Abstract
Soils are typically considered to be suboptimal environments for enteric organisms, but there is increasing evidence that Escherichia coli populations can become resident in soil under favorable conditions. Previous work reported the growth of autochthonous E. coli in a maritime temperate Luvic Stagnosol soil, and this study aimed to characterize, by molecular and physiological means, the genetic diversity and physiology of environmentally persistent E. coli isolates leached from the soil. Molecular analysis (16S rRNA sequencing, enterobacterial repetitive intergenic consensus PCR, pulsed-field gel electrophoresis, and a multiplex PCR method) established the genetic diversity of the isolates (n = 7), while physiological methods determined the metabolic capability and environmental fitness of the isolates, relative to those of laboratory strains, under the conditions tested. Genotypic analysis indicated that the leached isolates do not form a single genetic grouping but that multiple genotypic groups are capable of surviving and proliferating in this environment. In physiological studies, environmental isolates grew well across a broad range of temperatures and media, in comparison with the growth of laboratory strains. These findings suggest that certain E. coli strains may have the ability to colonize and adapt to soil conditions. The resulting lack of fecal specificity has implications for the use of E. coli as an indicator of fecal pollution in the environment.
Escherichia coli is a well-established indicator of fecal contamination in the environment. The organism's validity as an indicator of water pollution is dependent, among other factors, on its fecal specificity and its inability to multiply outside the primary host, the gastrointestinal tracts of humans and warm-blooded animals (9). While many pathogens and indicator organisms are considered to be poorly adapted for long-term survival, or proliferation, outside their primary hosts (24), there is increasing evidence that this view needs to be reconsidered with respect to E. coli (17, 38). In particular, questions remain about its fate and survival capacity in environmental matrices, such as soil. While the habitat within the primary host is characterized by constant warm temperature conditions and a ready availability of nutrients and carbon, that of soil is often characterized by oligotrophic and highly dynamic conditions, temperature and pH variation, predatory populations, and competition with environmentally adapted indigenous microflora (39). Soils are thus typically considered to be suboptimal environments for enteric organisms, and growth is thought to be negligible, with die-off of organisms at rates reported to be a function of the interaction of numerous factors, including the type and physiological state of the microorganism, the physical, chemical, and biological properties of the soil, atmospheric conditions (including sunlight, moisture, and temperature), and organism application method (10).
In recent years, the growth of E. coli in soils, sediments, and water in tropical and subtropical regions has been widely documented, and the organism is considered to be an established part of the soil biota within these regions (4, 5, 7, 12, 14, 19, 25, 32). The integration of E. coli as a component of the indigenous microflora in soils of tropical and subtropical regions may be attributable to the nutrient-rich nature and warm temperatures of these habitats (21, 39), combined with the metabolic versatility of the organism and its simple nutritional requirements (21). In addition to tropical and subtropical regions, the presence of autochthonous E. coli populations in the cooler soils of temperate and northern temperate regions has also been reported (6, 20, 22, 37), with one report on an alpine soil (34) and, most recently, a report on a maritime temperate grassland soil (3). The growth of E. coli within soils can act as a reservoir for the further contamination of bodies of water (20, 31, 32), compromising the indicator status of E. coli within these regions. As such, an understanding of the ecological characteristics of E. coli in soil is critical to its validation as an indicator organism. With respect to the input of pathogenic E. coli into the environment, this knowledge becomes essential for assessing the potential health risk to human and animal hosts from agricultural activities such as landspreading of manures and slurries (24).
It has been suggested that E. coli can sustain autochthonous populations within soils in temperate regions, wherever favorable conditions exist (21). The phenotypic traits of the organism (including its metabolic diversity and its ability to grow both aerobically and anaerobically in a broad temperature range) may assist the persistence, colonization, and growth of E. coli when conditions permit. The challenging nature of the soil environment and the disparity of conditions between the primary host and the secondary habitat raises the question of how these E. coli populations survive and compete for niche space among the highly competitive and diverse coexisting populations of the indigenous microflora (15, 21). There is some evidence that naturalized E. coli may form genetically distinct populations in the environment (17, 20, 34, 36). This suggests that autochthonous E. coli populations in soil may have increased environmental fitness, facilitating their residence in soil (20, 34, 38). Little is known, however, of the physiology of these organisms, and their capacity for survival in soil remains poorly understood (21).
