Abstract
Epithelial ovarian carcinoma (EOC) is a leading cause of death from gynecologic malignancy, due mainly to the prevalence of undetected metastatic disease. The process of cell invasion during intra-peritoneal anchoring of metastatic lesions requires concerted regulation of many processes, including modulation of adhesion to the extracellular matrix and localized invasion. Exploratory cDNA microarray analysis of early response genes (altered after 4 hours of 3-dimensional collagen culture) coupled with confirmatory real-time RT-PCR, multiple three-dimensional cell culture matrices, Western blot, immunostaining, adhesion, migration, and invasion assays were used to identify modulators of adhesion pertinent to EOC progression and metastasis. cDNA microarray analysis indicated a dramatic downregulation of connective tissue growth factor (CTGF) in EOC cells placed in invasion-mimicking conditions (3-dimensional type I collagen). Examination of human EOC specimens revealed that CTGF expression was absent in 46% of the tested samples (n=41), but was present in 100% of normal ovarian epithelium samples (n=7). Reduced CTGF expression occurs in many types of cells and may be a general phenomenon displayed by cells encountering a 3D environment. CTGF levels were inversely correlated with invasion such that downregulation of CTGF increased, while its upregulation reduced, collagen invasion. Cells adhered preferentially to a surface comprised of both collagen I and CTGF relative to either component alone using α6β1 and α3β1 integrins. Together these data suggest that downregulation of CTGF in EOC cells may be important for cell invasion through modulation of cell-matrix adhesion.
Keywords: ovarian carcinoma, metastasis, invasion, adhesion, connective tissue growth factor
INTRODUCTION
Epithelial ovarian carcinoma (EOC) has the poorest survival of all gynecologic malignancies 1, due predominantly to the presence of disseminated intraperitoneal metastasis 2. Successful metastatic spread requires concerted regulation of crucial cellular processes, including apoptosis, proliferation, cell-cell and cell-matrix adhesion, migration, and breakdown of the extracellular matrix 3. A detailed understanding of these mechanisms is a prerequisite for the design of therapeutics essential for retardation or prevention of metastatic spread. The tumor microenvironment plays a major role in orchestrating the cell behavior 4. The microenvironment in sites of EOC metastatic dissemination is predominantly comprised of interstitial collagens I and III 5, which can be modeled experimentally in order to distinguish effects of the microenvironment on cell behavior 6, 7. Experimentally created three-dimensional cultures recapitulate the in vivo cellular environment more closely than traditional cell culture on planar substrata. It has been demonstrated that these three-dimensional culture models and organotypic cultures can accurately and reliably replicate certain conditions in the living organs and, therefore, can be used as a preliminary model to more closely understand the consequences of interactions of cells with the surrounding microenvironment 8, 9. This is highlighted by a recent study comparing cDNA microarray expression profiles of cells cultured in 2-dimensional planar cell culture to 3-dimensional culture and murine xenografts. Results demonstrated that global gene expression profiles of the 3D cultures were more closely aligned with those of tumor xenografts 10. Our previous studies have shown that three-dimensional collagen I (3DCI) gels, a microenvironmental component relevant to that encountered by metastasizing ovarian carcinoma cells 11, 12, dramatically modulates cell behavior and promotes a pro-invasive phenotype 6, 7, 13-16. Specifically, our data demonstrate that 3DCI enhances the ability of cells to migrate through upregulation of membrane Wilms’ tumor gene product 1 17 and actinin alpha-4 13, and to digest extracellular matrix via upregulation of membrane type-1 matrix metalloproteinase 6, 7, 15. These findings led to the speculation that other cellular mechanisms pertinent to pro-invasive and migratory behavior, such as cell-matrix adhesion, may be altered through interaction of cells with 3DCI as well. Adhesion of cells to the extracellular matrix plays a key role in the mechanisms regulating migration and invasion, and often reduction of adhesive strength is required to achieve balance between the ability to migrate and survive for anchorage-dependent cells 18, 19.
In this study we have performed an exploratory cDNA microarray analysis to identify possible target genes regulating ovarian carcinoma matrix adhesion. A rapid and dramatic reduction in the gene encoding connective tissue growth factor (CTGF) was observed in three-dimensional collagen culture. Connective tissue growth factor (CTGF) is a secreted molecule with many functions. It has been shown to participate in fibrogenesis, migration, proliferation, and adhesion 20-22. Its function in EOC progression is not known, however it has been shown to be silenced in human ovarian carcinoma via epigenetic mechanisms 23. Our data demonstrate that CTGF is downregulated in cells cultured on 3DCI compared to those on thin layer collagen film. This downregulation of CTGF occurs in response to three-dimensional culture regardless of the biological composition. A variety of cell types including breast carcinoma, rat cortical neurons, fibrosarcoma cells, and endothelial cells respond to 3DCI culture by downregulation of CTGF, indicating that this downregulation may be a general phenomenon attributable to many different types of cells. We demonstrate that ovarian carcinoma cells adhere stronger to a collagen I –CTGF mixture than to collagen I or CTGF alone and that downregulation of CTGF enhanced, while its overexpression reduced collagen invasion. Together these data suggest that reduction of extracellular CTGF may be beneficial for pro-invasive behavior via weakening the adhesion to the matrix in metastasizing cells.
