Abstract
In agricultural cropping systems, crop residues are sources of organic carbon (C), an important factor influencing denitrification. The effects of red clover, soybean, and barley plant residues and of glucose on denitrifier abundance, denitrification gene mRNA levels, nitrous oxide (N2O) emissions, and denitrification rates were quantified in anoxic soil microcosms for 72 h. nosZ gene abundances and mRNA levels significantly increased in response to all organic carbon treatments over time. In contrast, the abundance and mRNA levels of Pseudomonas mandelii and closely related species (nirSP) increased only in glucose-amended soil: the nirSP guild abundance increased 5-fold over the 72-h incubation period (P < 0.001), while the mRNA level significantly increased more than 15-fold at 12 h (P < 0.001) and then subsequently decreased. The nosZ gene abundance was greater in plant residue-amended soil than in glucose-amended soil. Although plant residue carbon-to-nitrogen (C:N) ratios varied from 15:1 to 30:1, nosZ gene and mRNA levels were not significantly different among plant residue treatments, with an average of 3.5 × 107 gene copies and 6.9 × 107 transcripts g−1 dry soil. Cumulative N2O emissions and denitrification rates increased over 72 h in both glucose- and plant-tissue-C-treated soil. The nirSP and nosZ communities responded differently to glucose and plant residue amendments. However, the targeted denitrifier communities responded similarly to the different plant residues under the conditions tested despite changes in the quality of organic C and different C:N ratios.
Denitrification is the enzymatic, stepwise reduction of nitrate and nitrite to the gases nitric oxide (NO), nitrous oxide (N2O), and molecular nitrogen (N2). A series of reductases, including nitrate reductase (Nar), nitrite reductase (Nir), nitric oxide reductase (Nor), and nitrous oxide reductase (Nos), are in the denitrification respiratory chain. Nitrous oxide is a known greenhouse gas with a global warming potential 296 times that of carbon dioxide (CO2) (29), resulting in serious environmental concerns. Denitrification in agricultural soils is a leading source of anthropogenic N2O, especially in humid climates (4).
Crop residues are important sources of C and N in agricultural cropping systems (28). Crop residues affect the rate of denitrification (27), primarily by increasing organic C availability, one of the most important factors influencing denitrification (39). The rate of plant tissue decomposition depends on the physical characteristics of the plant residues, such as residue particle size and the quality of the crop residue. The amount of available N (25) and the C-to-N (C:N) ratio (2) are often the best indicators of the availability of C in plant residues; however, they do not explain the differences in plant residue decomposition rates in all studies (28).
Increases in rates of denitrification enzyme activity (DEA) and N2O emissions in soils have been observed after the incorporation of crop plant residues (17, 27), and the addition of plant residues has been found to influence the composition and diversity of the denitrifying community (11, 33). Plant residues also influence the partitioning of gaseous losses to N2O and N2; plant residues with lower C:N ratios increase the amount of N2O produced compared with N2, while N2 is the predominant gaseous product from soil amended with plant residues with higher C:N ratios (24).
Denitrifier community abundances in environmental samples, including agricultural soils (13, 26, 30, 31), estuarine sediment (16), and activated sludge (1, 19), have been quantified. Miller et al. (31) found a positive correlation between the abundance of Pseudomonas mandelii (a predominant culturable denitrifier from agricultural soil in New Brunswick, Canada [12]) and C addition. Miller et al. (30) also found increases in the abundances of P. mandelii and related species (cnorBP) and the nosZ-bearing community after the addition of plant residues to soil. In another study, denitrifier community abundance increased in soil microcosms after the addition of artificial rhizosphere exudates (AREs), with the exception of the nosZ-bearing community (23).
Quantitative reverse transcriptase (RT) PCR (qRT-PCR) has been used to quantify the mRNA levels of genes involved in denitrification, which provides insight into active denitrifier communities (6, 36). This technique has been used to evaluate denitrification gene expression in Thiobacillus denitrificans under denitrifying conditions (6) and to quantify the abundance of denitrification gene transcripts in estuarine sediments (36). To date, there have been no studies to assess the effect of complex C sources, such as crop plant residues, on denitrification gene expression in agricultural soils.
The objective of this research was to evaluate the effect of complex C sources on denitrifier abundance and mRNA levels in an agricultural soil. Our hypothesis was that the addition of different plant residues would result in different levels of abundance and/or denitrification mRNA levels due to differences in C availability, as indicated by differences in C:N ratios of the plant residues, and that greater C availability would stimulate greater denitrifier abundance and/or activity. We also hypothesized that the increase in N2O emissions observed after the addition of plant residues was due to an increase in the denitrifier abundance and/or denitrification mRNA levels. Glucose was used as a simple and readily metabolized organic C source for comparative purposes. Complex C sources included plant residues from two legume species, red clover (Trifolium pratense cultivar AC Endure) and soybean (Glycine max cultivar 90M101), and one nonlegume, barley straw (Hordeum vulgare cultivar Encore). The complex C sources were chosen to reflect representative rotation crops used in potato production in Atlantic Canada.
MATERIALS AND METHODS
Soil characteristics and sampling.
Soil was collected from a 0- to 15-cm depth from a field in potato (Solanum tuberosum)-spring wheat (Triticum aestivum L.) rotation in Fredericton, New Brunswick, Canada (45°52′N, 66°31′W). The soil was collected in September 2007 after potato harvesting. The soil was coarse loamy till and contained 111 g kg−1 clay, 345 g kg−1 sand, and 543 g kg−1 silt as determined by a pipette method with organic matter removal. The soil pH was 6.0, as determined by a 1:1 soil-water suspension. The organic C concentration was 25.5 g kg−1, and the total N concentration was 1.70 g kg−1, as determined by combustion (Leco CNS-1000) (9, 32). Soil was frozen at −20°C (6 months) to limit biological activity. Fields in New Brunswick, Canada, are snow covered/frozen for at least 4 months each winter, so soil communities are commonly subjected to similar conditions. At the start of the experiment, soil was thawed at room temperature, air dried to a gravimetric water content of 0.30 g g−1 dry soil, homogenized, and passed through a 2-mm sieve. The soil was stored in the dark at room temperature for 5 days prior to use.
Plant tissue preparation.
Soybean, red clover, and barley were seeded on 9 May 2007 and were sampled on 26 July 2007. After 78 days of growth, the red clover crop was ready for its first harvest, the soybean crop was flowering, and the barley crop was not yet mature. The plant tissues used as C treatments were an above-ground plant biomass of red clover and soybean and only stalks of barley straw. Collected plant tissues were dried at 55°C and ground for passage through a 2-mm screen. The total C and N contents of the plant tissue were determined by combustion with an Elementar varioMACRO apparatus (Elementar Americas Inc., Mt. Laurel, NJ). Total C concentrations were similar in the plant tissues, with values of 444, 422, and 432 g C kg−1 for barley straw, soybean, and red clover residues, respectively. However, the total N concentration varied among plant tissues, with values of 14.8, 18.8, and 36.8 g N kg−1 for barley straw, soybean, and red clover residues, respectively. The C:N ratios were 30:1, 22:1, and 15:1 for barley straw, soybean, and red clover, respectively. The C:N ratio of the barley straw was relatively lower than expected from data reported previously by Miller et al. (30) because the straw was not completely mature at the time of sampling.