Previous work (3) recorded continuous low-level leaching of viable E. coli from lysimeters of a poorly drained Luvic Stagnosol soil type, more than 9 years after the last application of fecal material. This finding was indicative of the growth of E. coli within the soil and suggested the presence of autochthonous E. coli populations within the soil that could be leached subsequently. To our knowledge, prior to this report, naturalized autochthonous E. coli populations persisting under the relatively oligotrophic, low-temperature conditions of maritime temperate soil environments had not been described previously. Growth within this soil was attributed chiefly to favorable characteristics of the soil, which include high clay and moisture contents, nutrient retention, and the presence of anaerobic zones. The objective of this work was to characterize, by molecular and physiological means, the genetic diversity and physiology of environmentally persistent E. coli isolates leached. In particular, we were interested in determining if the isolates possessed phenotypic characteristics that may enhance their capacity to survive and occupy niche space within the soil. This study tested the hypothesis that E. coli clones persisting in lysimeters of this soil form a genetically distinct grouping and possess a physiology tailored to the soil environment.
MATERIALS AND METHODS
Isolate preparation.
Environmental isolates were extracted from water leached from Luvic Stagnosol soil lysimeters installed in a lysimeter unit in Johnstown Castle, Wexford, Ireland (6°30′W, 52°17′N) under maritime temperate climatic conditions. The lysimeters comprise intact monoliths of soil (diameter, 0.6 m; depth, 1 m) contained in fiberglass cylinders. The soil type and lysimeter unit design were described by Ryan and Fanning previously (29). Leachate from natural rainfall was collected from the lysimeters, which had not received any fecal material for 8.5 years, over a period of 488 days. E. coli in lysimeter leachate was enumerated by the Idexx Colisure method (26). Luvic Stagnosol soils were found to leach low levels of viable E. coli continuously throughout the drainage period (3). Isolates were verified as E. coli by positive confirmation on MacConkey plates (Oxoid), UTI chromogenic plates (Oxoid), and API 20E (bioMérieux, Paris, France) strips. The isolates were then subcultured (5 times) to ensure axenic culture conditions. Two E. coli laboratory strains (K-12 strains MG1655 and ATCC 25922) were used for comparative purposes.
DNA extraction.
A phenol-chloroform method derived from the work of Delbès et al. (11) was used for DNA extraction. Luria-Bertani (LB) broth cultures (40 ml) of each isolate were grown overnight at 37°C. Two milliliters of each culture was centrifuged (Sigma 1-15 centrifuge) at 12,052 × g for 2 min. DNA was extracted from the pellets by first adding 0.1 g glass beads (acid washed; diameter, 150 to 212 μm; Sigma), 0.5 ml phosphate-buffered saline, 0.5 ml phenol, and 0.5 ml chloroform and then vortexing for 1 min. The DNA was then washed twice with 0.5 ml chloroform by vortexing for 1 min and centrifuging as described above for 5 min. The DNA was then removed from the top layer with a pipette and was stored.
PCR amplification of 16S rRNA.
16S rRNA genes were amplified from the extracted DNA using primers 27F (bacterial) and 1392R (universal) (1). The PCR mixture (50 μl) consisted of 5 μl 10× NH4 buffer, 1.5 μl 50 mM MgCl2, 1 μl of each primer (20 pmol), 1 μl 100 mM deoxynucleoside triphosphate (dNTP), 38.1 μl H2O, 2 U (0.4 μl) BioTaq (Bioline), and 2 μl of template DNA. The PCR was performed with an Eppendorf Mastercycler gradient (model 5331) with the following steps: denaturation for 3 min at 94°C; 30 cycles of 45 s at 94°C, 45 s at 55°C, and 1 min at 72°C; and a final extension step of 5 min at 72°C. Amplified rRNA gene restriction analysis (ARDRA) was carried out with 3 restriction enzymes: HaeIII, AluI, and HindIII (Promega). Digestion was modified from the manufacturer's directions: the amount of DNA was increased to 5 μl. On the basis of ARDRA profiles, the PCR products (approximately 1,400 bp) of representative isolates were cloned using a TOPO TA cloning kit as described by the manufacturer (Invitrogen). Clones with correctly sized inserts were sequenced. Phylogenetic analysis was conducted using MEGA4 software (33). A distance-based tree was built using the neighbor-joining method (30) with bootstrap analysis.
Multiplex PCR.