MATERIALS AND METHODS
Materials
The ovarian carcinoma cell line DOV13 was kindly provided by Dr. R. Bast, Jr. (M.D. Anderson Cancer Center, Houston, TX) and maintained as previously described 24 between passages 45 and 65. High-density cortical neuronal cultures were prepared from E18 rat embryos (under animal protocol approved by Northwestern University animal committee) as described before 25 and maintained in neurobasal media supplemented with glutamine and B27 (Invitrogen Corporation). Fibrosarcoma HT1080 cell line was generously provided by Dr. J. Jones (Northwestern University, Chicago, IL) and propagated in minimal essential media (Invitrogen Corporation) supplemented with 10% fetal bovine serum (Invitrogen Corporation) between passages 2 and 6. Human umbilical vein endothelial cells (HUVEC) were obtained from ATCC and maintained in media 199 (Sigma), supplemented with 3% fetal bovine serum, 10 mM HEPES, 2 mM glutamine, 30 μg/ml heparin, 50 μg/ml endothelial mitogen (Biomedical Technologies) between passages 3 and 6. MDA-MB231 was obtained from ATCC and maintained in minimal essential media supplemented with 10% fetal bovine serum. Rat tail type I collagen was obtained from BD Biosciences. Human collagen I, RGDS peptide, and bovine serum albumin (BSA) were purchased from Sigma (St.Louis, MO). 4-arm PEG-acryl (10K) was obtained from SunBio (Orinda, CA) and 2,2-Dimethyl-2-phenyl-acetophenone was a generous gift from Ciba (Tarrytown, NY). CTGF overexpressing plasmid (pCTGF) was a generous gift from Dr. M.-L. Kuo (National Taiwan University, Taipei, Taiwan). Human recombinant CTGF protein HPLC-purified from E.coli was purchased from Antigenix America (Huntington Sta., NY; www.antigenix.com). Polyclonal antibodies against CTGF, control and CTGF siRNA were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Function blocking antibodies against α2 (MAB, αV (MAB1980 and MAB2021Z), α3 (MAB2056), αVβ3 (MAB1976), α6 (MAB1378), β1 (MAB1959) integrins, as well as anti-mouse IgG, were obtained from Chemicon (Temecula, CA). TissueScan real-time ovarian cancer disease panel I containing 48 tissues covering four disease stages and normal tissues was obtained from Origene (Rockville, MD).
Three-Dimensional Matrix Cell Culture Models
Several three-dimensional cell culture models composed of various matrix components were used to characterize their interaction with ovarian carcinoma cells. Three-dimensional collagen I (3DCI) gel at 0.8 mg/ml, three-dimensional collagen III (3DCIII) at 0.25 mg/ml, and synthetic 10% 4-arm polyethylene glycol acryl (PEG-acryl) containing 0.3 mM RGDS, were used to mimic three-dimensional matrix conditions encountered by invading ovarian carcinoma cells. Synthetic 10% 4-arm polyethylene glycol acryl (PEG-acryl) containing 0.3 mM RGDS was prepared by photocrosslinking using 0.5% 2,2-Dimethyl-2-phenyl-acetophenone in polyvinylpyrrolidone (600mg/ml) as the photoinitiator. Cells were cultured atop three-dimensional matrices for various periods of time as described before 6. Control cells were plated either on 10 μg/ml thin layer collagen I (two-dimensional collagen I or 2DCI) or 10 μg/ml thin layer collagen III (2DCIII), or 0.3 mM RGDS (2D). MDA-MB231, HT1080, rat cortical neurons, and HUVEC were cultured atop 3DCI as described above.
Spheroidal Ovarian Carcinoma Cell Culture Model
Ovarian carcinoma multicellular aggregates (MCAs, spheroids) were prepared using the hanging drop method as previously described 26. Cells that formed multicellular aggregates were designated ‘spheroids’, and cells released from monolayers with trypsin were designated ‘single cells’.
cDNA Microarray
Cells were cultured atop 3DCI and 2DCI for 4 hours, experiments were performed in triplicate. Total RNA for cDNA microarray experiments was extracted using Trizol according to the previously published protocol 27, 28. All DNA microarray gene expression studies used human oligonucleotide arrays custom-printed by a dedicated core facility within the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center (Omaha, NE) as described previously 29, 30. Microarray slides were scanned with a ScanArray 4000 confocal laser system (Perkin-Elmer).
Statistical Analysis of Microarray data
Analysis of microarray gene expression data, accumulated from three independent experiments, was performed using the limma package 31, available through the Bioconductor project 32 for use with R statistical software 33 as described before 13. After pre-processing, the analysis of differential gene expression was based on moderated t-statistics on the replicated log 2 ratios for each gene. Statistical significance was assessed using an Empirical Bayes approach 31. Adjustment for multiple comparisons according to the false discovery rate method of Benjamini and Hochberg 34 was performed, and genes with adjusted p-values less than 0.05 were selected as differentially expressed. Log 2 ratios were transformed back to fold change values for interpretation purposes in this report.
mRNA Extraction and cDNA synthesis for Real Time RT-PCR
To perform real time RT-PCR cells were cultured on 3D matrices as described above. Total mRNA was purified from 1-2×106 cells using SV Total RNA Isolation kit (Promega) according to the manufacturer’s instructions. cDNA was synthesized from 5-10 μg of total RNA using qScript cDNA Synthesis Kit (Quanta Biosystems). mRNA purification and cDNA synthesis experiments were repeated three times for each condition and cell type.
