counts for Campylobacter species-positive carcasses were previously found to vary for different processing plants in New Zealand (4). However, these observations were not significant, which may have been a result of the study's design and power. Recently, similar supplier-specific observations were made in Belgium (23). Our findings support the view that the level of exposure of humans to Campylobacter spp. through retail poultry in New Zealand is high (1) and revealed that contamination probabilities and levels differ between the individual suppliers. The differences could originate from differences in the contamination of flocks from environmental sources during primary production or from differences in the use of control measures during further processing. Differences in mean contamination levels have previously been used to link poultry product lots to disease clusters (3), and, as a consequence, temporal investigation of contamination levels provided an opportunity for the development and implementation of control measures.
We examined the similarity and relatedness of genotypes of human and poultry isolates obtained from samples from the different suppliers to explore the diversity of Campylobacter STs in the New Zealand poultry supply and to investigate further the importance of poultry products as a contributor to the human disease burden, thereby extending the analysis of data gathered for risk attribution studies (34). A total of 51 different STs were detected in human samples in our study. Twenty-one of these STs were also isolated from poultry sources, and these STs accounted for 85.5% of human cases. This overlap between human- and poultry-associated genotypes is consistent with the hypothesis that poultry is important in disease transmission in New Zealand (1, 14). However, to make further inferences in this context about the individual poultry suppliers, the different sampling efforts have to be taken into account as the method used influences the probability of detecting different genotypes and the precision of attribution estimates. In this study whole fresh chicken carcasses were used, and we considered them representative of portioned products for our typing results. In New Zealand 51% of all slaughtered birds leave primary processing as whole carcasses, and the other 49% are portioned or used as value-added products.
This study revealed that there are both supplier-specific and ubiquitous strains, and a total of 22 STs were found to be unique to one of the suppliers. Although with a larger sample size and therefore higher detection rates this number could have been smaller, this is still an unexpected finding. Whereas the dominant human pathogen, ST-474 (33), was commonly found in samples from supplier A (20.6% of samples), it was isolated only rarely from other sources in the sentinel surveillance site, including samples from supplier B (1.7% of samples) and bovine, ovine, and environmental samples (34). As another example, ST-3609 was commonly isolated from samples from supplier B (12.6% of samples), but it has not been detected yet in samples from any other supplier or in any human clinical cases. On the other hand, ST-45, ST-48, ST-50, and ST-53 were ubiquitous in the poultry supply, and together these STs accounted for about one-quarter of human cases (26.6%). This suggests that both ubiquitous and poultry supplier-associated strains contribute to human campylobacteriosis in New Zealand. As a consequence, the relative contributions of individual suppliers to the public health burden may differ substantially. This supports the findings of previous studies from the same sentinel surveillance site which used risk attribution models to quantify the contributions of different sources to the human disease burden. These approaches estimated that 80% of human cases originate from poultry and that the contributions of the individual suppliers ranged from 7 to 58% (32, 34).
In an exploratory analysis using only a small subset of samples, we detected a high proportion of poultry samples that were contaminated with more than one ST. Similar findings have been obtained in other studies (25), and this further underlines the importance of considering the consequences of selecting and testing only one presumptive isolate per sample. In this study we assumed that, on average, by typing only one isolate per sample we would detect isolates at the relative frequencies at which they occur in all samples. However, this method does affect the probability of detecting all strains present in a source, and the presence of multiple strains in individual samples may decrease the likelihood of discovering all strains. The relationship between sample size and the probability of detecting all strains present in a sample was recently discussed by Dopfer et al. (13).
In this study, the PSI was calculated to assess formally the similarity between the distribution of bacterial subtypes in samples from the different poultry suppliers and the distribution of bacterial subtypes in human samples. The highest PSI was observed for genotypes for samples from supplier A and human cases, providing further evidence for the link between this supplier and human cases. This conclusion is supported by the finding that the prevalence of ST-474 was very low in samples from nonpoultry sources in New Zealand (29, 34). It is notable that the similarity of supplier A sample genotypes to human sample genotypes is higher than the similarity between poultry supplier sample genotypes. This observation may be explained both by the high market share of supplier A and hence the relatively high rate of exposure of the human population to strains from this supplier and by the vertically integrated structure of the industry, which is likely to reduce between-supplier transmission. In the sentinel surveillance site, genotypes from poultry samples shared a higher level of similarity with genotypes from human cases, as estimated by the PSI, compared to other sources, such as ruminants or wildlife (34).