A triplex method was used to group E. coli isolates into each of the 4 main clone groups using primer pairs ChuA.1-ChuA.2, YjaA.1-YjaA.2, and TspE4C2.1-TspE4C2.2 as described by Clermont et al. (8). The PCR mixture (50 μl) consisted of 5 μl 10× NH4 buffer, 2 μl 50 mM MgCl2, 1 μl of each of the 6 primers (20 pmol), 1 μl 100 mM dNTP, 33.6 μl H2O, 2.5 U (0.5 μl) BioTaq (Bioline), and 2 μl of template DNA. The PCR steps were as follows: denaturation for 4 min at 94°C, 30 cycles of 5 s at 94°C and 10 s at 59°C, and a final extension step of 5 min at 72°C. PCR products were separated on a 2% Tris-acetate-EDTA buffer (TAE) agarose electrophoresis gel stained with Sybr Safe (5 μl/100 ml). The isolates were assigned to genetic clusters on the basis of the presence or absence of 279-, 211-, and 152-bp fragments generated by the primer sets.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as described previously (28). The gel was stained with 1 μg/ml of ethidium bromide and was visualized with the aid of a Gel Doc 2000 system (Bio-Rad). Fingerprints were analyzed using Bionumerics software with the Dice coefficient at band migration tolerances of 1.5%. Clustering of patterns was performed by the unweighted-pair group method with arithmetic averages (UPGMA).
ERIC PCR.
Enterobacterial repetitive intergenic consensus (ERIC) PCR fingerprinting was carried out on isolates using primers ERIC1R and ERIC2 as described previously by Versalovic et al. (35). DNA amplification was carried out using the following program: denaturation for 3 min at 94°C; 35 cycles consisting of 94°C for 5 s, 48°C for 45 s, and 72°C for 1 min; and a final extension step of 72°C for 5 min. Each 50-μl reaction mixture contained 2 μl DNA, 2 U (0.4 μl) BioTaq polymerase (Bioline),1 μl 100 mM dNTP, 1 μl of primers ERIC1R and ERIC2 (20 pmol), 1.5 μl 50 mM MgCl2, 5 μl 10× NH4 buffer, and 38.1 μl of nuclease-free H2O. PCR products were electrophoretically separated on a 2% TAE agarose gel stained with GelRed (5 μl/100 ml) for visualization of DNA bands. A matrix was generated for the E. coli isolates on the basis of the presence or absence of PCR products of distinct sizes, and similarity was determined by means of multivariate cluster analysis (single linkage) with a Bray-Curtis measurement of similarity (expressed as a percentage) using PAST (18).
Growth medium preparation.
Isolates were grown both on McIlvaine's minimal medium [18 mM citric acid, 56.6 mM Na2HPO4, 5 mM K2HPO4, 0.4 mM MgSO4·7H2O, 7.6 mM (NH4)2SO4, 3 μM thiamine, 6 μM (NH4)2SO4·FeSO4·6H2O, and 0.4% (wt/vol) glucose] and on soil extract medium, which was prepared exactly like McIlvaine's minimal medium but using a soil extract instead of deionized H2O. Soil extract medium was used to mimic the nutrient, energy, and carbon sources that E. coli most likely encounters in soil environments. The soil used for the soil extract was a composite of soils from different depths from a Luvic Stagnosol soil site. Soil organic matter and dry matter contents for this soil composite were 8.0% (±2.0%) and 76.2% (±0.5%), respectively. A soil-water slurry was made up with distilled H2O at a soil/water ratio of 4:9, manually shaken by inverting for 5 min, autoclaved for 1 h at 121°C, and allowed to settle overnight. The supernatant was then removed, centrifuged (Beckman Coulter Avanti J-20XP) at 5,000 × g for 15 min, reautoclaved for 20 min at 121°C, and analyzed for nutrient and metal contents (13).
Growth curves.
Two environmental isolates (isolates 3 and 5) were selected (on the basis of the greatest difference in PFGE and ERIC fingerprinting patterns), along with the K-12 laboratory strain, for growth curve analysis. Anaerobic growth curves were generated on the two growth media at 37°C, 15°C, and 10°C; the latter two temperatures were chosen to mimic typical soil temperatures in Ireland. Anaerobic culture conditions were chosen, because it was suspected that autochthonous E. coli populations in this soil type would likely be inhabiting an anaerobic zone within the soil (3). All cultures were inoculated to obtain a starting optical density at 600 nm (OD600) of ∼0.05 (Implen NanoPhotometer) using 18-h overnight aerobic cultures as inocula. A 3-by-3-by-2 factorial (temperature, strain, medium) analysis of variance (ANOVA) was performed on mean specific growth rates (SGR) using Proc Glimmix (SAS, version 9.1). Mixed-model analysis was used to allow for heterogeneous variance within the structure of the experiment, and Proc Glimmix was used to examine interactions with simple effects. The data assumptions of the statistical test were met. Multiple-comparison posthoc analyses were performed with the Tukey honestly significant difference test.