Quantitative Real Time PCR
Real time PCR was carried out with the ABI Prizm (Applied Biosystems) according to the manufacturer’s instructions as described before 6, 13. SYBR Green was used for quantitative PCR as a double-stranded DNA-specific fluorophore. Expression of CTGF in human cells, DOV13, HT1080, HUVEC, MDA-MB231, was evaluated using previously published sequences of primers 35, RPL-19 was used a housekeeping gene control 6. Expression of CTGF in rat cortical neurons was evaluated using primers for rat Ctgf and rat Rpl19 as a housekeeping control (Supplementary Material, Table 1). Efficiency of amplification was determined using the standard curves method. Relative quantification of gene expression between experimental (3DCI) and control (2DCI) samples was measured by normalization against endogenous RPL-19 using the ΔCT method 36. Prior to using RPL-19 as a control, it has been established that its expression correlated well with the total RNA concentration and did not change with the time and treatment used in our studies. Fold changes were quantified as 2-(ΔCt sample-ΔCt control) as described previously 36.
Real time RT-PCR was also used to detect the levels of CTGF, α6, β1, α3, β4, αV mRNAs in samples from tissues of normal ovary and ovarian carcinoma patients commercially available from Origene according to the manufacturer’s suggestions using the amount of cDNA provided, 2 ng (for detailed description of the samples go to www.origene.com or 37). Primers for α6, β1, α3, β4, αV and RPL-19 mRNA detection are presented in Supplementary Material, Table 1, and primers for beta-actin detection (ACTNB) were supplied with the cDNA samples panel (Origene). Ct values for CTGF, α6, β1, α3, β4, and αV were obtained using ABI Prizm (Applied Biosystems) according to the manufacturer’s instructions as described above in Methods. RPL19 and ACTNB were expressed in each sample as reported before 37.
Immunohistochemistry
Immunohistochemical analysis was done retrospectively on tumor tissue microarrays prepared with Institutional Review Board approval by the Pathology Core Facility of the Robert H. Lurie Comprehensive Cancer Center at Northwestern University assembled from tissue originally taken for postoperative diagnostic purposes as described before 6, 13, 38. The microarray tissue specimens included 16 primary ovarian cancer tissues (15 serous, 1 endometroid). Breast carcinoma tissue was used as positive control, and human normal lymph node as a negative control for CTGF. Analysis of tissue sections was done by light microscopy by an anatomic pathologist (B.P.A.) without prior knowledge of the clinical variables. Scoring of CTGF was assigned according to the average overall intensity of the staining and was graded as follows: 0 = no staining, 1 = fine granular staining, 2 = somewhat coarse staining, but less than positive control tissue (human testis), 3 = very coarse staining, similar to positive control tissue. Staining <10% of tumor cells, regardless of intensity, was considered negative. Staining of between 10% and 75% of tumor cells was considered focal positive, and staining of greater than 75% of tumor cells was considered diffuse positive.
Transient Transfections
Transient transfections were performed using the lipofection method with Lipofectamine (Invitrogen) as a vehicle. 60 pmole of CTGF and control siRNA, as well as 1 mg of pCTGF, were transiently transfected into DOV13 cells growing in 6-well tissue culture plates according to the manufacturer’s instructions.
Collagen Invasion
Invasion assays were performed using Transwell chambers (0.8 μm, Becton Dickinson, Bedford, MA) as described before 7.
Scratch wound assay
DOV13 cells were cultured in standard conditions, as described above in the Methods (Cell Culture), until 70-80% confluence following transient transfection with either CTGF siRNA or control siRNA, as indicated in Methods (Transient Transfections). Wounds were introduced into the confluent monolayers by a pipette tip. Wound closure was monitored over time and photographed using Zeiss Axiovert microscope at 5× magnification on the objective. The distance between the monolayers was measured using Zeiss Axiovert software in 10 random places, averaged, and calculated into the percentage of the wound healing at a given time compared to the initial wound width.
Western Blotting
Cells incubated under various conditions were collected as described before 6. The antibodies were used at the following dilutions: 1:200 for anti-human CTGF polyclonal antibody in 3% bovine serum in TBST, 1:1000 for anti-β-tubulin monoclonal antibody in 5% skim milk in TBST. Immunoreactive bands were visualized with an anti-(rabbit-IgG)-peroxidase, anti-goat-IgG)-peroxidase, or anti-(mouse-IgG)-peroxidase (1:1000 in 5% skim milk in TBST) and enhanced chemiluminescence using LAS3000 (Fujifilm) and LAS3000 ImageReader software. Band intensities were determined using LAS3000 ImageGauge software according to the manufacturer’s instructions.
Cell Adhesion
Ovarian carcinoma cell adhesion to collagen I (0.5 μg), CTGF (0.8 μg), and a mixture of collagen I (0.5 μg) and CTGF (0.8 μg) was evaluated by seeding cells (3,000) on planar substrata deposited by passive adsorption in 96 well plates for 1 or 3 hours. Non-adherent cells were removed, adherent cells were washed once with phosphate buffered saline, fixed and stained with Diff-Quik kit, counted in five random fields and averaged. To determine the involvement of specific integrin receptors in ovarian carcinoma adhesion to CTGF, DOV13 cells were seeded on CTGF-coated surfaces in the presence and absence of α2, αV, α6, αVβ3, α3, β1 function blocking antibodies. Anti-mouse IgG was used as a control. Cells were pre-incubated with the antibodies for 20 min at 37 °C prior to plating. After plating, cells were allowed to attach for 3 hours, followed by the removal of non-adherent cells, and processing as described above.
