Abstract
In this study, we examined the effect of various pooling strategies on the characterization of soil microbial community composition and phylotype richness estimates. Automated ribosomal intergenic spacer analysis (ARISA) profiles were determined from soil samples that were (i) unpooled (extracted and amplified individually), (ii) pooled prior to PCR amplification, or (iii) pooled prior to DNA extraction. Regression analyses suggest that the less even the soil microbial community (i.e., low Shannon equitability, EH), the greater was the impact of either pooling strategy on microbial detection (R2 = 0.766). For example, at a tropical rainforest site, which had the most uneven fungal (EH of 0.597) and bacterial communities (EH of 0.822), the unpooled procedure detected an additional 67 fungal and 115 bacterial phylotypes relative to either of the pooled procedures. Phylotype rarity, resulting in missed detection upon pooling, differed between the fungal and bacterial communities. Fungi were typified by locally abundant but spatially rare phylotypes, and the bacteria were typified by locally rare but spatially ubiquitous phylotypes. As a result, pooling differentially influenced plot comparisons, leading to an increase in similarity for the bacterial community and a decrease in the fungal community. In conclusion, although pooling reduces sample numbers and variability, it could mask a significant portion of the detectable microbial community, particularly for fungi due to their higher spatial heterogeneity.
Microbial communities in soils are extremely complex, with heterogeneity expressed on a wide variety of scales (6-9, 16). Therefore, soil sampling strategies typically combine multiple small samples, obtained from various locations within the area of interest, into a single homogenized sample that is then subsampled for analysis. Previous studies (5, 11, 15) have compared the sizes of the subsamples to best represent the microbial diversity in the pooled samples. Larger sample sizes are typically recommended for community profiling (5, 11, 15) because they can reduce variability in the subsample and appear to adequately capture the dominant members of the community (3, 11). Conversely, multiple small subsamples have been proposed to be better suited for identifying rare community members and estimating species richness (10-11). While previous studies have largely been conducted to determine the variability of the subsample—and, hence, its ability to represent the larger, homogenized sample—the impact of soil sample size and pooling to best represent the site of interest and its influence on plot comparisons has not been adequately explored. For example, “rare” species in the pooled, homogenized sample may arise from two different scenarios: (i) species are found in high abundance at fine scales but are heterogeneously spaced, and (ii) species are found in low abundance but are ubiquitously distributed. Furthermore, detection of rare species could be problematic with molecular approaches that rely on PCR for amplification and detection.
Although molecular techniques can detect many microbial species missed by traditional culturing (20), they suffer from several potential biases (14, 17) that may limit successful PCR amplification and detection. For example, because PCR is a competitive process, species with a low relative abundance will be amplified to a lesser degree and may not reach detection threshold levels. This process is routinely utilized in competitive PCR to analyze starting template concentrations in mixed nucleic acid samples (17, 19). Furthermore, this effect would be seen by any process that could dilute rare phylotypes, such as pooling DNA extracts prior to amplification. Therefore, if the microbial community in the starting template is too complex, reducing the soil sample size will increase the likelihood that less abundant species are successfully amplified and detected.
In this study, we analyzed the influence of three different sampling strategies on microbial community profiling using automated ribosomal intergenic spacer analysis (ARISA) and the following types of samples: (i) unpooled, (ii) pooled prior to PCR amplification, or (iii) pooled prior to DNA extraction. This sampling scheme was designed to test the effects of different common sampling strategies on microbial community profiles of samples containing equal soil volumes. Studies were conducted at three different field sites with various types of plant overstory complexity: an agricultural corn field, a ponderosa pine forest, and a tropical rainforest.
MATERIALS AND METHODS
Site selection.
Three field sites were selected for analysis to represent a wide range of complexity and aboveground plant biodiversity. The first site was an agricultural corn field located at the Colorado State University Agricultural Research Development and Education Center (ARDEC) near Wellington, CO; the second site was a tropical rainforest in the Tambopata National Reserve (TNR), Peru; and the third site was a ponderosa pine forest located at Young's Gulch YG in the Roosevelt National Forest near Mishawaka, CO (Table 1).
TABLE 1.