RESULTS
A total of 7 isolates extracted from positive Quanti-Tray wells from 6 separate leachate sampling days were confirmed as E. coli and were selected for further analysis. A biochemical profile was generated for each isolate using 20 reactions of the API 20E strip and a filter paper oxidase test. In all but 4 tests, all isolates reacted similarly, suggesting similar metabolic capacities among the isolates (Table 1). All isolates except isolate 5 could utilize sorbitol, while isolate 2 was the only isolate that was able to ferment or oxidize the glycoside amygdalin. Out of the 7 isolates tested, 4 (isolates 1, 3, 6, and 7) did not utilize saccharose, while only 3 isolates (isolates 3, 4, and 5) tested positive for the ornithine decarboxylase enzyme. The results of metal and nutrient content analysis of the soil extract medium are as follows: total oxidized nitrogen (TON), 1.97 mg/liter; Cl, 5.4 mg/liter; P, 0.062 mg/liter; NH4-N, 0.834 mg/liter; NO2-N, 0.017 mg/liter; total nitrogen (TN), 13.63 mg/liter; total phosphorus (TP), 0.43 mg/liter; nonpurgeable organic carbon (NPOC), 96.7 mg/liter; Ca, 11.48 mg/liter; Mg, 5 mg/liter; Na, 4.71 mg/liter; K, 3.51 mg/liter; Cu, 32 μg/liter; Fe, 31 mg/liter; Mn, 171 μg/liter; Zn, 42 μg/liter.
TABLE 1.
Biochemical testing of environmental E. coli isolates with the API 20E biochemical test strip
| Enzymatic testb | Test resulta for isolate: |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| β-Galactosidase (ONPG) | + | + | + | + | + | + | + |
| Arginine dihydrolase (ADH) | − | − | − | − | − | − | − |
| Lysine decarboxylase (LDC) | + | + | + | + | + | + | + |
| Ornithine decarboxylase (ODC) | − | − | + | + | + | − | − |
| Citrate utilization (CIT) | − | − | − | − | − | − | − |
| H2S production (H2S) | − | − | − | − | − | − | − |
| Urease (URE) | − | − | − | − | − | − | − |
| Tryptophan deaminase (TDA) | − | − | − | − | − | − | − |
| Indole production (IND) | + | + | + | + | + | + | + |
| Acetoin production (VP) | − | − | − | − | − | − | − |
| Gelatinase (GEL) | − | − | − | − | − | − | − |
| Fermentation/oxidation of: | |||||||
| Glucose (GLU) | + | + | + | + | + | + | + |
| Mannitol (MAN) | + | + | + | + | + | + | + |
| Inositol (INO) | − | − | − | − | − | − | − |
| Sorbitol (SOR) | + | + | + | + | − | + | + |
| Rhamnose (RHA) | + | + | + | + | + | + | + |
| Saccharose (SAC) | − | + | − | + | + | − | − |
| Melibiose (MEL) | + | + | + | + | + | + | + |
| Amygdalin (AMY) | − | + | − | − | − | − | − |
| Arabinose (ARA) | + | + | + | + | + | + | + |
| Cytochrome oxidase (OX) | − | − | − | − | − | − | − |
| Nitrate reduction (GLU) | + | + | + | NT | NT | NT | + |
+, positive result; −, negative result; NT, not tested. Results are given in boldface when they differ for different isolates.
Biochemical test, with letters in parentheses representing the respective reaction/enzyme.
16S rRNA analysis.
Three different ARDRA profiles were identified within the 7 environmental isolates, with the greatest diversity obtained on HindIII restrictions. E. coli isolates were phylogenetically closely clustered as determined by 1,399-bp 16S rRNA clone analysis. Isolates were also closely clustered with additional E. coli and Shigella sequences downloaded from the RDP database.
Multiplex analysis.
Multiplex analyses classified the isolates into E. coli clone clusters A, B1, B2, and D on the basis of the presence or absence of multiplex PCR products (8) (Fig. 1). The results indicated that isolates from at least 3 clonal groups (A, B1, and D) were leaching from Luvic Stagnosol lysimeter soils.
FIG. 1.