RESULTS
Expression of CTGF Is Downregulated by Three-Dimensional Culture
Our previous data demonstrate that 3DCI culture of ovarian carcinoma cells leads to acquisition of the invasive phenotype, which is accompanied by changes in expression of genes regulating invasion and migration 6, 7, 15, 17, 37. cDNA microarray analysis of early changes in gene expression showed a 3.8-fold downregulation of CTGF mRNA (p<0.05) in cells cultured for 4 hours atop 3DCI gel compared to those cultured on thin layer collagen I film (Supplementary Material, Table 2), in addition to 21 other significantly downregulated genes. Expression of CTGF in 2-dimensional cultures remained constant at the 1, 2, and 4 hour time points. Since cell:substratum adhesion is an important parameter in cell invasion, and CTGF has been shown to function as an adhesion regulator 20-22, additional experiments were performed to evaluate the potential functional significance of CTGF downregulation by 3DCI. Real time RT-PCR data confirmed the cDNA microarray results, demonstrating a significant loss of CTGF mRNA after 4 h in 3DCI culture (Figure 1A). Prolonged ovarian carcinoma cell culture atop 3DCI also resulted in sustained downregulation of secreted (Figure 1B) and total cellular CTGF protein for 24 – 48 h (Figure 1C, D). Culturing cells in 3DCI results in multivalent clustering of collagen binding integrins 39 in addition to altering cytoskeletal architecture, both of which may ultimately influence gene expression 16. To differentiate the relative contribution of cell shape vs integrin engagement, CTGF expression was evaluated by real time RT-PCR following culture in a variety of three-dimensional culture model systems. Expression of CTGF was significantly downregulated in both RGD-seeded polyethylene glycol (PEG) gels (Figure 1E) and in 3D gels comprised of type III collagen (Figure 1F) relative to the corresponding 2D cultures. Metastatic ovarian carcinoma cells exist in ascites as both multicellular aggregates (MCAs) and single cells 40, 41, with MCAs representing a 3-dimensional cellular structure. Interestingly, MCA culture also downregulated CTGF expression (Figure 1F), supporting the hypothesis that CTGF is downregulated in ovarian carcinoma cells in a three-dimensional microenvironment regardless of its biochemical composition.
Figure 1. Expression of CTGF is downregulated by three-dimensional environment.
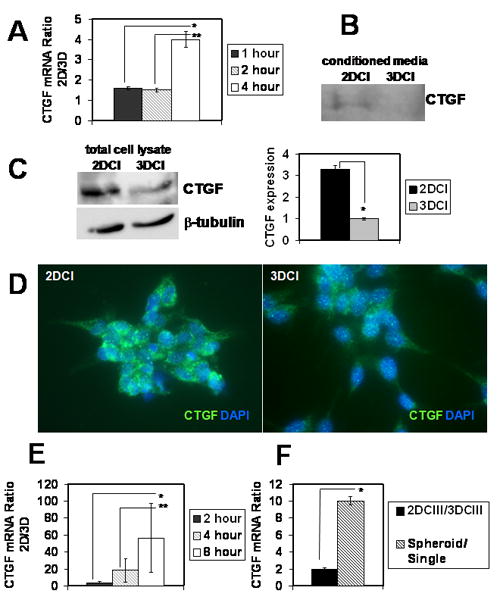
(A) DOV13 cells respond to three-dimensional collagen I culture with downregulation of CTGF mRNA after 4 hours of culture. Cells were cultured for indicated periods of time on 3DCI and 2DCI and collected for extraction of total RNA and subsequent cDNA synthesis. Real time RT-PCR was performed to detect CTGF expression. Histogram depicts ratios of CTGF mRNA in cells cultured on 2DCI compared to those on 3DCI using 2-ΔΔCt method. Ratio >1 reflects downregulation of CTGF expression on 2DCI versus 3DCI. *, **, p<0.05 Secreted (B) (into conditioned media) and total (C) CTGF protein is downregulated in cells cultured on 3DCI compared to those on 2DCI for 24-48 hours. Anti-human CTGF antibody (1:200 dilution) was used to visualize CTGF-reactive bands, β-tubulin (1:1000 dilution) was used as a loading control for detection of CTGF in total cell lysates. For samples prepared from conditioned media, loading was normalized to the amount of seeded cells (amount of collected cells did not change significantly). Conditioned media were collected, centrifuged at 12,000 rpm for 5 min, and concentrated 5 times using 10K Centricon filters. *, p<0.05 (D) Immunofluorescent staining of cells cultured on 2DCI and 3DCI for 24 h. Rabbit anti-CTGF (H-55) and goat anti-rabbit Alexa488 antibodies used at 1:50 and 1:500 dilutions, respectively. Histogram shows quantitative analysis of CTGF expression in total lysates of cells cultured atop 3DCI and 2DCI; *, p<0.05 (E) Three-dimensional PEG gel culture downregulates CTGF mRNA. Cells were cultured atop 3D 10% PEG, containing 0.3 mM RGD, as well as on 0.3 mM RGD-coated tissue culture plates (2D) for indicated periods of time. Ratios of CTGF mRNA expression on 2D compared to 3D were found with 2-ΔΔCt method. *, **, p<0.05 (F) DOV13 cells were cultured atop 3D collagen III and thin-layer collagen III for 8 hours. Ratio of CTGF expression on 2D vs 3D collagen III is shown by the black bar. Expression of CTGF mRNA in spheroids (hatched bar) was compared to that in cells released from monolayers (single). *, p<0.05 All experiments were performed three times; data represent mean ± standard deviation.
To evaluate whether downregulation of CTGF is unique to ovarian carcinoma cells, real time RT-PCR analysis was performed to quantify CTGF mRNA in other migratory/invasive cell types interacting with 2D vs 3D collagen matrices. Invasive breast carcinoma cells (MDA-MB231), primary rat cortical neurons, HT1080 fibrosarcoma cells and HUVEC all responded to 3DCI culture by a 2-10-fold downregulation of CTGF (Figure 2A-D), demonstrating that downregulation of CTGF by 3DCI is a common mechanism shared by many types of invasive cells. Time courses of the experiments were chosen based on specific optimal response from each cell type to 3DCI.