Experimental design.
The experiment was a randomized complete-block design, with incubation time and C treatment as fixed factors and six replicates. The experimental blocks were three time periods (1 week each), and in each block, two completely randomized replicates of the experiment were conducted. There were duplicate soil jars, one incubated with the addition of 10% (vol/vol) acetylene (C2H2; inhibits the reduction of N2O to N2) to quantify total denitrification and one incubated without the addition of C2H2 to quantify N2O emissions, for each treatment combination. Treatments included a control with no C amendment (C0) and amendment with 1,000 mg C kg−1 dry soil as glucose (G1000), red clover (RC1000), soybean (S1000), or barley straw (B1000). Each treatment included 500 mg N kg−1 dry soil as potassium nitrate (KNO3) to ensure a nonlimiting nitrate supply. The required quantity of C treatment was added to approximately 180 g soil (quantity required for an individual jar) and incorporated into the soil by mixing. Nitrate solution was added to obtain a water content equivalent to a water-filled pore space (WFPS) of 70%. The soil was then packed into 250-ml canning jars (Bernardin, Toronto, ON, Canada) with a 2-cm headspace. The jars were sealed with a screw lid fitted with a rubber septum, evacuated to 2 torr, flushed with nitrogen (N2) gas, and equilibrated to atmospheric pressure. The sealed jars were placed immediately into an incubation chamber at 25°C for 72 h. Each jar was packed in a time series so that there was less than 2 min between the addition of carbon treatment and the start of incubation. Treated soils were destructively sampled at 0, 2, 4, 8, 12, 24, 48, and 72 h of incubation. A 20-ml headspace gas sample was taken from jars that were incubated with and without C2H2 at the end of the incubation periods and injected into a previously evacuated Exetainer (Labco, United Kingdom) containing the desiccant magnesium perchlorate. The jars were opened, and 25 g of soil was sampled to perform analytical measurements, including measurements of extractable organic carbon (EOC), nitrate (NO3−), and ammonium (NH4+) concentrations. A soil sample comprised of a composite of six subsamples amounting to a total weight of 10 g was flash-frozen using liquid nitrogen and kept at −80°C for nucleic acid extraction.
Analytical methods.
Headspace gas was analyzed for N2O and CO2 concentrations by using a Varian Star 3800 gas chromatograph (Varian, Walnut Creek, CA) fitted with an electron capture detector (to measure N2O), a thermal conductivity detector (TCD; to measure CO2), and a Combi-PAL Autosampler (CTC Analytics, Zwingen, Switzerland). The electron capture detector was operated at 300°C with 90% Ar and 10% CH4 carrier gas at 20 ml min−1 with a Haysep N 80/100-mesh precolumn (0.32 cm in diameter and 50 cm in length) and Haysep D 80/100-mesh analytical columns (0.32 cm in diameter and 200 cm in length) in a column oven operated at 70°C. The precolumn was used in combination with a 4-port valve to remove water from the sample. The TCD was operated at 130°C with prepurified He carrier gas at 30 ml min−1 and a Haysep N 80/100-mesh (0.32-cm diameter by 50-cm length) precolumn followed by a Porapak QS 80/100-mesh (0.32-cm diameter by 200-cm length) analytical column maintained at 70°C.
Concentrations of EOC, NO3−, nitrite (NO2−), and NH4+ were quantified from 25 g of soil previously shaken for 45 min with 50 ml of 0.5 M K2SO4. The extracts were vacuum filtered and stored at −20°C until they were colorimetrically analyzed with a Technicon Auto Analyzer II system (Technicon Industrial Systems, Terrytown, MA). Technicon industrial methods 455-76W/A and 97-70W were used to determine the concentrations of EOC and NH4+, respectively. Technicon Industrial Method 100-70W was used to determine the concentrations of NO3− plus NO2− and NO2−, where the latter was performed without the cadmium reduction coil.
Nucleic acid extraction.
DNA and RNA were coextracted from 1.5 g soil and split equally into two 2-ml tubes according to a method described previously by Griffiths et al. (20). Genomic DNA was obtained from half of the nucleic acids from each duplicate soil extraction and combined into a single tube. From the other half of the extracted nucleic acids, RNA was further purified by using 50 U DNase (127 U/μl) (Invitrogen, Burlington, ON, Canada) in 200 μl of buffer (1.1 mM CaCl2 and 1.1 mM MgCl in Tris-HCl [pH 7.5]) and then incubated for 1 h at 37°C. RNA was extracted using phenol-chloroform-isoamyl alcohol (25:24:1), incubated at room temperature for 5 min, and centrifuged for 5 min. RNA from the aqueous phase was precipitated by incubation overnight at −20°C with twice the volume of 97% ethyl alcohol (EtOH) and a 1/10 volume of 5 M NaCl. The RNA was pelleted by centrifugation at 16,000 × g for 45 min and resuspended in nuclease-free, RT-PCR-grade water. RNA from duplicate extractions was combined before being purified using an RNeasy Cleanup column (Qiagen, Mississauga, ON, Canada). The quality of DNA and RNA was assessed by agarose (1.0% [wt/vol]) gel electrophoresis. The lack of DNA in the RNA samples was confirmed by performing a PCR without reverse transcriptase. The RNA and DNA were quantified by using the fluorescent dyes Ribogreen and Picogreen, respectively, as recommended by the manufacturer (Invitrogen, Burlington, ON, Canada).
Primers used in this study.
Primers targeting several denitrifier communities were tested on DNA and cDNA synthesized from total RNA isolated from soil. Primers for nirS (8, 26, 38), nirK (21), and nosZ (22) were tested by using quantitative PCR (qPCR) conditions and cycling parameters as described previously.
Primers were designed to target the nirS gene of P. mandelii and closely related species (nirSP). Pseudomonas lini and P. migulae have been isolated from soil (bulk and rhizosphere) and were shown to be taxonomically close to P. mandelii (15). Primers were designed in the identified conserved region found among P. mandelii (GenBank accession number DQ518190) strains and the closely related species P. lini (accession number DQ518197) and P. migulae (accession number DQ518195) by using Primer Select (Lasergene 7) based on standard conditions for quantitative PCR as recommended by Applied Biosystems (Foster City, CA). The designed primers nirSsh2F (ACC GCC GCC AAC AAC TCC AAC A) and nirSsh4R (CCG CCC TGG CCC TTG AGC) amplified a 244-bp fragment using PCR. The specificity of the primers was tested by comparing their sequences with sequences from the GenBank database and by PCR with genomic DNA from several pure cultures of denitrifying and nondenitrifying strains as templates (Table 1). Eleven amplification products from the primer pair using extracted soil genomic DNA as a template were cloned into the vector pCR2.1 TOPO (Invitrogen), sequenced (Robarts Research Institute Sequencing Facility, London, ON, Canada), and then compared to sequences from the GenBank database.
TABLE 1.