The Structure program was used as an exploratory method to identify genotypic clusters among isolates from the different poultry suppliers. This analysis suggested that genotype distribution is structured by the individual suppliers; for example, the majority of isolates from supplier A belonged to cluster 1, the cluster which contained the major human pathogen, ST-474. A different cluster was associated with each company, and this further supports the hypothesis that there are supplier-specific transmission pathways, most likely as a consequence of the vertically integrated nature of the New Zealand poultry industry, in which suppliers control all stages from primary production to processing with no importation and very little or no overlap between companies. The differences in the diversity of genotypes in the three clusters could be a result of differences in the levels of biosecurity between suppliers, where presumably low biosecurity by a supplier would lead to higher diversity of genotypes present in the cluster associated with that supplier. Such differences could originate from differences in the contamination of flocks from environmental sources during primary production or from differences in the use of control measures during further processing. It would be interesting to see if similar findings can be obtained in other countries with a different industry structure and to investigate further the reasons for this diversity.
It is striking that the majority of human cases were caused by an internationally rare ST, ST-474, which is highly prevalent in one of New Zealand's poultry suppliers but was rarely found elsewhere in the sentinel surveillance site. ST-474 was the genotype most commonly isolated in the winter epidemic in 2006 in New Zealand (29) and along with the other major human STs contributed to the human disease burden throughout the study period (33). To date, this ST has been submitted only once to the Campylobacter PubMLST database (10) (from a chicken sample in the Czech Republic) and has been reported only sporadically (2, 5, 50) outside of New Zealand (29, 34, 47). This is in contrast to ST-45, which was prevalent in samples from all poultry suppliers and other animal sources tested in the region, both livestock and wildlife, but accounted for far fewer human cases (a total of 8.2%). Studies by Taboada et al. (47) and Pope et al. (35) showed that some Campylobacter strains, including ST-474 strains, are more virulent for humans than other strains. Further research into the pathogenicity and virulence genes associated with dominant clones could reveal determinants for human infection and commensal colonization and may further explain the molecular epidemiology of C. jejuni in New Zealand.
Other STs detected in our study are also rare in other countries or have never been found elsewhere (34). This may be due in part to reporting bias, since it is likely that not all detected strains are described in published studies or submitted to the MLST database, particularly since MLST has only recently emerged as new approach for typing of Campylobacter spp. However, researchers in many countries have recently submitted isolates to the shared database and conducted large-scale studies using MLST (6, 44, 50). In addition, many of the internationally common Campylobacter STs, such as ST-45 and ST-48, can be found in human and poultry samples in New Zealand. These STs are widespread in other countries (7, 22, 30), which is reflected by a wide range of isolates included in the Campylobacter PubMLST database. The introduction of a variety of animals and animal products into a geographically isolated country during early settlement in combination with the present tight border biosecurity would provide a framework for a distinct ecosystem of Campylobacter spp. in New Zealand, which may have originated from imported pathogens during early settlement. In support of this hypothesis, French et al. (18) isolated C. jejuni genotypes from wild bird feces in New Zealand that were associated with wild bird populations in the northern hemisphere.
A structured longitudinal sampling approach was used in this study, which included simultaneous collection of data from human clinical cases and the corresponding disease source to represent exposure. This sampling strategy has several advantages, such as greater representativeness, over commonly used approaches, such as cross-sectional surveys, which may not be concurrent in space and time. This study revealed the unique molecular epidemiology of C. jejuni in the Manawatu region of New Zealand, provided valuable information concerning the link between poultry and human campylobacteriosis, and aided the development of control strategies in the poultry industry. Comparative studies conducted in Christchurch and Auckland showed a similar distribution of genotypes in both humans and poultry (19), suggesting that the results of the Manawatu study are likely to be similar to the results of studies performed in other regions, allowing extrapolation of the results to all of New Zealand (19, 34). This study extended the work previously reported from the sentinel surveillance site (33, 34) by providing an in-depth investigation of the epidemiology of C. jejuni in New Zealand poultry sources. In addition, New Zealand's simply structured poultry industry provides a unique background for understanding the epidemiology of this pathogen in the poultry supply. In countries with a more diverse supply and a less vertical structure, where poultry may be imported from several countries, such complexity provides strong challenges for both understanding and controlling the disease (23, 40).