Figure 2. Three-dimensional collagen I culture downregulated CTGF in many cell types.
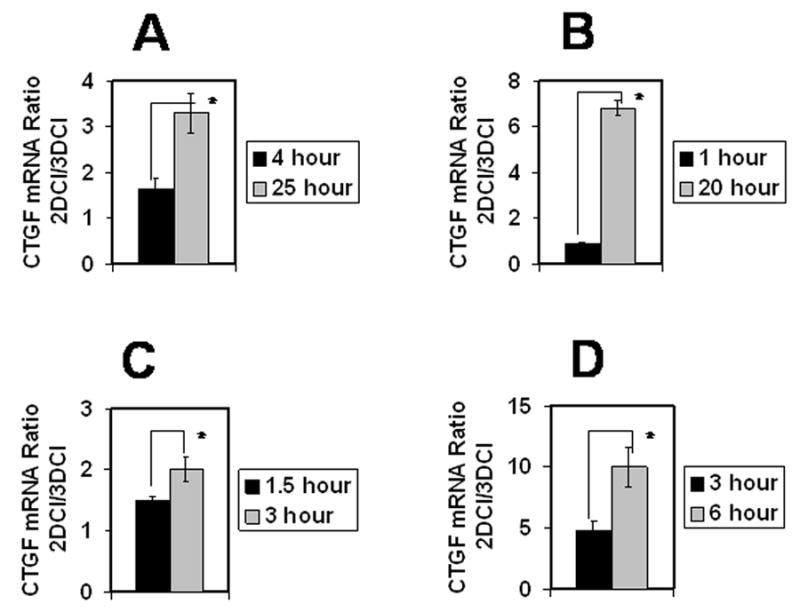
Breast carcinoma MDA-MB231 (A), rat cortical neurons (B), fibrosarcoma HT1080 (C), and HUVEC (D) were cultured atop 3DCI and on thin layer collagen I for indicated periods of time. Real time RT-PCR was performed to evaluate CTGF mRNA expression. Ratio of CTGF mRNA expression on 2DCI versus 3DCI was quantified with 2-ΔΔCt method. Ratio equal to 1 represents no change, greater than 1 denotes an increase, and lower than 1 shows a decrease between the compared conditions. Experiments for each cell type were performed twice, averaged and plotted as mean ± standard deviation, *, p<0.05.
Expression of CTGF in Human Ovarian Carcinoma
Expression of CTGF mRNA was evaluated in normal human ovarian epithelium and ovarian carcinoma specimens from multiple stages of disease progression using real time RT-PCR (Table 1). Whereas normal ovarian specimens exhibited 100% positivity for CTGF expression, 46% of tumors were negative for CTGF mRNA (Table 1), suggesting a loss of expression with tumor progression. Similar results were obtained from immunohistochemical analysis of 15 late stage (III/IV) ovarian carcinoma specimens, with 7% exhibiting negative staining, 20% weak reactivity, 60% moderate reactivity and only 6% with strong reactivity (Figure 3, Table 2).
Table 1.
Expression of CTGF, α6, α3, β1, β4, αV integrin RNA in samples of ovarian carcinoma and non-malignant ovary.
| FIGO stage | # | Ovarian Tissue from Patients with the Following Diagnosis | * Expression of | |||||
|---|---|---|---|---|---|---|---|---|
| CTGF | α6 | α3 | β1 | β4 | αV | |||
| 0 | 1 | Adenocarcinoma of endometrium | +(30.9) | - | +(33.7) | - | - | +(33.8) |
| 2 | Carcinoma of cervix | +(33.8) | - | - | +(34.8) | - | +(33) | |
| 3 | Abscess | +(30.9) | - | - | +(32) | - | - | |
| 4 | Endometriosis | +(33.3) | - | - | +(35) | - | - | |
| 5 | Endometriosis | +(30.9) | - | +(33.4) | - | - | - | |
| 6 | Endometriosis | +(33.7) | - | - | - | - | - | |
| 7 | Endometriosis | +(33.4) | - | - | +(35) | - | +(32) | |
| I | 8 | Carcinoma of ovary, endometrioid | - | +(32.8) | - | - | - | - |
| 9 | Adenocarcinoma of ovary, papillary serous | - | +(32.8) | - | +(35) | - | - | |
| 10 | Tumor of ovary, papillary serous, borderline | - | +(32.2) | - | _ | - | - | |
| 11 | Tumor of ovary, papillary serous, borderline | +(31.3) | +(30.7) | - | +(33.5) | - | - | |
| 12 | Carcinoma of ovary, endometrioid | +(34) | +(32.6) | - | +(34.5) | - | - | |
| 13 | Tumor of ovary, serous, borderline | +(32.3) | +(32.8) | +(34.5) | - | - | - | |
| 14 | Tumor of ovary, borderline | - | +(31.8) | +(33.2) | +(30.5) | - | - | |
| 15 | Tumor of ovary, mucinous, borderline | +(34) | +(31.6) | +(32.1) | - | - | +(34.6) | |
| 16 | Adenocarcinoma of ovary, mucinous | +(33.7) | - | +(34) | +(33.4) | - | - | |
| 17 | Adenocarcinoma of ovary, endometrioid | - | +(33.7) | - | - | +(34.2) | - | |
| 18 | Tumor of ovary, borderline | +(34.4) | - | - | - | - | - | |
| 19 | Tumor of ovary, mucinous, borderline | +(32.6) | - | - | - | - | - | |