Bacterial strains used to test specificity of primers
| Speciesa | Classb | qPCR result with primersc |
|---|---|---|
| Nondenitrifiers | ||
| Achromobacter cycloclastes ATCC 21921 | Betaproteobacteria | − |
| Alcaligenes faecalis ATCC 35655 | Betaproteobacteria | − |
| Bradyrhizobium japonicum USDA 110 | Alphaproteobacteria | − |
| Escherichia coli ATCC 29425 | Gammaproteobacteria | − |
| nirK gene-bearing species | ||
| Ochrobactrum anthropi ATCC 49187 | Alphaproteobacteria | − |
| nirS gene-bearing species | ||
| Azoarcus tolulyticus ATCC 51758 | Betaproteobacteria | − |
| Paracoccus denitrificans ATCC 17741 | Alphaproteobacteria | − |
| Paracoccus denitrificans ATCC 19367 | Alphaproteobacteria | − |
| Pseudomonas fluorescens ATCC 13525 | Gammaproteobacteria | − |
| Pseudomonas mandelii PD 30 | Gammaproteobacteria | + |
| Pseudomonas mandelii PD 8 | Gammaproteobacteria | + |
| Pseudomonas mandelii PD 2 | Gammaproteobacteria | + |
| Pseudomonas stutzeri ATCC 14405 | Gammaproteobacteria | − |
| Pseudomonas stutzeri ATCC 17588 | Gammaproteobacteria | − |
| Thauera aromatica DSMZ 14793 | Betaproteobacteria | − |
ATCC, American Type Culture Collection (Manassas, VA); USDA, U.S. Department of Agriculture (Beltsville, MD); DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany).
Data from the literature.
+, PCR product of the expected size; −, no PCR product.
Quantitative PCR.
Gene copy number and transcript abundance for the nirSP- and nosZ-bearing communities were quantified via qPCR and qRT-PCR using an Applied Biosystems (Streetsville, ON, Canada) ABI Prism 7000 thermal cycler and SYBR green detection. Quantitative PCR for nirSP gene numbers was performed by using SYBR green PCR master mix (Invitrogen), primers nirSsh2F and nirSsh4R at 300 nM each, 0.25 ng T4 gene 32 protein (New England Biolabs, Pickerington, ON, Canada), and 5 μl of a 1:100 dilution of DNA extracted from soil (about 10 ng DNA) in a 25-μl reaction mixture. Cycling conditions were 1 cycle of 50°C for 2 min followed by 1 cycle of 95°C for 10 min and then 40 cycles of 95°C for 15 s, 68.4°C for 30 s, 72°C for 30 s, and 83°C for 30 s. Fluorescence was measured at 83°C to remove signal from any nonspecific products or primer-dimers. qPCR for nosZ-bearing community gene numbers was performed in a 25-μl final volume with the same reaction mix components as those described above for the nirSP gene except that the primers used were nosZ1F and nosZ1R (22) at concentrations of 300 nM and 400 nM, respectively, and 2 μl of a 1:50 dilution of DNA extracted from soil (about 10 ng DNA) was used. The cycling conditions were 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, and 6 cycles of 95°C for 15 s, 68°C to 62°C (−1°C per cycle) for 1 min, and 81.5°C for 30 s, followed by 40 cycles of 95°C for 15 s, 62°C for 1 min, and 81.5°C for 30 s. Fluorescence was read at 81.5°C. No template controls were included in triplicate for both nirSP and nosZ primer sets.
nirSP transcripts were quantified by using the SuperScript III Platinum One-Step qRT-PCR kit (Invitrogen) in a 25-μl reaction mixture according to the manufacturer's recommendations. Primers nirSsh2F and nirSsh4R were used at 300 nM and 500 nM, respectively, and the reaction mixture contained 5 μl of a 1:100 dilution of RNA extracted from soil (about 500 pg RNA). The cycling conditions were identical to those previously described for measurements of nirSP gene numbers with the exception that the initial incubation at 50°C was extended to 30 min to facilitate reverse transcriptase activity. nosZ transcripts were quantified as described above for the nirSP transcripts by using primers nosZ1F and nosZ1R at same concentrations as those used for qPCRs. The reaction mixture contained 2 μl of a 1:50 dilution of RNA extracted from soil (about 500 pg RNA), and cycling conditions were identical to those described above for nosZ gene copy number determinations with the exception that the initial incubation at 50°C was extended to 30 min. No template controls and no reverse transcription controls (nine random samples) were included in triplicate to ensure that contaminating DNA was not present.
Standard curves were constructed for the absolute quantification of nirSP and nosZ gene numbers and transcripts. For a standard curve of nirSP, genomic DNA from P. mandelii strain PD30 (13) was used to amplify a nirSP gene fragment by using primers nirSsh2F and nirSsh4R, which was then cloned in the TOPO vector and verified by sequencing. The plasmid was linearized by digestion with SacI (Roche, Laval, Quebec, Canada) and quantified by using Picogreen. The copy number of the nirSP-containing plasmid was calculated from the concentration and size (base pairs) of the extracted plasmid. A standard curve was generated by using three replicates of 10-fold serial dilutions (from 101 to 106 copies) of linearized plasmid containing the nirSP sequence as a template and the cycling conditions described above. For the standard curve of nosZ, genomic DNA from Pseudomonas brassicacearum strain PD5 (13) was used to amplify a nosZ gene fragment by using primers nosZ1F and nosZ1R. The gene fragment was cloned and sequenced as previously described (30). The plasmid carrying the nosZ fragment was used to generate a standard curve of nosZ as described above for the nirSP standard curve and using the PCR conditions described above for nosZ.
Soil DNA and RNA extracts were tested for the presence of coextracted inhibitory substances. A known quantity of nirSP gene fragment bearing plasmid or RNA from a pure culture of P. mandelii cells grown under anoxic conditions was used as a control reaction. A concentration of the nirSP gene fragment or RNA identical to that used for the controls was added to soil DNA or RNA extracts that were serially diluted 10-fold. The nirSP product was amplified by using qPCR or qRT-PCR. Gene copy or transcript numbers were compared with those of the control reactions. The presence of inhibitors in soil at 100 and 10−1 dilutions resulted in no amplification or in lower cycle threshold values than those of the control reactions. A 10−2 dilution of soil DNA or RNA templates was not inhibitory to the qPCR or qRT-PCRs.
Data analysis.
All statistical analyses were conducted by using Systat software 12 (Systat Software Inc., Chicago, IL). Homogeneity of variance within the data was tested by using Levene tests. Data were tested for normality by using the Shapiro-Wilk test, and all nonnormal data were log transformed. A mixed-model three-way analysis of variance was performed based on a randomized complete-block design with treatment and time as fixed factors and block as a random factor. Interactions were tested by using Tukey-adjusted least-squares means. Simple main effects were determined by performing post hoc Tukey honestly significant difference tests. Regression analyses were performed to determine possible relationships between NO3−, EOC, CO2, N2O, or total denitrification and gene copies and transcripts of nosZ and nirSP at individual time points. Treatment means and standard errors presented in the figures were calculated from untransformed data. Significance was accepted at a P value of <0.05.
RESULTS
Changes in soil analyte concentrations in response to organic carbon treatment.