Site descriptions and soil characteristics
| Site characteristic |
Soil characteristic |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Description | Latitude | Longitude | Elevation (ft) | Texture | pH | Relative lime content | OM (%)a | NO3-N (ppm) | P (ppm) | K (ppm) |
| ARDEC | Agricultural corn field | N40°39′07" | W104°59′58" | 5,094 | Clay | 8.1 | Very high | 3.1 | 6.2 | 19.9 | 399 |
| N40°39′04" | W104°59′58" | 5,091 | Clay | 8.1 | Very high | 3.3 | 8.0 | 13.1 | 357 | ||
| N40°39′03" | W104°59′58" | 5,090 | Clay | 7.8 | Very high | 3.3 | 36.4 | 36.1 | 390 | ||
| TNR | Tropical forest | S77°18′55" | W70°11′47" | 663 | Clay | 3.5 | Low | 5.2 | 12.9 | 6.2 | 92 |
| S77°18′54" | W70°11′52" | 656 | Clay loam | 3.3 | Low | 17.3* | 14.6 | 8.7 | 147 | ||
| S77°18′54" | W70°11′60" | 656 | Clay | 3.5 | Low | 11.6* | 17.1 | 18.7 | 110 | ||
| YG | Ponderosa pine | N40°41′17" | W105°20′49" | 5,862 | Loam | 5.8 | Low | 10.9* | 2.5 | 9.9 | 289 |
| N40°40′37" | W105°20′55" | 6,181 | Sandy loam | 5.8 | Low | 5.2 | 0.7 | 2.5 | 235 | ||
| N40°40′20" | W105°20′50" | 6,362 | Sandy loam | 7.1 | Medium | 11.3 | 6.4 | 16.8 | 641 | ||
OM, organic matter. *, expressed as weight loss on ignition.
Soil sampling.
At each site, soil was collected from the top 0 to 5 cm from three different plots. Each plot consisted of three transects radiating out from a single plant at 90° angles, with sampling locations at 90, 120, and 240 cm from the base of the plant. To address the impact of pooling procedures on microbial detection, three different procedures were used to assess microbial richness at each plot. For unpooled samples (procedure I), nine 0.5-g soil samples per plot were extracted (total volume, 4.5 g) and amplified individually (180 ng of DNA in nine biological replicates); for samples pooled prior to PCR amplification (procedure II), aliquots (10 μl) from each of the nine DNA extracts generated in the unpooled procedure were combined to generate a single, pooled DNA extract, which was used in nine separate reactions (180 ng of DNA in nine technical replicates); for samples pooled prior to DNA extraction (procedure III), nine 0.5-g soil samples per plot were combined and extracted as a single 4.5-g soil sample, and the DNA was used in nine separate PCRs (180 ng of DNA in nine technical replicates). Technical replicates were included in procedures II and III to account for the effects of multiple runs inherent in procedure I and to standardize the total amount of PCR-amplified DNA in all three procedures. In addition, procedure II has the added benefit of examining the effect of pooling without introducing potential differences due to the use of a different DNA extraction kit, as was necessary for the soil sample in procedure III. DNA extraction of 0.5-g samples was performed using a MoBio UltraClean-htp 96-well Soil DNA Isolation Kit (Carlsbad, CA), whereas DNA extraction from 4.5-g samples was performed using a MoBio UltraClean Mega Soil DNA Isolation Kit using the manufacturer's recommendations plus an additional purification step with AMPure beads (Agencourt) to allow additional removal of humic acids or other PCR inhibitors. All DNA extractions were quantified spectrophotometrically and diluted to a final concentration of 10 ng μl−1.
ARISA.
PCRs were conducted for each plot/sampling procedure as follows. Fungi were amplified using the primers 2234C (5′ hexachlorofluorescein [HEX] label) and 3126T (18), and bacteria were amplified with the primers S-D-Bact-1522-b-S-20 (5′ tetrachloro-6-carboxyfluorescein [TET] label) and L-D-Bact-132-a-A-18 (13). PCR mixtures contained 2 μl (10 ng μl−1) of soil DNA, 10 μl of 2× Jumpstart reaction mixture (Sigma, St. Louis, MO), 2.4 μl of 25 mM MgCl2, 0.2 μl of 1 mM fluorescein, and 0.4 μl of 10 mM forward and reverse primers and were brought to 20 μl with distilled H2O (dH2O). The PCR products were amplified for 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s. PCRs were diluted with 80 μl of distilled water, and a 2-μl aliquot from each fungal and bacterial reaction mixture was added to 10 μl of a loading buffer (1,250 μl of formamide and 50 μl of Genescan 2500 [6-carboxytetramethylrhodamine (TAMRA)] size standard) and analyzed directly by capillary electrophoresis (ABI Prism 310; Applied Biosystems, Foster City, CA) without further modification, i.e., denaturation heating. Electrophoresis conditions were as follows: 47-cm capillary, Genescan POP4 polymer, 15-s injection for 15 kV, and 45-min electrophoresis at 15 kV. Amplicons were assigned to 1.5-bp bins using the automated binning procedure contained in the Genemapper software (version 4). Phylotype richness of each sample was determined as the number of amplicons (bins) above a threshold of 50 relative fluorescence units (RFU), with the understanding that any given peak may contain amplicons from multiple species (12). Phylotype richness (observed; Sobs), total estimated phylotype richness (Schao), Shannon diversity (H′), and Jaccard similarity indices (presence/absence data; Jclass) were determined for each location using the nine replicate ARISA profiles from rarefaction curves constructed with the aid of EstimateS (4). Statistical differences in community characteristics were analyzed using a two-way repeated measures analysis of variance (ANOVA), with site and sampling as the main effects and plots as the subject using PROC MIXED in SAS software, version 9.1 (Cary, NC). All pairwise comparisons were adjusted using Tukey's honestly significant differences (HSD) test.