| 20 | Tumor of ovary, serous, borderline | +(33.7) | +(33) | - | - | - | - | |
| 21 | Tumor of ovary, serous, borderline | - | - | +(32.1) | - | - | - | |
| 22 | Adenocarcinoma of ovary, mucinous | - | - | - | +(33.2) | - | - | |
| 23 | Adenocarcinoma of ovary, endometrioid, squamous features | - | +(34.6) | +(33) | - | - | - | |
| II | 24 | Adenocarcinoma of ovary, serous | - | - | - | +(33.6) | - | - |
| 25 | Adenocarcinoma of ovary, endometrioid | - | - | - | +(35) | - | - | |
| 26 | Adenocarcinoma of ovary, endometrioid | +(34.5) | - | - | +(35) | - | - | |
| III | 27 | Adenocarcinoma of ovary, papillary serous | - | +(30.9) | - | +(26) | - | - |
| 28 | Adenocarcinoma of ovary, serous | - | +(32.3) | - | - | - | - | |
| 29 | Carcinoma of ovary, endometrioid | +(33) | - | - | - | - | - | |
| 30 | Tumor of ovary, serous, borderline | +(32.5) | +(31.4) | - | - | - | +(35) | |
| 31 | Adenocarcinoma of ovary, papillary serous | +(34.4) | +(32.1) | - | - | - | - | |
| 32 | Adenocarcinoma of ovary, serous | +(31.8) | +(32) | - | - | - | - | |
| 33 | Adenocarcinoma of ovary, endometrioid | - | +(33) | - | +(32.6) | - | - | |
| 34 | Adenocarcinoma of ovary, papillary serous | +(34) | - | - | +(34.6) | - | - | |
| 35 | Adenocarcinoma of ovary, papillary serous | - | - | +(34.4) | - | - | - | |
| 36 | Tumor of ovary, serous, borderline | +(34) | +(32.6) | +(32.3) | +(33.4) | - | - | |
| 37 | Adenocarcinoma of ovary, metastatic | - | +(31.8) | +(34) | - | +(34) | - | |
| 38 | Adenocarcinoma of ovary, papillary serous | +(33.9) | +(34.2) | +(33.5) | - | - | - | |
| 39 | Adenocarcinoma of ovary, papillary serous | - | - | +(33.6) | - | - | - | |
| 40 | Adenocarcinoma of ovary, papillary serous | +(32.7) | +(31.6) | +(34.1) | - | - | - | |
| 41 | Adenocarcinoma of ovary, papillary serous | +(34.3) | - | - | - | - | - | |
| 42 | Adenocarcinoma of ovary, papillary serous | +(34.7) | +(31.4) | - | - | - | - | |
| 43 | Carcinoma of ovary | +(33.7) | +(32.5) | +(34.7) | +(33.7) | - | - | |
| 44 | Adenocarcinoma of ovary, papillary serous | - | - | - | - | - | - | |
| 45 | Adenocarcinoma of ovary, serous | - | - | - | +(31.1) | - | - | |
| IV | 46 | Adenocarcinoma of ovary, papillary serous | +(33.9) | - | +(33.5) | +(33.6) | - | + |
| 47 | Adenocarcinoma of ovary, metastatic | - | +(35) | - | - | - | - | |
| 48 | Adenocarcinoma of ovary, papillary serous | +(34.4) | - | - | - | - | - | |
Positive (+) expression is defined by Ct value lower or equal to 35, and the exact parameter of Ct is indicated in the brackets. Negative (-) expression of the genes of interest observed when either no product accumulation was detected, or Ct was higher then 35.
Figure 3. Immunohistochemical analysis of CTGF expression in human normal ovary and EOC.
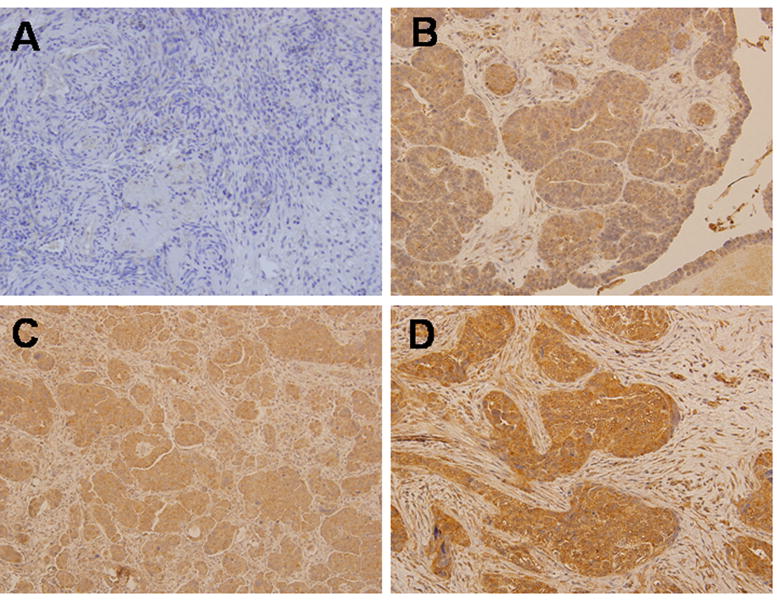
Representative examples of CTGF expression in normal ovarian stroma (A) and primary epithelial serous adenocarcinoma with the intensity of staining 1+ (B), 2+ (C), 3+ (D). Tissues were stained with CTGF specific antibodies as indicated in Methods. Cytoplasmic staining with CTGF antibodies in EOC was detected (CTGF – brown, nuclear DNA – hematoxylin&eosin); magnification = 200×.
Table 2.