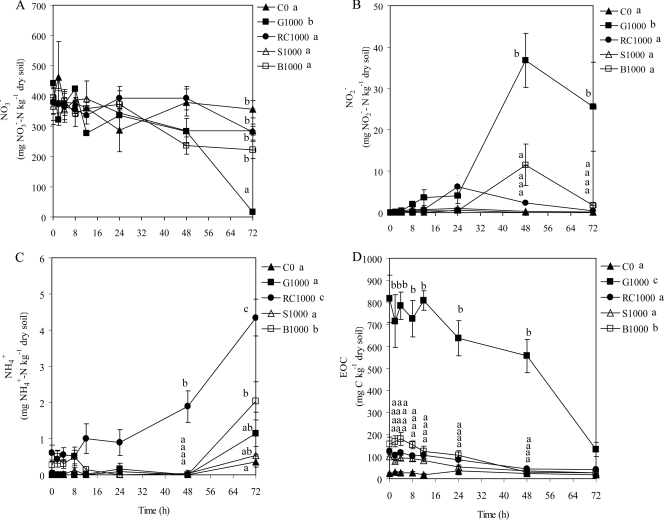
The C treatments had a significant effect on the soil nitrate (NO3−) concentration (P < 0.001) (Fig. 1A). Following the application of 1,000 mg C kg−1 dry soil as glucose (G1000), the soil NO3− concentration decreased from 443 mg NO3−-N kg−1 dry soil at 0 h to 15 mg NO3−-N kg−1 dry soil at 72 h. In contrast, there was no significant difference among soils amended with 1,000 mg C kg−1 dry soil as red clover (RC1000), as soybean (S1000), and as barley (B1000) and for the nonamended soil (C0) (Fig. 1A), and soil NO3− concentrations did not change significantly over time for these treatments. The C treatments also had a significant effect on the soil nitrite (NO2−) concentration (P < 0.001) (Fig. 1B). The nitrite concentration significantly increased with the G1000 treatment at 48 h and 72 h (average of 36 mg NO2−-N kg−1 dry soil) and with the B1000 treatment at 48 h (11 mg NO2−-N kg−1 dry soil) compared with the nitrite concentration at the beginning of the incubation, whereas the NO2− concentrations with C0, RC1000, and S1000 did not increase significantly during the incubation period (Fig. 1B).
FIG. 1.
Soil concentrations of NO3− (A), NO2− (B), NH4+ (C), and EOC (D). Treatments consisted of 0 mg C kg−1 dry soil (C0), 1,000 mg glucose-C kg−1 dry soil (G1000), 1,000 mg red clover-C kg−1dry soil (RC1000), 1,000 mg soybean-C kg−1 dry soil (S1000), and 1,000 mg barley straw-C kg−1 dry soil (B1000). Values are means (n = 6). Error bars are ±1 standard error. Significant differences among mean values are represented in two ways based on Tukey's test (P < 0.05): (i) differences among treatment means for individual time points are represented by letters adjacent to the time points but only for sampling dates in which significant differences among treatments means exist, and (ii) differences among treatment means averaged across time points are shown by letters in the figure legend.
The soil ammonium (NH4+) concentration at the start of the incubation period was very low, <0.1 mg NH4+-N kg−1 dry soil. For the C0, G1000, S1000, and B1000 treatments, the NH4+ concentration remained very low and reached concentrations ranging from 0.8 to 2.0 mg NH4+ kg−1 dry soil at 72 h (Fig. 1C). The NH4+ concentration of the RC1000 treatment was significantly higher than those for all other treatments (P < 0.001) and averaged 0.6 mg NH4+-N kg−1 dry soil for the first 24 h of incubation and then increased over time to a final concentration of 4.4 mg NH4+-N kg−1 dry soil at 72 h.
EOC concentrations in soil were significantly affected by soil treatments (P < 0.001). The soil EOC concentration with the C0 treatment did not change over time, while the EOC concentration significantly decreased over the incubation period in all C-amended soils (Fig. 1D). The EOC concentration was not significantly different among the RC1000, S1000, and B1000 treatments (average of 127 mg EOC kg−1) at 0 h; however, it was significantly higher for the G1000 treatment (816 mg EOC kg−1 dry soil) at 0 h. The EOC concentrations of the plant-amended soils reached similar values at the end of the incubation, with an average of 31.7 mg EOC kg−1 dry soil, whereas the EOC concentration of the G1000 treatment was 123 mg EOC kg−1 dry soil at 72 h.
Effect of carbon treatment on N2O emissions, denitrification activity, and respiration.
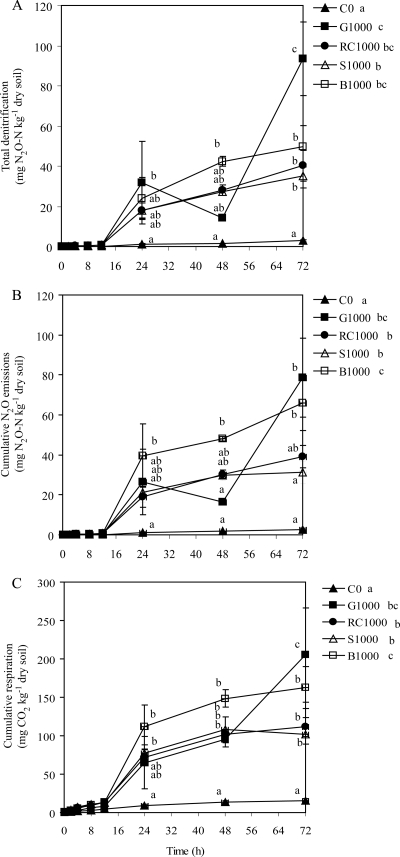
Cumulative denitrification was quantified from soils incubated with acetylene (C2H2), and cumulative N2O emissions were quantified from soil incubated without C2H2. There was no significant difference between denitrification and N2O emissions (P = 0.697), indicating that almost all gaseous emissions from denitrification occurred as N2O (Fig. 2A and B). However, some differences were noted between total denitrification and N2O emission data sets. For example, there was significantly more total denitrification with the G1000 treatment at 72 h than with other treatments, but cumulative N2O emissions with G1000 at 72 h were different only for the C0 treatment at 72 h. Discrepancies between the two data sets are probably due to the use of two different jar sets to measure total denitrification and N2O emissions. Cumulative N2O emissions and denitrification significantly increased over 72 h in all treatments (P < 0.001) (Fig. 2A and B). Cumulative denitrification and N2O emissions from C0, G1000, RC1000, S1000, and B1000 after 72 h averaged 3, 94, 39, 36, and 60 mg N2O-N kg−1 dry soil, respectively (Fig. 2A and B). The cumulative N2O emissions and denitrification from nonamended soil were negligible. The cumulative denitrification at the end of the incubation was equivalent to 10, 26, 39, 50, and 33% of the reduction in soil NO3 and NO2 concentrations for the C0, G1000, RC1000, S1000, and B1000 treatments, respectively.
FIG. 2.
Total denitrification (N2O plus N2) (A), cumulative N2O emissions (B), and respiration (C). Treatment mixtures were incubated with C2H2 to measure cumulative denitrification or without C2H2 to measure cumulative N2O emissions. Respiration was calculated from the average between soils incubated with and those incubated without C2H2. Treatments consisted of 0 mg C kg−1 dry soil, 1,000 mg glucose-C kg−1 dry soil, 1,000 mg red clover-C kg−1 dry soil, 1,000 mg soybean-C kg−1 dry soil, and 1,000 mg barley-C kg−1 dry soil. Values are means (n = 6). Error bars are ±1 standard error. Significant differences among mean values are represented in two ways based on Tukey's test (P < 0.05): (i) differences among treatment means for individual time points are represented by letters adjacent to the time points but only for sampling dates in which significant differences among treatments means exist, and (ii) differences among treatment means averaged across time points are shown by letters in the figure legend.