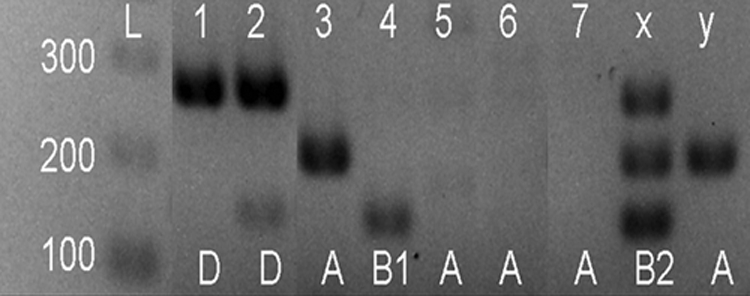
Triplex PCR method indicating genotypic groupings of environmental E. coli isolates (isolates 1 to 7) and control strains x (ATCC 25922) and y (MG1655). Lane L, ladder.
Fingerprinting.
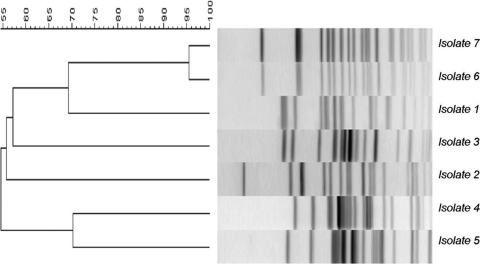
Fingerprinting methods suggested that a wide diversity of E. coli genotypes were present in leachate from the lysimeter soils. PFGE cluster analysis of the 7 environmental isolates showed similarity ranges of 100 to 55% (Fig. 2), while ERIC cluster analysis showed similarity ranges of 100 to 43%.
FIG. 2.
PFGE similarity cluster analysis, showing percentages of similarity of environmental E. coli isolates (isolates 1 to 7) leached from Luvic Stagnosol lysimeter soils.
Growth curves.
The environmental isolates tested (isolates 3 and 5) grew well across a range of temperatures under the conditions tested. They outperformed the K-12 laboratory strain at all temperatures and in all media tested (Table 2). This was particularly apparent at lower temperatures, where the environmental isolates had mean specific growth rates 2.3 to 5.4 times greater than those of K-12 on soil extract medium. Statistical analysis demonstrated that the interaction term for temperature, media, and strain was significant (P, <0.0001). Growth at 37°C was significantly greater than growth at the other two temperatures, while the growth of the environmental isolates (isolates 3 and 5) was significantly greater than that of the K-12 laboratory strain under all the conditions tested. Multiple-comparison procedures within each temperature showed that the maximum SGR for K-12 on both media were significantly (α, 0.05) lower than those of the environmental isolates in all cases except one (K-12 versus isolate 5 on soil extract medium at 37°C).
TABLE 2.
Maximum specific growth rates of two environmental isolates and a laboratory straina in anaerobic cultures at 10, 15, and 37°C
| Temp and organism | Maximum specific growth rate (h−1) ± SEb |
|
|---|---|---|
| Minimal medium | Soil extract medium | |
| 37°C | ||
| K-12 | 0.379 ± 0.029 B | 0.394 ± 0.029 B |
| Isolate 3 | 0.528 ± 0.029 A | 0.524 ± 0.029 A |
| Isolate 5 | 0.555 ± 0.029 A | 0.505 ± 0.029 A |
| 15°C | ||
| K-12 | 0.061 ± 0.031 F | 0.036 ± 0.031 H |
| Isolate 3 | 0.166 ± 0.031 D | 0.194 ± 0.031 C |
| Isolate 5 | 0.140 ± 0.031 E | 0.148 ± 0.031 E |
| 10°C | ||
| K-12 | 0.016 ± 0.017 I | 0.019 ± 0.017 I |
| Isolate 3 | 0.063 ± 0.017 F | 0.061 ± 0.017 F |
| Isolate 5 | 0.042 ± 0.017 GH | 0.044 ± 0.017 G |
Isolates 3 and 5 are environmental isolates; K-12 is a laboratory strain.
Capital letters next to growth rates represent Bonferroni groupings for within-temperature comparisons. Growth rates followed by the same letter are not significantly different.
DISCUSSION
This study was designed to characterize, by molecular and physiological means, the genetic diversity and physiology of environmentally persistent E. coli isolates leached from a maritime temperate Luvic Stagnosol soil. Molecular analysis determined the genetic diversity of the isolates, while physiological methods aimed at comparing the metabolic capacity and environmental fitness of the isolates to those of laboratory strains by examining specific growth rates under the conditions tested. We tested the hypothesis that E. coli clones persisting in the lysimeter soils would form a genetically distinct grouping. In addition, we investigated whether the environmental E. coli isolates would possess phenotypic characteristics that would confer on them a level of environmental fitness superior to that of laboratory strains under oligotrophic and low-temperature anaerobic conditions.