Soil characteristics.
For each sampling location (nine per plot), an approximately 2-g subsample was randomly drawn and combined to generate a single soil sample for each plot and was then analyzed for routine soil characteristics by the Soil, Water, and Plant Testing Laboratory at Colorado State University.
RESULTS
Treatment-specific effects.
There was a significant main effect for sampling procedure on estimates of phylotype richness (Sobs and Schao) for both fungi and bacteria (Table 2; see also Tables S1 and S2 in the supplemental material). For Sobs, pooling resulted in the missed detection of at least 20 fungal and 78 bacterial phylotypes, and for Schao pooling resulted in the missed detection of at least 51 fungal and 111 bacterial phylotypes (Table 2). A visual inspection of the raw chromatograms reveals a number of amplicons that are unique to individual samples with the unpooled procedure (see Fig. S1A in the supplemental material) while the main variability between samples for either of the two pooling procedures is largely related to differences in peak heights (Fig. S1B). Because the strategy of pooling samples prior to PCR used the exact same soil samples and DNA extracts, no differences can be attributed to the selection of soil samples or DNA extraction.
TABLE 2.
Phylotype richness and Jaccard similarity between replicate soil fungal and bacterial ARISA profiles at three different field sites using three sampling procedures
| Group and procedurea | Procedure effectb |
||
|---|---|---|---|
| Sobs | Schao | Jclass | |
| Bacteria | |||
| I | 286 A | 361 A | 0.363 B |
| II | 208 B | 235 B | 0.582 A |
| III | 203 B | 250 B | 0.583 A |
| Fungi | |||
| I | 60 A | 98 A | 0.321 B |
| II | 39 B | 44 B | 0.638 A |
| III | 40 B | 42 B | 0.635 A |
I, no pooling; II, pooled prior to PCR; III, pooled prior to DNA extraction.
Reported values are the LSmean. Means with different letters are significantly different (Tukey's HSD, P < 0.05).
Site-specific observations.
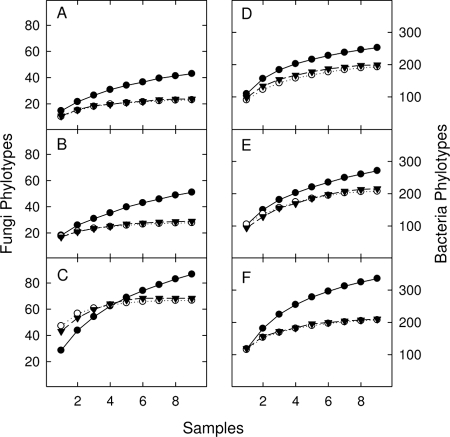
Both pooling strategies negatively affected the total number of observed phylotypes for both bacteria and fungi and masked variability in phylotype richness between sites compared to the unpooled method (Table 3). Estimates of phylotype richness for each site were determined from rarefaction curves constructed from the nine biological (procedure I, unpooled) and nine technical (procedure II, pooled prior to PCR, or procedure III, pooled prior to DNA extraction) replicates (Fig. 1). For Schao, there was a significant interaction for both fungi and bacteria (see Table S2 in the supplemental material). In both cases, only the unpooled sampling procedure was able to detect significant differences between sites (Table 3), with the lowest levels of both fungal and bacterial richness detected at the agricultural ARDEC sites.
TABLE 3.