Soil respiration was measured over the experiment by quantifying cumulative CO2 emissions. There was no difference in respiration between soils incubated with and those incubated without C2H2 (P = 0.139), indicating that C2H2 was negligible as a C source for soil microbes. Respiration significantly increased (P < 0.001) over the incubation period for all treatments (Fig. 2C) and averaged 16, 199, 106, 99, and 142 mg CO2-C kg−1 dry soil for the C0, G1000, RC1000, S1000, and B1000 treatments at 72 h, respectively (Fig. 2C). Respiration was significantly higher in C-amended soil than in nonamended soil. Within the C-amended soils, the CO2 emissions from G1000 were significantly greater than that from the plant tissue C-amended soils at 72 h (Fig. 2C). The CO2 emissions from the B1000 treatment were significantly greater than the CO2 emissions from the legume-amended soil (RC1000 and S1000). When corrected for CO2 emissions from the nonamended soil, the increases in cumulative CO2 emissions measured at 72 h were the equivalents of 29, 11, 10, and 15% of the C added for the G1000, RC1000, S1000, and B1000 treatments, respectively. Therefore, C concentrations in red clover, soybean, and barley straw were estimated to be about 38% (RC1000 CO2/G1000 CO2), 34% (S1000 CO2/G1000 CO2), and 51% (B1000 CO2/G1000 CO2), respectively, as available for microbial respiration as glucose C.
Specificity of nirSP primers.
Alignment of nirS nucleotide sequences from the related Pseudomonas species P. mandelii, P. lini, and P. migulae showed a high degree of identity (96 to 98% identity) among these species; thus, primers were designed to quantify this group of denitrifiers. The specificity of the primers was tested by PCR using genomic DNA from four nondenitrifying bacteria, one nirK-bearing denitrifier, and 10 nirS-bearing bacterial species as templates (Table 1). Amplification with the nirSP primers resulted in the expected 244-bp fragment for all P. mandelii strains tested (Table 1). No PCR products were obtained for nondenitrifiers, the nirK-bearing denitrifier, and other nirS-bearing denitrifiers (Table 1). When soil genomic DNA and cDNA (synthesized from total RNA) were used as templates, the nirSP primers amplified a single DNA fragment of the expected size. The 11 clones bearing the nirS fragment obtained by PCR amplification of soil genomic DNA showed between 97 and 99% sequence identity with P. mandelii nirS (GenBank accession number DQ518190), P. lini (accession number DQ518197), and P. migulae (accession number DQ518195) (GenBank accession numbers GU353322 to GU353332). Of the 11 clones sequenced, no other known Pseudomonas sp. nirS genes were amplified by using these primers.
Denitrifier abundance and denitrification gene mRNA levels.
Primers targeting nirS- or nirK-bearing denitrifiers did not successfully amplify both genomic DNA and cDNA from soil even after exhaustive optimization of the PCR conditions and cycling parameters. Primers targeting nosZ (22) and nirSP were successful in amplifying both DNA and cDNA from soil nucleic acid extractions.
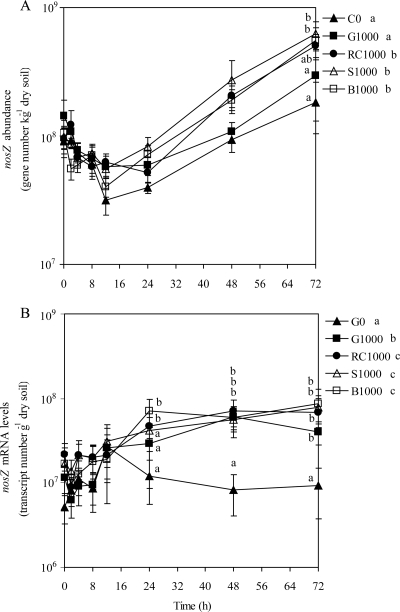
The nosZ abundance decreased numerically for all treatments between 0 and 12 h, but this change in the nosZ abundance was not statistically significant. The nosZ abundance during the first 12 h of the incubation did not differ among treatments and averaged 7.7 × 107 gene copies g−1 dry soil (Fig. 3A). The nosZ abundance significantly increased after 12 h for all treatments (P < 0.001), reaching 1.8 × 108, 3.0 × 108, 5.1 × 108, 6.3 × 108, and 5.5 × 108 gene copies g−1dry soil for the C0, G1000, RC1000, S1000, and B1000 treatments, respectively, at 72 h. The nosZ abundance with the RC1000, S1000, and B1000 treatments averaged over time were significantly higher than those with the G1000 and C0 treatments. G1000 and C0 had similar mRNA levels when averaged over time (Fig. 3A). The nosZ mRNA levels increased significantly over the 72 h of incubation in C-amended soils (P = 0.001), with averages of 1.7 × 107 transcripts g−1 dry soil at 0 h and of 6.9 × 107 transcripts g−1 dry soil at 72 h, representing a 4-fold increase (Fig. 3B). The nosZ mRNA levels did not change over time with the C0 treatment, with an average of 1.1 × 107 gene transcripts g−1 dry soil. There were significantly more nosZ transcripts in the C-amended soils than in the nonamended soil at 48 h and 72 h (Fig. 3B). The nosZ mRNA levels for plant residue treatments averaged over time were significantly higher than those with G1000 and C0. There was a significantly higher nosZ mRNA level with G1000 averaged over time than with the C0 treatment. Similar results were observed when ratios of nosZ gene mRNA levels/abundance were calculated, with averages of 0.32 ± 0.41 for C0, 0.45 ± 0.32 for G1000, and 0.51 ± 0.37 to 0.56 ± 0.38 for plant residue treatments.
FIG. 3.
Quantification of nosZ gene numbers (A) and nosZ transcripts (B) using qPCR and RT-qPCR, respectively. Treatments consisted of 0 mg C kg−1 dry soil (C0), 1,000 mg glucose-C kg−1 dry soil (G1000), 1,000 mg red clover-C kg−1 dry soil (RC1000), 1,000 mg soybean-C kg−1 dry soil (S1000), and 1,000 mg barley straw-C kg−1 dry soil (B1000). Values are means (n = 6). Error bars are ±1 standard error. Standard curve descriptors and detection levels are as follows: for nosZ gene numbers, y = −3.33x + 39.14, R2 = 0.999, E = 99.9%, and NTC = undetected; for nosZ transcripts, y = −3.27x + 38.41, R2 = 0.999, E = 102%, and NTC = undetected. Significant differences among mean values are represented in two ways based on Tukey's test (P < 0.05): (i) differences among treatment means for individual time points are represented by letters adjacent to the time points but only for sampling dates in which significant differences among treatments means exist, and (ii) differences among treatment means averaged across time points are shown by letters in the figure legend.