The tight clustering of the 16S rRNA clones and the close association with Shigella sp. strains were not unexpected and reflect the limited power of 16S rRNA sequence analysis to resolve within-species phylogenetic diversity. The wide genotypic diversity of the isolates, as determined, according to the conditions of our experiment, by both the PFGE and ERIC fingerprinting methods, provides some evidence against the first hypothesis. E. coli is known to be subject to much horizontal genetic transfer (21), so the diversity observed by both these methods may be a function of this, as the methods will concurrently detect a range of genomic change processes (36). As previously reported by others (27), we also found difficulties in reproducibility between different PCR runs with the ERIC method. The mixture of genetic groups in the soils by the multiplex method would also suggest that isolates do not form a single genetic grouping but that multiple clonal groups (A, B1, and D) are capable of surviving and proliferating in this environment. This is relevant from a health risk perspective, since it is known that the majority of Shiga toxin-producing E. coli strains belong to groups A and B1, while the majority of extraintestinal pathogenic E. coli strains belong to group B2 and, to a lesser extent, to group D (8, 21).
The rapid growth of the environmental isolates under oligotrophic conditions at 10 and 15°C was unexpected, as was the discrepancy between the specific growth rates of the laboratory strain and environmental isolates at 37°C. The results of the growth curves strongly support the hypothesis that the environmental isolates have a favorable physiology which confers on them a level of environmental fitness superior to that of the laboratory strains under the conditions tested. The survival of E. coli in soil implies the ability to overcome the challenges presented by low carbon and nutrient availability combined with temperature variability (39). If organisms can respond quickly to nutrient/carbon availability, this ability may confer a competitive advantage where these resources become available. At all temperatures, the growth of the environmental isolates far exceeded that of their laboratory counterpart. The greatest relative differences were observed at the lower temperatures. Growth at 37°C was significantly greater than growth at 10 and 15°C, suggesting that 37°C remains optimal for E. coli growth, even for environmentally adapted organisms. The two environmental isolates had similar specific growth rates. Additional work at 37°C (results not shown) found that the environmental isolates grew significantly faster than laboratory strains (K-12, ATCC 25922) on McIlvaine's minimal medium, soil extract medium, and Luria-Bertani medium. The faster growth of the environmental isolates on the latter medium suggests that, while these isolates may have adapted to soil conditions via ecological specialization, they still retain their environmental fitness under the optimal-temperature and nutrient-rich conditions characteristic of the primary host.
Our findings contrast with those of other authors who have reported the presence of genetically distinct E. coli populations in soils. Ishii et al. (20) found that naturalized E. coli in northern temperate forest soils had horizontal fluorophore-enhanced repetitive extragenic palindromic PCR (HFERP) DNA fingerprints unique to specific soils and locations, while Byappanahalli et al. (6) reported that E. coli strains isolated from similar soils had repetitive extragenic palindromic (Rep)-PCR profiles genetically distinct from those tested from animal sources. Texier et al. (34) observed environmentally distinct UidA sequence clusters in E. coli strains persisting in alpine grassland soils. Walk et al. (36) found that persistent naturalized E. coli strains on freshwater beaches belonged to the E. coli clone group B1 and suggested that the persistent genotypes had an adaptive advantage in the environment. In addition to reports of genetically distinct populations in soil, there is some direct evidence that E. coli communities can alter considerably in transition from the primary host to the environment (16). Whittam (38) found that only 10% of E. coli clones were common to domestic birds and their litter and that the environmental isolates were genetically distinct. Gordon et al. (17) subsequently found that strains recovered from a household septic tank were genetically distinct from strains removed from contributing humans.
Very little work has been reported on the physiology of autochthonous soil E. coli. Klein and Casida (23) reported that E. coli cells recovered after residence in soil appeared to have a lower growth rate and could later revert to a higher growth rate after acclimatization to laboratory media. This could, however, be considered a lag phase in which the synthesis of RNA and suitable proteins is occurring. In our work, the isolates were acclimatized to media prior to the conduct of the growth curve experiments. Gordon et al. (17) demonstrated that septic tank strains grew better than strains isolated directly from the contributing humans at lower temperatures, while at higher temperatures the opposite was apparent. This suggests ecological specialization, combined with a loss of fitness in the primary host. In contrast to these findings, we found that our isolates had a higher growth rate than laboratory strains both under conditions characteristic of the soil environment (low temperature, oligotrophic) and under conditions characteristic of the primary host (high temperature, nutrient rich).