Phylotype richness and Jaccard similarity between replicate soil fungal and bacterial ARISA profiles at three different field sites using three sampling procedures
| Group and site | Value for the parameter with the indicated procedurea |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Sobs |
Schao |
Jclass |
|||||||
| I | II | III | I | II | III | I | II | III | |
| Bacteria | |||||||||
| ARDEC | 252 BC | 193 D | 199 D | 295 BC | 234 C | 240 C | 0.405 B | 0.597 A | 0.590 A |
| TNR | 271 B | 207 CD | 215 CD | 368 AB | 253 C | 252 C | 0.362 B | 0.592 A | 0.584 A |
| YG | 336 A | 208 CD | 210 CD | 419 A | 219 C | 257 C | 0.323 B | 0.557 A | 0.575 A |
| Fungi | |||||||||
| ARDEC | 43 BC | 23 C | 24 C | 52 C | 30 C | 30 C | 0.315 B | 0.594 A | 0.608 A |
| TNR | 51 BC | 28 C | 29 C | 103 AB | 29 C | 29 C | 0.321 B | 0.641 A | 0.633 A |
| YG | 87 A | 67 AB | 68 AB | 139 A | 67 BC | 72 BC | 0.260 B | 0.679 A | 0.665 A |
I, no pooling: II, pooled prior to PCR; III, pooled prior to DNA extraction. Reported values are the LSmean for the site and procedure interaction. Means with different letters are significantly different (Tukey's HSD, P < 0.05).
FIG. 1.
Site-averaged rarefaction curves for fungi (A to C) and bacteria (D to F) determined using three sampling strategies. Filled circles, no pooling; open circles, pooled prior to PCR; filled triangles, pooled prior to DNA extraction. At each site, three separate rarefaction curves were generated using the nine biological (no pooling) or nine technical (pooled procedures) replicates from each plot. Sites represented in the panels are as follows: ARDEC corn field (A and D), TNR tropical rainforest (B and E), and YG ponderosa pine forest (C and F).
As community evenness decreased, the unpooled sampling procedure showed a greater ability to detect additional phylotypes in the sample. We observed a good linear relationship (R2 = 0.766) between Shannon's equitability index and the percent increase in total phylotype richness detected at each plot (see Fig. S2 in the supplemental material). In particular, at the site with the lowest evenness, TNR, Schao increased by approximately 250% and 50% for the fungi and bacteria, respectively.
The similarity between replicate ARISA profiles increased with pooling (Table 2; see also Table S3 in the supplemental material), which is consistent with the associated loss of phylotype detection. The effect of pooling on within-plot Jclass estimates appears to be greater for the fungi than for the bacteria, with an increase of 0.314 and 0.219, respectively. Furthermore, based on Jclass estimates with the more sensitive unpooled procedure, there appears to be higher variability in the fungal community than in bacteria. For example, Jclass values were lower in the fungal than in the bacterial community by 0.090, 0.041, and 0.063 units for ARDEC, TNR, and YG, respectively (Table 3).
Between-plot comparisons.
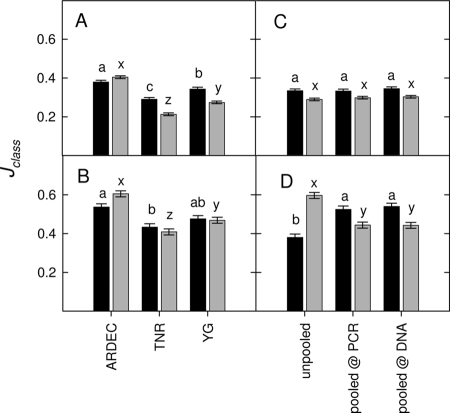
How does the additional detection of microbial phylotypes influence the relatedness of different samples? We hypothesized that if the additional phylotypes are rare but ubiquitous, then greater phylotype detection would cause relatedness to increase, whereas if the additional phylotypes are heterogeneously spaced, then relatedness will decrease. In this study, we found evidence of both cases. Jclass indices were calculated between each of the three replicate plots at each site. When each individual ARISA profile was compared, we saw a significant difference between sites (for fungi, P < 0.001; for bacteria, P = 0.002) but not between sampling procedures (for fungi, P = 0.640; for bacteria, P = 0.395) (see Table S4 in the supplemental material). ARDEC had the highest similarity between plots, followed by YG and finally TNR (Fig. 2A). When a composite profile was determined for each plot (i.e., average of all nine replicates), the similarity increased but still revealed a similar pattern such that similarity was greatest at ARDEC, followed by YG and TNR (Fig. 2B). Interestingly, similarity between plots was differentially affected for the fungi and bacteria due to the sampling procedure chosen (Table S4). For example, the unpooled procedure resulted in a significant decline in similarity for the fungi but an increase in similarity for the bacteria, and this effect was seen only when plots were compared using the composite profile for each plot (Fig. 2C and D).