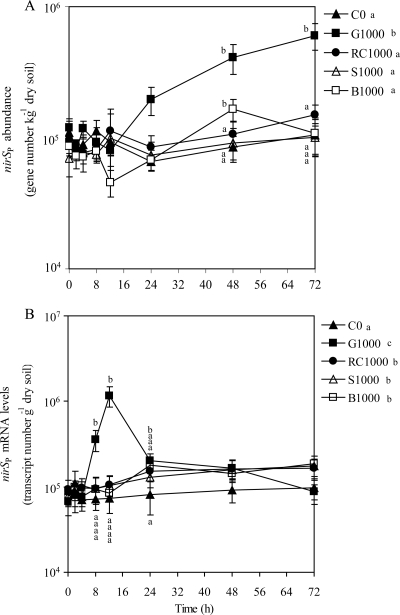
The nirSP abundance responded significantly to the treatments (P < 0.001) (Fig. 4A). The nirSP gene copy numbers in the G1000 treatment were significantly higher than those for all other treatments after 24 h and remained higher for the remainder of the incubation period. The nirSP gene copy numbers with the G1000 treatment increased about 7.5-fold, from 8.1 × 104 gene copies g−1 dry soil at 12 h to 6.1 × 105 gene copies g−1 dry soil at 72 h. The nirSP abundances with the C0, RC1000, S1000, and B1000 treatments averaged over time did not differ from each other and did not change over time (average of 9.1 × 104 gene copies g−1 dry soil) (Fig. 4A).
FIG. 4.
Quantification of nirSP gene numbers (A) and nirSP transcripts (B) using qPCR and RT-qPCR, respectively. Treatments consisted of 0 mg C kg−1 dry soil (C0), 1,000 mg glucose-C kg−1 dry soil (G1000), 1,000 mg red clover-C kg−1 dry soil (RC1000), 1,000 mg soybean-C kg−1 dry soil (S1000), and 1,000 mg barley straw-C kg−1 dry soil (B1000). Values are means (n = 6). Error bars are ±1 standard error. Standard curve descriptors and detection levels are as follows: for nirSP gene numbers, y = −3.50x + 36.01, R2 = 0.999, E = 93.4%, and NTC = undetected; for nirSP transcripts, y = −3.45x + 37.28, R2 = 0.993, E = 95.7%, and NTC = undetected. Significant differences among mean values are represented in two ways based on Tukey's test (P < 0.05): (i) differences among treatment means for individual time points are represented by letters adjacent to the time points but only for sampling dates in which significant differences among treatments means exist, and (ii) differences among treatment means averaged across time points are shown by letters in the figure legend.
Soil treatments had a significant effect on mRNA levels of nirSP (P < 0.001) (Fig. 4B). For the G1000 treatment, the nirSP mRNA levels did not change during the first 4 h of incubation, averaging 7.5 × 104 transcripts g−1 dry soil, and then increased significantly more than 15-fold, to a maximum of 1.2 × 106 transcripts g−1 dry soil at 12 h. The mRNA levels of nirSP subsequently decreased between 12 and 24 h and then did not change from 24 to 72 h, averaging 1.5 × 105 transcripts g−1 dry soil. The nirSP transcript abundances at 8 and 12 h with the G1000 treatment were significantly different from the nirSP mRNA levels at all other time points. nirSP mRNA levels of plant residue-amended soils averaged over time were significantly higher than levels with the C0 treatment. nirSP mRNA levels with the C0, RC1000, S1000, and B1000 treatments did not change over the 72 h of incubation, averaging 1.1 × 105 transcripts g−1 dry soil (Fig. 4B). The ratio of nirSP gene mRNA levels/abundance of G1000 averaged over the measurement period was higher (3.46 ± 5.64) than those for plant residue-amended soils (1.68 ± 0.52 to 1.86 ± 0.78) and C0 (1.34 ± 0.52).
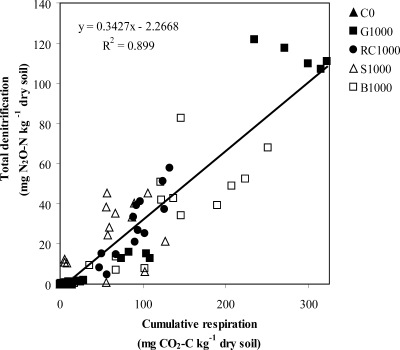
Possible relationships between analytical measurements (NO3− and EOC), respiration, cumulative N2O emissions, denitrification, and the abundance or denitrification gene mRNA levels of denitrifier communities were explored. There was a significant positive relationship between cumulative denitrification and respiration over the 72-h incubation period (P = 0.001) (Fig. 5). There were no other significant relationships between NO3−, EOC, respiration, cumulative N2O emission, and denitrification. Using regression analyses, no significant relationships were identified between denitrification gene abundance and denitrification gene mRNA levels of nirSP- or nosZ-bearing denitrifiers and denitrification as well as N2O emissions and respiration.
FIG. 5.
Relationship between denitrification and respiration over the 72-h incubation period. Treatments consisted of 0 mg C kg−1 dry soil (C0), 1,000 mg glucose-C kg−1 dry soil (G1000), 1,000 mg red clover-C kg−1 dry soil (RC1000), 1,000 mg soybean-C kg−1 dry soil (S1000), and 1,000 mg barley straw-C kg−1 dry soil (B1000). The line of best fit indicates the linear relationship described by the following equation: y = 0.342x − 2.27 (R2 = 0.90).
DISCUSSION
Effect of organic C on N2O emissions, denitrification activity, and respiration.
This study evaluated the effects of simple (glucose) and complex (plant residues) C treatments on the abundances and gene mRNA levels of broad (nosZ) and specific (nirSP) denitrifier communities, and on cumulative N2O emissions and denitrification, in anoxic agricultural soil microcosms. The addition of simple and complex C sources increased microbial activity, as indicated by increased soil respiration, over that of nonamended soil. The addition of simple and complex C sources also increased total denitrification, and there was a positive relationship between denitrification and respiration regardless of the C treatment. Previous microcosm studies also reported strong positive relationships between denitrification and respiration across a wide variety of C sources (30, 31).
Red clover, soybean, and barley straw were chosen as carbon amendments to obtain a range of C:N values for the plant tissues. Crop residues with a high C:N ratio commonly decompose more slowly than residues with a low C:N ratio (28). In this study, the availabilities of C in red clover, soybean, and barley straw were estimated to be 37%, 34%, and 51% as available as glucose C based on soil respiration. Surprisingly, the barley straw was the tissue with the highest C:N ratio (30:1) but also the highest C availability. Thus, in this study, the availability of C in the plant tissue was poorly related with the C:N ratio of the tissue. The C:N ratio alone, however, may not be a good indicator of the decomposition of crop residues, and other components of the residues, for example, lignin and polyphenols, may influence decomposition rates (37). The higher C availability for the barley straw than those for red clover and soybean did not result in significantly higher total denitrification rates at 72 h. Previous incubation studies reported an inverse relationship between residue C:N ratios and both respiration and total denitrification (2, 3). The greater C availability of glucose C than plant tissue C resulted in increased respiration and total denitrification, as was previously reported (18).