This raises the question of how these E. coli populations with an advantageous physiology arise. They may be minor populations within the primary host, which display the favored physiology in the environment, becoming dominant by natural selection or by the die-off of less competitive strains. If this is the case, do they exist primarily in the environment, or do they retain the capacity to be returned to the primary host? Another possibility is that the organisms adapt upon arrival in the soil either by turning on different metabolic pathways or by picking up genetic elements in the soil, which are integrated into the genome. Baur et al. (2) found that marine E. coli strains were able to develop natural competency, which may enhance survival in the environment by the uptake of advantageous genetic elements. Soil E. coli strains may be able to do likewise. In addition to horizontal transfer, mutation rates of E. coli in soil could be increased by the induction of error-prone DNA polymerase under starvation conditions, increasing the genotypic and phenotypic diversity for natural selection to act on (21). If the strains do adapt upon arrival in the soil, they would have to be able to survive for the adaptation period, suggesting that the environmental conditions would have to be favorable.
In tropical soils, the growth of fecal bacteria is believed to be limited by the capacity of the indigenous soil microbiota to outcompete them for the limited supply of nutrients present in the system. However, the bacteria can proliferate sporadically when conditions are favorable (4) and thereby maintain themselves as minor populations in such soils. In the study (3) that reported the leaching of these autochthonous E. coli populations, out of four soil types studied, only the Luvic Stagnosol soil type was found to be continuously leaching E. coli, which would suggest that the characteristics of that soil type are particularly suited for the survival and growth of E. coli. Our results suggest that when conditions are favorable in the soil environment, adapted strains may be in a better position to react to available niche space, thereby maintaining their populations and facilitating their integration into the autochthonous soil microflora. Although we have not shown growth in soil itself, the capacity to grow at low temperatures under oligotrophic conditions, combined with the presence of viable cells in the leachate of control soils, suggests that growth is possible in the soil, and a physiology tailored to the soil environment may partially explain the survival and growth of E. coli under soil conditions.
This work adds further support to the hypothesis that E. coli populations can become naturalized in soil where conditions are favorable, forming a reservoir of E. coli in the environment. The resulting lack of fecal specificity has serious implications for the use of this organism as an indicator of fecal pollution in the environment. The health implications of this, where input organisms may be pathogenic, are unknown, but the presence of virulence factors, for example, in environmentally persistent E. coli strains should now be investigated. Microbiologically, these findings raise numerous interesting questions about the physiological, biochemical, and pathogenic capabilities of these environmentally isolated organisms. Further work is needed to characterize the biology of these organisms, including determination of clonal population dynamics in soil and investigation of the genetic response to growth in the secondary soil environment. In addition, the adaptive capabilities of other enteric bacteria, including pathogens, for persistence and growth in soil remain largely unexplored.
Acknowledgments
This work was carried out with the financial support of the Irish Research Council for Science, Engineering and Technology (IRCSET) and of Science Foundation Ireland.
We thank Sandra Galvin and Martin Cormican, Department of Bacteriology, National University of Ireland, Galway, for the PFGE analysis, and Denny Brennan, Teagasc, for analysis of the soil extract. We are also grateful to Jim Grant, Teagasc Kinsealy, and Laura Kirwan, Waterford Institute of Technology, for statistical assistance.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Baertsch, C., T. Paez-Rubio, E. Viau, and J. Peccia. 2007. Source tracking aerosols released from land-applied class B biosolids during high-wind events. Appl. Environ. Microbiol. 73:4522-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baur, B., K. Hanselmann, W. Schlimme, and B. Jenni. 1996. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl. Environ. Microbiol. 62:3673-3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan, F. P., V. O'Flaherty, G. Kramers, J. Grant, and K. G. Richards. 2010. Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl. Environ. Microbiol. 76:1449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byappanahalli, M., and R. Fujioka. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50(1):27-32. [PubMed] [Google Scholar]
- 5.Byappanahalli, M. N., and R. S. Fujioka. 1998. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 38(12):171-174. [Google Scholar]
- 6.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 7.Carrillo, M., E. Estrada, and T. C. Hazen. 1985. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl. Environ. Microbiol. 50:468-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Committee on Indicators for Waterborne Pathogens, National Research Council. 2004. Indicators for waterborne pathogens. National Academies Press, Washington, DC.