FIG. 2.
Average Jaccard similarity index (Jclass) between the three plots at each site (ARDEC corn field, TNR tropical rainforest, and YG ponderosa pine forest). Jclass (phylotype presence/absence) was calculated between the three plots at each site using each individual ARISA profile (A and C) or a single composite ARISA profile (i.e., average of all nine biological [no pooling] or nine technical [pooled procedures] replicates) for each plot (B and D). Averages are reported by site (A and B) or sampling procedure (C and D). Black bars, fungi; gray bars, bacteria. Bars are the LSmeans with the pooled standard error (SE); bars with different letters denote significant Tukey's HSD test results (P < 0.05).
DISCUSSION
Due to the complexity and heterogeneous nature of soil microbial communities, multiple, small samples taken from throughout a plot are frequently pooled and homogenized to generate a soil sample that is then subsampled and assumed to be representative of the original plot of interest. In a variety of studies, the appropriate size of the subsample has been examined in detail (5, 11, 15), and the sample size that results in the lowest variation is often assumed to best represent the underlying plot. As a result of these studies, two types of samples are typically recommended: (i) small, multiple samples appear to be beneficial for species richness estimates (9, 11); (ii) larger, pooled samples are better suited to minimize plot/treatment variability and to estimate the dominant microbial community structure (5, 11, 15). In this study, we examined the effect of two common pooling strategies, pooling prior to DNA extraction or prior to PCR amplification, at three different sites to further examine how pooling may influence microbial community estimates.
As expected, we found that pooling at any step in the preparation of soil DNA for ARISA significantly reduced the detectable phylotype richness and that this effect differed between sites. Based on a significant regression analysis, the data suggest that site differences are best related to differences in the evenness of the microbial community. Furthermore, this effect is consistent with the competitive nature of PCR. In mixed-template PCR (e.g., competitive PCR), the level of amplification achieved is positively correlated with the amount of starting template DNA; thus, as relative abundance is reduced, amplification of rare templates/phylotypes may not be sufficient for detection. Thus, as evenness decreases, a greater portion of phylotypes will become undetectable, particularly when samples are pooled to generate large, homogenized soil samples.
When estimates of total phylotype richness are not the primary research objective, it has been suggested that larger, homogenized samples can adequately capture the heterogeneity and dominant microbial community (11). Interestingly, we observed two different scenarios when we compared fungal or bacterial communities between plots located within a single site. For example, it was observed that pooling resulted in a decline in the similarity of bacterial communities between plots, whereas pooling led to an increase in similarity of fungal communities between plots. We suggest that this effect is due to a differential pattern of rare fungal and bacterial species in the pooled, homogenized sample, where the bacterial species are ubiquitously distributed but of a low abundance and where the fungi are heterogeneously spaced with a higher local abundance. To support this conclusion, we observed that the average abundance of the additional phylotypes detected with the unpooled procedure was 480 ± 128 and 72 ± 59 RFU for the fungi and bacteria, respectively.
Although pooling may be employed to reduce sample numbers and variability, it can clearly influence estimates of microbial phylotype richness and diversity. However, based on our results, because pooling removes the spatial heterogeneity of the underlying sample, many locally dominant but spatially rare phylotypes will become “rare” in the final sample, rendering them undetectable. Although ARISA has a fairly coarse resolution compared to rRNA sequencing, this effect is consistent with the competitive nature of PCR and will be present to some degree in all PCR-dependent methods of microbial detection. As a result, we recommend the use of multiple small samples, as opposed to fewer large samples, to characterize microbial communities, particularly when the community is suspected to be highly uneven and spatially variable. Unfortunately, this recommendation cannot easily be applied without a priori knowledge of the underlying soil microbial community. Therefore, based on the current study, we further suggest the use of the unpooled procedure, particularly for the analysis of fungal communities or at sites with high plant overstory complexity (i.e., the TNR site in this study). In regard to the latter, plant-microbial communities are highly linked, and plants have been shown to cultivate their own microbial communities (1-2). Thus, as plant richness and heterogeneity increase, so should the richness and heterogeneity of the underlying soil microbial community; however, unless the sampling scheme can account for this heterogeneity, locally dominant species will become rare and undetectable.