The residue C:N ratio may also influence denitrification through the supply of N from the residue. For example, soil treatment with a legume (alfalfa) residue resulted in significantly more denitrification than treatment with simple organic C sources (glucose and sucrose), a finding attributed to nitrate depletion following the addition of simple organic C sources, whereas N was supplied by the legume residue (14). However, the addition of NO3− to all treatments and the inhibition of nitrification by anoxic conditions would be expected to minimize any influence of the N content of the plant tissue as a controlling factor in decomposition in this study. A greater depletion of soil NO3− following treatment with glucose than with other organic treatments that contain N was previously reported and was attributed to net immobilization (30).
Nitrite rarely accumulates in soil since its metabolism in soil can be extremely rapid (10). Interestingly, 7 and 23% of the reduced NO3− accumulated as nitrite in the B1000 and G1000 treatments, respectively, at 48 h. The increase in the soil NO2− concentration can be attributed to two competing processes under anoxic conditions, denitrification or dissimilatory nitrate reduction to ammonium (DNRA) (39). Ammonium concentrations remained low in soil, and there was a positive correlation between denitrification and respiration, suggesting that denitrification was more important than DNRA under the conditions used. A difference in denitrification enzyme activities between the nitrate reductase and nitrite reductase could result in nitrite accumulation (7, 34). It is possible that the remainder of the metabolized soil NO3− was assimilated by soil microorganisms.
Effect of organic C on denitrifier abundance and denitrification gene mRNA levels.
The addition of simple and complex carbon sources resulted in an increase in the nosZ-bearing community abundance. In contrast, nosZ-bearing communities did not increase significantly over time in soil amended with glucose or plant residues in studies described previously by Miller et al. (30); however, those studies used lower concentrations of organic C treatments. These results suggest that under anoxic conditions, a high concentration of glucose or plant residues, i.e., 1,000 mg C kg−1 dry soil, is required to increase nosZ gene abundance. In this study, there was no difference in the abundance of the nosZ-bearing community among plant residue-amended soils, but the abundance of nosZ was higher following treatment with plant residues than treatment with glucose. While the C from plant residues used in this study was only 34 to 51% as available as glucose C, plant tissues contain water-soluble components, including simple sugars, amino acids, vitamins, and aliphatic acids (5), that may have been beneficial for bacterial growth.
The absence of an increase in the nirSP-bearing community abundance with the addition of plant residues suggests that the nirSP-bearing community could not effectively compete for the nutrients under the conditions used, although glucose was readily utilized. Limited information on organic C utilized by P. mandelii is currently available in the literature. Verhille et al. (40) previously reported that several simple sources of C were assimilated by P. mandelii under oxic conditions. P. mandelii cells could also grow in pure culture under low-oxygen conditions using glucose as a sole source of carbon (our unpublished data). Miller et al. (30) also found an increase in the abundance of P. mandelii and closely related species in glucose-amended soil compared with red clover-amended soil, although the increase was greater than that observed in this study. However, Miller et al. (30) used a different soil microcosm system with different incubation parameters than those used for the current study, and properties of the red clover tissue might have differed due to differences in the plant growth stage.
This study found a significant increase in nosZ mRNA levels in C-amended soil compared with levels in unamended soil. There were no differences in nosZ mRNA levels among the different plant residue treatments. These results suggest that mRNA levels of nosZ were stimulated by all C sources used in this study by increasing the nosZ mRNA levels per cell and by increasing the abundance of nosZ-bearing denitrifiers. In a previous study, transcripts of the nosZ gene were not detected in agricultural soil following C release from freeze-thaw events (35), and there have been no other studies that have examined the mRNA levels of nosZ transcripts in soils amended with complex C sources. In contrast to the increase of nosZ mRNA levels in complex C-amended soil over 72 h, there was no increase in nirSP mRNA levels in unamended or amended soils, with the exception of a nirSP expression peak at 12 h in glucose-amended soil. The absence of an increase of nirSP mRNA levels in response to plant residues further supports that the nirSP guild is not well adapted to using plant residues.
This study did not observe relationships between the abundance of nosZ- or nirSP-bearing denitrifiers and denitrification or N2O emissions. Previous studies also did not find any relationships between denitrifier abundance in soils and N2O emissions (13, 30); however, nirS gene abundance was correlated with the denitrification rate in sediment (16). A relationship between denitrifier abundance and gas emissions in the current study may not have been found due to the taxonomic diversity of denitrifiers; the N2O emissions were from the entire denitrifying community and not just the denitrifiers quantified in this study. No relationships were observed between mRNA levels of nirSP or nosZ and denitrification activity or N2O emissions. The expression of the nosZ gene might be expected to be negatively correlated with N2O emissions, as increased numbers of nosZ transcripts would be expected to lead to more N2O reduction to N2. Under the conditions used in this study, N2O accumulated but was not reduced to N2 even when nosZ gene expression was detected, indicating either a discrepancy in activity of the NosZ enzyme compared with other denitrification enzymes or the preferential use of the more-oxidized N-oxides over N2O. It is possible that a significant relationship between denitrification gene mRNA levels and N2O emissions was not found because denitrification gene mRNA levels do not necessarily indicate the activity of the resulting enzyme.
In conclusion, denitrification and respiration increased in soil amended with organic carbon, including plant residues, compared to unamended soil, although denitrification levels were not different among the plant residue treatments. The nirSP and nosZ communities responded differently to organic carbon amendments, and the response of these communities was not influenced by the type of plant residue applied under the conditions tested.
Acknowledgments
Funding for this work was provided by a Natural Sciences and Engineering Research Council of Canada (NSERC) strategic grant and by Agricultural and Agri-Food Canada.
We thank Jan Zeng, Drucie Janes, Ginette Decker, and Karen Terry for providing technical laboratory assistance and Myriam Barbeau for her assistance with the statistical analysis.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Araki, N., Y. Tsukamoto, A. Nagano, T. Yamaguchi, and H. Harada. 2006. Real time PCR quantification of nitrite reductase (nirS) genes in nitrogen removing fluidized bed reactor. Water Sci. Technol. 53:59-65. [DOI] [PubMed] [Google Scholar]
- 2.Aulakh, M. S., J. W. Doran, D. T. Walters, A. R. Mosier, and D. D. Francis. 1991. Crop residue type and placement effects on denitrification and mineralization. Soil Sci. Soc. Am. J. 55:1020-1025. [Google Scholar]
- 3.Baggs, E. M., R. M. Rees, K. A. Smiths, and A. J. A. Vinter. 2000. Nitrous oxide emissions from soils after incorporating crop residues. Soil Use Manage. 16:82-87. [Google Scholar]
- 4.Bateman, E. J., and E. M. Baggs. 2005. Contributions of nitrification and denitrification to N2O emissions from soils at different water-filled pore space. Biol. Fertil. Soils 41:379-388. [Google Scholar]
- 5.Beauchamp, E. G., J. T. Trevors, and R. W. Paul. 1989. Carbon substrates for bacterial denitrification, p. 113-142. In B. A. Stewart (ed.), Advances in soil science, vol. 10. Springer-Verlag Inc., New York, NY. [Google Scholar]
- 6.Beller, H. R., T. E. Letain, A. Chakicherla, S. R. Kane, T. C. Legler, and M. A. Coleman. 2006. Whole-genome transcriptional analysis of chemolithoautotrophic thiosulfate oxidation by Thiobacillus denitrificans under aerobic versus denitrification conditions. J. Bacteriol. 188:7005-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betlach, M. R., and J. M. Tiedje. 1981. Kinetic explanation for accumulation of nitrite, nitric oxide, and nitrous oxide during bacterial denitrification. Appl. Environ. Microbiol. 42:1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bremner, J. M. 1996. Nitrogen—total, p. 961-1010. In D. L. Sparks (ed.), Methods of soil analysis: chemical methods, part 3. Soil Science Society of America, Madison, WI.