Immunohistochemical evaluation of CTGF expression in specimens of primary human ovarian carcinoma.
| Sample # | Histotype | Score * |
|---|---|---|
| 1 | Serous | 2+d |
| 2 | Serous | 2+d |
| 3 | Serous | 2+d |
| 4 | Serous | neg |
| 5 | Serous | 2+d |
| 6 | Serous | 2+d |
| 7 | Serous | 1+d |
| 8 | Serous | 3+d |
| 9 | Serous | 2+d |
| 10 | Serous | 2+d |
| 11 | Endometrioid | 2+d |
| 12 | Serous | 1+d |
| 13 | Serous | 2+d |
| 14 | Serous | 2+f |
| 15 | Serous | 2+d |
| 16 | Serous | 1+d |
CTGF staining intensity is described as negative (neg), 1+ (weakly positive), 2+ (positive), 3+ (strong positive), and as focal (f) or diffuse (d), as described in Methods.
CTGF Modulates Collagen Adhesion and Invasion of EOC cells
CTGF has been characterized as both an adhesion molecule and a motility modulator 20-22. To evaluate its potential function in regulation of ovarian carcinoma cell adhesion, binding of cells to CTGF and collagen was evaluated. Ovarian cancer cells adhered preferentially to a substratum comprised of both CTGF and collagen, relative to either component individually (Figure 4A). To evaluate the role of CTGF in cellular invasion, a modified Boyden chamber assay was used to assess invasion of 3D collagen gels. Cells in which CTGF expression is downregulated, using an siRNA knockdown approach, exhibited a significant increase in 3D collagen invasion relative to cells transfected with a control siRNA (Figure 4B, black bar). In support of these data, transfection of cells with a CTGF expression plasmid resulted in a significant inhibition of invasion (Figure 4B, grey bar). CTGF-specific siRNAs resulted in 40% reduction of CTGF expression relative to the control, while transfection with pCTGF yielded in about 3.5-fold increase in CTGF expression (Figure 4C). Motility, as assessed using a wound healing cell migration assay, was unaffected by modulation of CTGF levels (Figure 4D).
Figure 4. CTGF affects ovarian carcinoma collagen adhesion to and invasion of collagen.
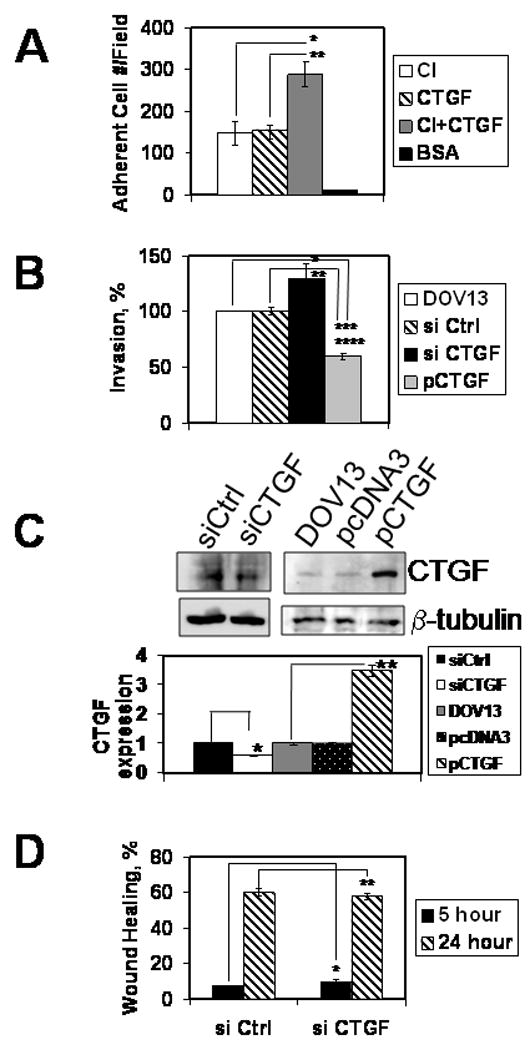
(A) Cells were allowed to adhere to surfaces coated with CTGF, collagen I, collagen I and CTGF mixture, or BSA for 1 hour as described in Methods. Adherent cells were quantified, averaged, and plotted as a number of adherent cells per field, as indicated. Data represent mean ± standard deviation and are an average of two independent experiments, each performed in triplicate. *, **, p<0.05 (B) Three-dimensional collagen I invasion assay was performed for 18 hours as detailed in Methods with DOV13 cells transfected with control siRNA, CTGF siRNA, plasmid overexpressing CTGF, as indicated. Experiments were performed four times, each in triplicate. *, **, p<0.05 (C) Downregulation of CTGF expression by specific siRNA and overexpression in cells transfected with pCTGF was examined with Western blot. Histogram shows quantitative analysis of CTGF expression. Data are an average of three independent experiments. *, **, p<0.05 (D) Scratch wound assay was performed in monolayers of DOV13 cells transfected with CTGF or control siRNA as indicated. Healing wounds were observed 5 and 24 hours following the wounding and the data depicted as % wound healing. Data are an average of two independent experiments performed in duplicate and shown as mean ± standard deviation. Student’s t-test analysis demonstrated that differences in wound repair are not significant; *, **, p>0.05.