Supplementary Material
Acknowledgments
We thank Rachel Stong and Brittelle Bowers for their help with the DNA extractions and laboratory analysis. We also thank Max Gunther and the staff of the Explorer's Inn for assisting with logistics related to Tambopata sample collection.
Funding in J.M.V.'s laboratory was provided by the National Science Foundation (MCB-0542642). J.M.V. also acknowledges additional funding provided for the studies in Tambopata by NSF International Programs, the Fulbright Commission, and the Guggenheim Memorial Foundation.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print on 5 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Broeckling, C. D., A. K. Broz, J. Bergelson, D. K. Manter, and J. M. Vivanco. 2008. Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 74:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broz, A. K., D. K. Manter, and J. M. Vivanco. 2007. Soil fungal abundance and biodiversity: another victim of the invasive plant Centaurea maculosa. ISME J. 1:763-765. [DOI] [PubMed] [Google Scholar]
- 3.Chandler, D. P., S. M. Li, C. M. Spadoni, G. R. Drake, D. L. Balkwill, J. K. Fredrickson, and F. J. Brockman. 1997. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol. Ecol. 23:131-144. [Google Scholar]
- 4.Colwell, R. K. 2006. EstimateS: statistical estimation of species richness and shared species from samples, version 8. http://viceroy.eeb.uconn.edu/estimates.
- 5.Ellingsoe, P., and K. Johnsen. 2002. Influence of soil sample sizes on the assessment of bacterial community structure. Soil Biol. Biochem. 34:1701-1707. [Google Scholar]
- 6.Franklin, R. B., L. K. Blum, A. McComb, and A. L. Mills. 2002. A geostatistical analysis of small-scale spatial variability in bacterial abundance and community structure in salt-marsh creek bank sediments. FEMS Microbiol. Ecol. 42:71-80. [DOI] [PubMed] [Google Scholar]
- 7.Franklin, R. B., and A. L. Mills. 2003. Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 44:335-346. [DOI] [PubMed] [Google Scholar]
- 8.Green, J. L., A. J. Holmes, M. Westoby, I. Oliver, D. Briscoe, M. Dangerfield, M. Gillings, and A. J. Beattie. 2004. Spatial scaling of microbial eukaryote diversity. Nature 432:747-750. [DOI] [PubMed] [Google Scholar]
- 9.Grundmann, L. G., and D. Debouzie. 2000. Geostatistical analysis of the distribution of NH4+ and NO2− oxidizing bacteria and serotypes at the millimeter scale along a soil transect. FEMS Microbiol. Ecol. 34:57-62. [DOI] [PubMed] [Google Scholar]
- 10.Grundmann, L. G., and F. Gourbière. 1999. A microsampling approach to improve the inventory of bacterial diversity in soil. Appl. Soil Ecol. 13:123-126. [Google Scholar]
- 11.Kang, S., and A. L. Mills. 2006. The effect of sample size in studies of soil microbial community structure. J. Microbiol. Methods 66:242-250. [DOI] [PubMed] [Google Scholar]
- 12.Manter, D. K., and J. M. Vivanco. 2007. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Methods 71:7-14. [DOI] [PubMed] [Google Scholar]
- 13.Normand, P., C. Ponsonnet, X. Nesme, M. Neyra, and P. Simonet. 1996. ITS analysis of prokaryotes, p. 1-12. In D. L. Akkermans, J. D. van Elsas, and E. I. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic, Amsterdam, Netherlands.
- 14.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjard, L., D. P. Lejon, C. Mougel, L. Schehrer, D. Merdinoglu, and R. Chaussod. 2003. Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ. Microbiol. 5:1111-1120. [DOI] [PubMed] [Google Scholar]
- 16.Ranjard, L., and A. Richaume. 2001. Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152:707-716. [DOI] [PubMed] [Google Scholar]
- 17.Reysenbach, A. L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sequerra, J., R. Marmeisse, G. Valla, P. Normand, A. Capellano, and A. Moiroud. 1997. Taxonomic position and intraspecific variability of the nodule forming Penicillium nodositatum inferred from RFLP analysis of the ribosomal intergenic spacer and random amplified polymorphic DNA. Mycol. Res. 101:465-472. [Google Scholar]
- 19.Siebert, P. D., and J. W. Larrick. 1992. Competitive PCR. Nature 359:557-558. [DOI] [PubMed] [Google Scholar]
- 20.Torsvik, V. J., J. Gokoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.