- 10.Burns, L. C., R. J. Stevens, and R. J. Laughlin. 1995. Determination of the simultaneous production and consumption of soil nitrite using 15N. Soil Biol. Biochem. 27:839-844. [Google Scholar]
- 11.Clays-Josserand, A., J. F. Ghiglion, L. Philippot, P. Lemanceau, and R. Lensi. 1999. Effect of soil types and plant species on the fluorescent pseudomonads nitrate dissimilating community. Plant Soil 209:275-282. [Google Scholar]
- 12.Dandie, C. E., D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Analysis of denitrification genes and comparison of nosZ, cnorB, and 16S rDNA from culturable denitrifiying bacteria in potato cropping systems. Syst. Appl. Microbiol. 30:128-138. [DOI] [PubMed] [Google Scholar]
- 13.Dandie, C. E., M. N. Miller, D. L. Burton, B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Nitric oxide reductase-targeted real-time PCR quantification of denitrifier populations in soil. Appl. Environ. Microbiol. 73:4250-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Catanzaro, J. B., and E. G. Beauchamp. 1985. The effect of some carbon substances on denitrification rates and carbon utilization in soil. Biol. Fertil. Soils 1:183-187. [Google Scholar]
- 15.Delorme, S., P. Lemanceau, R. Christen, T. Corberand, J. M. Meyer, and L. Garden. 2002. Pseudomonas lini sp. nov., a novel species from bulk and rhizospheric soils. Int. J. Syst. Evol. Microbiol. 52:513-523. [DOI] [PubMed] [Google Scholar]
- 16.Dong, L. F., C. J. Smith, S. Papaspyrou, A. Stott, A. M. Osborn, and D. B. Nedwell. 2009. Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne Estuary, United Kingdom). Appl. Environ. Microbiol. 75:3171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drury, C. F., X. M. Yang, W. D. Reynolds, and C. S. Tan. 2004. Influences of crop rotation and aggregate size on carbon dioxide production in denitrification. Soil Tillage 79:87-100. [Google Scholar]
- 18.Gillam, K. M., B. J. Zebarth, and D. L. Burton. 2008. Nitrous oxide emissions from denitrification and the partitioning of gaseous losses as affected by nitrate and carbon addition and soil aeration. Can. J. Soil Sci. 88:133-143. [Google Scholar]
- 19.Greets, J., M. de Cooman, L. Wittebolle, K. Heylen, S. Vanparys, P. de Vos, W. Verstraete, and N. Boon. 2007. Real time PCR assays for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 95:211-221. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry, S., E. Baudoin, J. C. Lopez-Gutierrez, F. Martin-Laurent, A. Brauman, and L. Philippot. 2004. Quantification of denitrifying bacteria in nirK gene targeted real time PCR. J. Microbiol. Methods 59:327-335. [DOI] [PubMed] [Google Scholar]
- 22.Henry, S., D. Bru, B. Stres, S. Hallet, and L. Philippot. 2006. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry, S., S. Texier, S. Hallet, D. Bru, C. Dambreville, D. Cheneby, F. Bizouard, J. C. Germon, and L. Philippot. 2008. Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10:3082-3092. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Y., J. Zou, X. Zheng, Y. Wang, and X. Xu. 2004. Nitrous oxide emissions as influenced by amendment of plant residues with different C:N ratios. Soil Biol. Biochem. 36:973-981. [Google Scholar]
- 25.Janzen, H. H., and R. M. N. Kucey. 2006. C, N, and S mineralization of crop residues as influences by crop species and nutrient regime. Plant Soil 106:35-41. [Google Scholar]
- 26.Kandeler, E., K. Deiglmayr, D. Tscherko, D. Bru, and L. Philippot. 2006. Abundance of narG, nirS, nirK, and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72:5957-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemedtsson, L., S. Simkins, B. H. Svensson, H. Johnsson, and T. Rosswall. 1991. Soil denitrification in three cropping systems characterized by differences in nitrogen and carbon supply. II. Water and NO3 effects on the denitrification process. Plant Soil 138:273-286. [Google Scholar]
- 28.Kumar, K., and K. M. Goh. 2000. Crop residues and management practices: effects on soil quality, soil nitrogen dynamics, crop yield and nitrogen recovery. Adv. Agron. 68:167-319. [Google Scholar]
- 29.Lashof, D. A., and D. R. Ahuja. 1990. Relative contributions of greenhouse gas emissions to global warming. Nature 334:529-531. [Google Scholar]
- 30.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevor. 2008. Crop residue influence on denitrification, N2O emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 40:2553-2562. [Google Scholar]
- 31.Miller, M. N., B. J. Zebarth, C. E. Dandie, D. L. Burton, C. Goyer, and J. T. Trevor. 2009. Influence of liquid manure on soil denitrifier abundance, denitrification and N2O emissions. Soil Sci. Soc. Am. J. 73:760-768. [Google Scholar]
- 32.Nelson, E. W., and L. E. Sommers. 1996. Total carbon, organic carbon, and organic matter, p. 1085-1122. In D. L. Sparks (ed.), Methods of soil analysis: chemical methods, part 3. Soil Science Society of America, Madison, WI.
- 33.Nijburg, J. W., M. J. L. Coolen, S. Gerads, P. J. A. K. Gunnewiek, and H. J. Laanbroek. 1997. Effects of nitrate availability and the presence of Glyceria maxima on the composition and activity of the dissimilatory nitrate-reducing bacterial community. Appl. Environ. Microbiol. 63:931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh-Lakha, S., K. E. Shannon, S. L. Henderson, B. J. Zebarth, D. L. Burton, C. Goyer, and J. T. Trevors. 2009. Effect of nitrate and acetylene on nirS, cnorB, and nosZ expression and denitrification activity in Pseudomonas mandelii. Appl. Environ. Microbiol. 75:5082-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma, S., Z. Szele, R. Schilling, J. C. Munch, and M. Schloter. 2006. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2007. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 73:3612-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorup-Kristensen, K., J. Magid, and J. S. Jensen. 2003. Catch crops and green manures as biological tools in nitrogen management in temperate zones. Adv. Agron. 79:227-302. [Google Scholar]
- 38.Throbäck, I. N., K. Enwall, A. Javis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirS, and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 39.Tiedje, J. M., A. J. Sexstone, D. D. Myrold, and J. A. Robinson. 1982. Denitrification: ecological niches, competition and survival. Antonie von Leeuwenhoek 48:569-583. [DOI] [PubMed] [Google Scholar]
- 40.Verhille, S., N. Baida, F. Dabboussi, D. Izard, and H. Leclerc. 1999. Taxonomic study of bacteria isolated from natural mineral waters: proposal of Pseudomonas jessenii sp. nov. and Pseudomonas mandelii sp. nov. Syst. Appl. Microbiol. 22:45-58. [DOI] [PubMed] [Google Scholar]