The findings obtained in this and other studies for the surveillance site provided significant input for the New Zealand Food Safety Authority's (NZFSA) Campylobacter Management Strategy in Poultry (http://www.nzfsa.govt.nz/consumers/food-safety-topics/food-borne-illnesses/campylobacter/strategy/index.htm), and the strategic introduction of a range of industry and regulatory control measures, including intensification of microbiological monitoring programs and setting of mandatory targets in the production chain, coincided with a >50% reduction in the number of human cases in 2008 to the lowest notification rate in 16 years.
Acknowledgments
We thank all members of the Hopkirk Molecular Epidemiology Team, Environmental Science and Research (ESR), MidCentral Health (in particular, Tui Shadbolt), Public Health Services, NZFSA (in particular, the Science Group, Roger Cook, Donald Campbell, and Sharon Wagener), and MedLab Central for their contributions. In particular, we thank Sarah Moore for maintaining the database and Lynn Rogers for enabling cooperation with MedLab Central.
Financial support was provided through New Zealand Food Safety Authority projects FDI/236/2005 (Enhancing Surveillance of Potentially Food-borne Enteric Diseases in New Zealand: Human Campylobacteriosis in the Manawatu) and FSPD/83/2004 (Ph.D. Human Campylobacteriosis [P.M.]).
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Baker, M., N. Wilson, R. Ikram, S. Chambers, P. Shoemack, and G. Cook. 2006. Regulation of chicken contamination urgently needed to control New Zealand's serious campylobacteriosis epidemic. N. Z. Med. J. 119:1243. [PubMed] [Google Scholar]
- 2.Best, E. L., A. J. Fox, R. J. Owen, J. Cheesbrough, and F. J. Bolton. 2007. Specific detection of Campylobacter jejuni from faeces using single nucleotide polymorphisms. Epidemiol. Infect. 135:839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callicott, K. A., H. Haroardottir, F. Georgsson, J. Reiersen, V. Frioriksdottir, E. Gunnarsson, P. Michel, J. R. Bisaillon, K. G. Kristinsson, H. Briem, K. L. Hiett, D. S. Needleman, and N. J. Stern. 2008. Broiler campylobacter contamination and human campylobacteriosis in Iceland. Appl. Environ. Microbiol. 74:6483-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrystal, N. D., S. J. Hargraves, A. C. Boa, and C. J. Ironside. 2008. Counts of Campylobacter spp. and prevalence of Salmonella associated with New Zealand broiler carcasses. J. Food Prot. 71:2526-2532. [DOI] [PubMed] [Google Scholar]
- 5.Clark, C. G., L. Bryden, W. R. Cuff, P. L. Johnson, F. Jamieson, B. Ciebin, and G. H. Wang. 2005. Use of the Oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J. Clin. Microbiol. 43:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colles, F. M., T. A. Jones, N. D. McCarthy, S. K. Sheppard, A. J. Cody, K. E. Dingle, M. S. Dawkins, and M. C. J. Maiden. 2008. Campylobacter infection of broiler chickens in a free-range environment. Environ. Microbiol. 10:2042-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowden, J. 1992. Campylobacter—epidemiologic paradoxes. Br. Med. J. 305:132-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump, J. A., D. R. Murdoch, and M. G. Baker. 2001. Emerging infectious diseases in an island ecosystem: the New Zealand perspective. Emerg. Infect. Dis. 7:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dopfer, D., W. Buist, Y. Soyer, M. A. Munoz, R. N. Zadoks, L. Geue, and B. Engel. 2008. Assessing genetic heterogeneity within bacterial species isolated from gastrointestinal and environmental samples: how many isolates does it take? Appl. Environ. Microbiol. 74:3490-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eberhart-Phillips, J., N. Walker, N. Garrett, D. Bell, D. Sinclair, W. Rainger, and M. Bates. 1997. Campylobacteriosis in New Zealand: results of a case-control study. J. Epidemiol. Community Health 51:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falush, D., M. Stephens, and J. Pritchard. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7:574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinsinger, P., E. E. Spears, and R. W. Poole. 1981. A simple measure of niche breadth. Ecology 62:27-32. [Google Scholar]
- 17.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherborrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 18.French, N., A. Midwinter, B. Holland, J. Collins-Emerson, R. Pattison, F. M. Colles, and P. Carter. 2009. Molecular epidemiology of Campylobacter jejuni isolated from wild bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French, N., and Molecular Epidemiology and Veterinary Public Health Group, Hopkirk Institute. 2008. Enhancing surveillance of potentially foodborne enteric diseases in New Zealand: human campylobacteriosis in the Manawatu. http://www.nzfsa.govt.nz/science/research-projects/Campy_Attribution_Manawatu.pdf.