- 10.Crane, S. R., and J. A. Moore. 1986. Modeling enteric bacterial die-off: a review. Water Air Soil Pollut. 27:411-439. [Google Scholar]
- 11.Delbès, C., R. Moletta, and J.-J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 12.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, A. D., L. S. Clesceri, E. W. Rice, and A. E. Greenberg (ed.). 2005. Standard methods for the examination of water and wastewater, 21st ed. American Public Health Association, Washington, DC.
- 14.Fujioka, R. S., and M. N. Byappanahalli. 2001. Microbial ecology controls the establishment of fecal bacteria in tropical soil environment, p. 273-283. In T. Matsuo, K. Hanaki, S. Takizawa, and H. Satoh (ed.), Advances in water and wastewater treatment technology: molecular technology, nutrient removal, sludge reduction and environmental health. Elsevier, Amsterdam, Netherlands.
- 15.Garcia, M. M., and K. A. McKay. 1970. Pathogenic microorganisms in soil: an old problem in a new perspective. Can. J. Comp. Med. 34:105-110. [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079-1085. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 18.Hammer, Ø., D. A. T. Harper, and P. D. Ryan. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:4-9. [Google Scholar]
- 19.Hartz, A., M. Cuvelier, K. Nowosielski, T. D. Bonilla, M. Green, N. Esiobu, D. S. McCorquodale, and A. Rogerson. 2008. Survival potential of Escherichia coli and enterococci in subtropical beach sand: implications for water quality managers. J. Environ. Qual. 37:898-905. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii, S., and M. J. Sadowsky. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23:101-108. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, S. C., W. Chu, B. H. Olson, J. W. He, S. Choi, J. Zhang, J. Y. Le, and P. B. Gedalanga. 2007. Microbial source tracking in a small southern California urban watershed indicates wild animals and growth as the source of fecal bacteria. Appl. Microbiol. Biotechnol. 76:927-934. [DOI] [PubMed] [Google Scholar]
- 23.Klein, D. A., and L. E. Casida, Jr. 1967. Escherichia coli die-out from normal soil as related to nutrient availability and the indigenous microflora. Can. J. Microbiol. 13:1461-1470. [DOI] [PubMed] [Google Scholar]
- 24.Lang, N. L., S. R. Smith, D. M. Bellett-Travers, and C. L. Rowlands. 2003. Decay of Escherichia coli in soil following the application of biosolids to agricultural land. Water Environ. J. 17:23-28. [Google Scholar]
- 25.López-Torres, A. J., T. C. Hazen, and G. A. Toranzos. 1987. Distribution and in situ survival and activity of Klebsiella pneumoniae and Escherichia coli in a tropical rain forest watershed. Curr. Microbiol. 15:213-218. [Google Scholar]
- 26.McFeters, G. A., S. C. Broadaway, B. H. Pyle, M. Pickett, and Y. Egozy. 1997. Comparative performance of Colisure. J. Am. Water Works Assoc. 89:112-120. [PubMed] [Google Scholar]
- 27.Rasschaert, G., K. Houf, H. Imberechts, K. Grijspeerdt, L. De Zutter, and M. Heyndrickx. 2005. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol. 43:3615-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, M., and A. Fanning. 1996. Effects of fertiliser N and slurry on nitrate leaching—lysimeter studies on 5 soils. Irish Geogr. 29:126-136. [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Sjogren, R. E. 1995. Thirteen-year survival study of an environmental Escherichia coli in field mini-plots. Water Air Soil Pollut. 81:315-335. [Google Scholar]
- 32.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 34.Texier, S., C. Prigent-Combaret, M. H. Gourdon, M. A. Poirier, P. Faivre, J. M. Dorioz, J. Poulenard, L. Jocteur-Monrozier, Y. Moenne-Loccoz, and D. Trevisan. 2008. Persistence of culturable Escherichia coli fecal contaminants in dairy Alpine grassland soils. J. Environ. Qual. 37:2299-2310. [DOI] [PubMed] [Google Scholar]
- 35.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 37.Whitman, R. L., M. B. Nevers, and M. N. Byappanahalli. 2006. Examination of the watershed-wide distribution of Escherichia coli along Southern Lake Michigan: an integrated approach. Appl. Environ. Microbiol. 72:7301-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittam, T. 1989. Clonal dynamics of Escherichia coli in its natural habitat. Antonie Van Leeuwenhoek 55:23-32. [DOI] [PubMed] [Google Scholar]
- 39.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]