- 20.Garrett, N., M. L. Devane, J. A. Hudson, C. Nicol, A. Ball, J. D. Klena, P. Scholes, M. G. Baker, B. J. Gilpin, and M. G. Savill. 2007. Statistical comparison of Campylobacter jejuni subtypes from human cases and environmental sources. J. Appl. Microbiol. 103:2113-2121. [DOI] [PubMed] [Google Scholar]
- 21.Gilpin, B., A. Cornelius, B. Robson, N. Boxall, A. Ferguson, C. Nicol, and T. Henderson. 2006. Application of pulsed-field gel electrophoresis to identify potential outbreaks of campylobacteriosis in New Zealand. J. Clin. Microbiol. 44:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gormley, F. J., M. MacRae, K. J. Forbes, I. D. Ogden, J. F. Dallas, and N. J. C. Strachan. 2008. Has retail chicken played a role in the decline of human campylobacteriosis? Appl. Environ. Microbiol. 74:383-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habib, I., I. Sampers, M. Uyttendaele, D. Berkvens, and L. De Zutter. 2008. Baseline data from a Belgium-wide survey of Campylobacter species contamination in chicken meat preparations and considerations for a reliable monitoring program. Appl. Environ. Microbiol. 74:5483-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Habib, I., I. Sampers, M. Uyttendaele, L. De Zutter, and D. Berkvens. 2008. A Bayesian modelling framework to estimate Campylobacter prevalence and culture methods sensitivity: application to a chicken meat survey in Belgium. J. Appl. Microbiol. 105:2002-2008. [DOI] [PubMed] [Google Scholar]
- 25.Kramer, J. M. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from humans infection. J. Food Prot. 63:1654-1659. [DOI] [PubMed] [Google Scholar]
- 26.Lake, R. J., M. G. Baker, N. Garrett, W. G. Scott, and H. M. Scott. 2000. Estimated number of cases of foodborne infectious disease in New Zealand. N. Z. Med. J. 113:278-281. [PubMed] [Google Scholar]
- 27.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 28.Little, C. L., J. F. Richardson, R. J. Owen, E. de Pinna, and E. J. Threlfall. 2008. Campylobacter and Salmonella in raw red meats in the United Kingdom: prevalence, characterization and antimicrobial resistance pattern, 2003-2005. Food Microbiol. 25:538-543. [DOI] [PubMed] [Google Scholar]
- 29.McTavish, S. M., C. E. Pope, C. Nicol, K. Sexton, N. French, and P. E. Carter. 2008. Wide geographical distribution of internationally rare Campylobacter clones within New Zealand. Epidemiol. Infect. 136:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mickan, L., R. Doyle, M. Valcanis, K. E. Dingle, L. Unicomb, and J. Lanser. 2007. Multilocus sequence typing of Campylobacter jejuni isolates from New South Wales, Australia. J. Appl. Microbiol. 102:144-152. [DOI] [PubMed] [Google Scholar]
- 31.Miller, W. G., S. L. W. On, G. L. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullner, P., G. Jones, A. Noble, S. Spencer, S. Hathaway, and N. French. 2009. Source attribution of food borne zoonoses in New Zealand: a modified Hald model. Risk Anal. 29:970-984. [DOI] [PubMed] [Google Scholar]
- 33.Mullner, P., T. Shadbolt, J. M. Collins-Emerson, A. C. Midwinter, S. E. F. Spencer, J. Marshall, P. E. Carter, D. M. Campbell, D. J. Wilson, S. Hathaway, R. Pirie, and N. P. French. 9 February 2010. Molecular and spatial epidemiology of human campylobacteriosis: source association and genotype-related risk factors. Epidemiol. Infect. doi: 10.1017/S0950268809991579. [DOI] [PubMed]
- 34.Mullner, P., S. E. F. Spencer, D. Wilson, G. Jones, A. D. Noble, A. C. Midwinter, J. Collins-Emerson, P. Carter, S. Hathaway, and N. P. French. 2009. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect. Genet. Evol. 9:1311-1319. [DOI] [PubMed] [Google Scholar]
- 35.Pope, C., J. Wilson, E. N. Taboada, J. MacKinnon, C. A. F. Alves, J. H. E. Nash, K. Rahn, and G. W. Tannock. 2007. Epidemiology, relative invasive ability, molecular characterization, and competitive performance of Campylobacter jejuni strains in the chicken gut. Appl. Environ. Microbiol. 73:7959-7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard, J., M. Stephens, and P. Donnelly. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, S. E., P. E. Brown, E. J. Wright, C. A. Hart, and N. P. French. 2009. Quantifying within- and between-animal variation and uncertainty associated with counts of Escherichia coli O157 occurring in naturally infected cattle faeces. J. R. Soc. Interface 6:169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosef, O., G. Kapperud, S. Lauwers, and B. Gondrosen. 1985. Serotyping of Campylobacter jejuni, Campylobacter coli and Campylobacter laridis from domestic and wild animals. Appl. Environ. Microbiol. 49:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenberg, N. A. 2004. Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4:137-138. [Google Scholar]
- 40.Rosenquist, H., N. L. Nielsen, H. M. Sommer, B. Norrung, and B. B. Christensen. 2003. Quantitative risk assessment of human campylobacteriosis associated with thermophilic Campylobacter species in chickens. Int. J. Food. Microbiol. 83:87-103. [DOI] [PubMed] [Google Scholar]
- 41.Savill, M. G., J. A. Hudson, A. Ball, J. D. Klena, P. Scholes, R. J. Whyte, R. E. McCormick, and D. Jankovic. 2001. Enumeration of Campylobacter in New Zealand recreational and drinking waters. J. Appl. Microbiol. 91:38-46. [DOI] [PubMed] [Google Scholar]
- 42.Scott, W. G., H. M. Scott, R. J. Lake, and M. G. Baker. 2000. Economic cost to New Zealand of foodborne infectious disease. N. Z. Med. J. 113:281-284. [PubMed] [Google Scholar]
- 43.Sheppard, S. K., J. F. Dallas, N. J. C. Strachan, M. Macrae, N. D. McCarthy, D. Falush, I. D. Ogden, M. C. J. Maiden, and K. J. Forbes. 2009. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 48:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strachan, N. J. C., F. J. Gormley, O. Rotariu, I. D. Ogden, G. Miller, G. M. Dunn, S. K. Sheppard, J. F. Dallas, T. M. S. Reid, H. Howie, M. C. J. Maiden, and K. J. Forbes. 2009. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J. Infect. Dis. 199:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stucki, U., J. Frey, J. Nicolet, and A. P. Burnens. 1995. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J. Clin. Microbiol. 33:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan, C. B., M. A. Diggle, and S. C. Clarke. 2005. Multilocus sequence typing—data analysis in clinical microbiology and public health. Mol. Biotechnol. 29:245-254. [DOI] [PubMed] [Google Scholar]
- 47.Taboada, E. N., J. M. MacKinnon, C. C. Luebbert, V. P. J. Gannon, J. H. E. Nash, and K. Rahn. 2008. Comparative genomic assessment of multi-locus sequence typing: rapid accumulation of genomic heterogeneity among clonal isolates of Campylobacter jejuni. BMC Evol. Biol. 8:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urwin, R., and M. C. J. Maiden. 2003. Multilocus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 49.Wagenaar, J. A., D. J. Mevius, and A. H. Havelaar. 2006. Campylobacter in primary animal production and control strategies to reduce the burden of human campylobacteriosis. Rev. Off. Int. Epizoot. 25:581-594. [PubMed] [Google Scholar]
- 50.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, T. L., L. Hollis, A. Cornelius, C. Nicol, R. Cook, and J. A. Hudson. 2007. Prevalence, numbers, and subtypes of Campylobacter jejuni and Campylobacter coli in uncooked retail meat samples. J. Food Prot. 70:566-573. [DOI] [PubMed] [Google Scholar]
- 52.Woolhouse, M. E. J. 2002. Population biology of emerging and re-emerging pathogens. Trends Microbiol. 10:S3-S7. [DOI] [PubMed] [Google Scholar]