Ovarian Carcinoma Cells Adhere to CTGF via α3β1 and α6β1 Integrins
Several integrin molecules have been implicated in cell-CTGF adhesion and downstream signaling 42-46. To identify integrins that may participate in ovarian cancer cell adhesion to CTGF, adhesion was evaluated in the presence of function-blocking integrin subunit-specific antibodies (Figure 5). Whereas α2 integrin function blocking antibody had little effect on adhesion to CTGF, either αV, α6 or α3 integrin blocking antibodies alone slightly reduced CTGF adhesion. However, a combination of αV and α3, αV and α6, α3 and α6 integrin or αV, α3, and α6 integrin blocking antibodies significantly inhibited CTGF adhesion, as did β1 integrin function blocking antibody alone, suggesting the participation of multiple integrins in ovarian cancer cell adhesion to CTGF. Expression of these integrin subunits was evaluated in human ovarian tumor tissues using real time RT-PCR (Table 1). α6 integrin was the most abundantly expressed (58%) in EOC specimens, while α3 and β1 integrins were expressed in 34 and 39% of the tested EOC samples, respectively. Very low levels of β4 and αV integrins were detected (5 and 7% of the EOC specimens, respectively). None of the normal ovarian tissues (no pathologic disease) expressed both β4 and α6 integrins, while 50, 25, and 38% of those were positive for β1, α3, and αV integrins, respectively.
Figure 5. Effect of integrin subunit-specific blocking antibodies on ovarian carcinoma cell adhesion to CTGF.
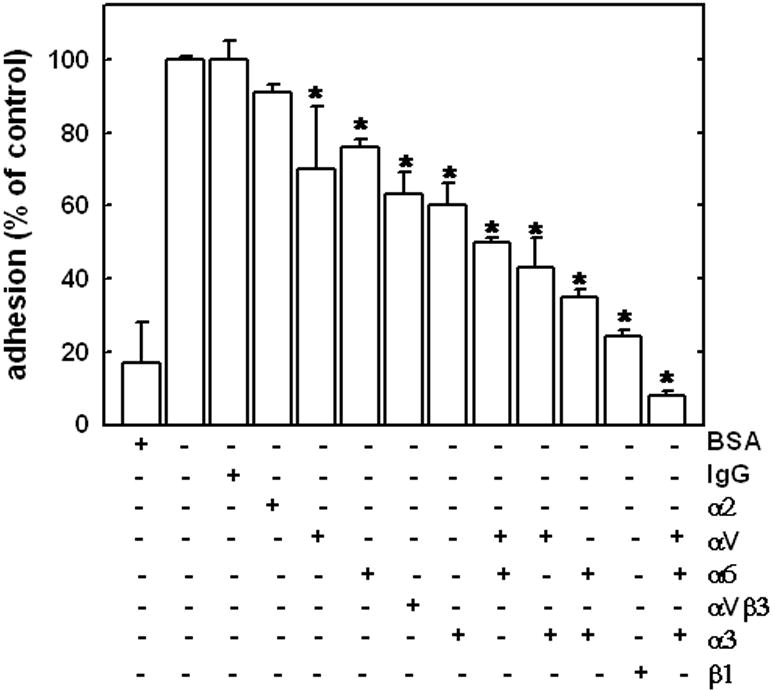
Adhesion to BSA (first bar from the left) or CTGF (0.8 μg; remaining bars) was evaluated as described in Materials and Methods. Adhesion to CTGF in the absence of any antibody (second bar from the left) was arbitrarily set as 100%, and remaining data are reported as a percentage relative to adhesion to CTGF. Total concentration of function blocking antibodies, where applicable, was 10 μg/ml. In the conditions where 2 or 3 different function blocking antibodies were used, individual concentrations were 5 μg/ml and 3.3 μg/ml, respectively. Two different αV-integrins were used, MAB1980 and MAB2021Z, with similar results, however, only the data for MAB2021Z are presented. In experiments containing a mixture of αV-integrin and other integrin function blocking antibodies, MAB1980 was used. Data are presented as mean ± standard deviation. Differences in cell adhesion to CTGF in the presence of function blocking antibodies were statistically significant, as indicated * p < 0.05. Experiments were performed thrice, each in duplicate, quantified, and averaged as detailed in Methods.
DISCUSSION
Epithelial ovarian carcinoma claims more than 14,000 lives a year 1. Most of EOC cases are diagnosed at late stages of the disease, against which current treatment regimens have somewhat modest efficacy. Understanding of the biology of ovarian carcinoma metastasis is limited; therefore, the development of effective molecular target-based anti-metastatic treatments is difficult. Genetic, epigenetic, and microenvironmental factors may modulate the behavior of the malignant cell 4, 8, 47. While several critical pathways have been demonstrated to play a role in EOC tumor etiology and progression, the influence of the microenvironment on malignant behavior and metastasis remains poorly defined.
EOC metastatic spread is uniquely confined largely to the peritoneal cavity mediated by intra-peritoneal shedding of cells from the primary tumor on the ovarian surface 2, 40. Adhesion of EOC cells into the collagen-rich submesothelial matrix of peritoneal organs anchors secondary lesions 2. This unique microenvironment of the metastatic sites is extremely permissive for the EOC spread. Cell migration and invasion, necessary for intra-peritoneal anchoring, requires complicated cellular machinery to modulate adhesion, cytoskeletal rearrangement, extracellular matrix breakdown, and cellular contractility 18, 19, 48, 49. Our previous data suggest that the mechanisms of cell motility and matrix degradation are upregulated in metastasizing ovarian carcinoma cells through their interaction with three-dimensional collagen matrix 6, 7, 17, 37, 50, a microenvironment relevant to the ovarian cancer metastatic process.
In this report, the influence of 3D collagen on the adhesive and invasive behavior of EOC cells was investigated. Successful invasion requires multiple cellular processes including adhesion, proteolysis of matrix barriers and motility. Our findings suggest that 3D collagen is instrumental in downregulating CTGF, a secreted protein to which many cells adhere. This loss of adhesive capacity, rather than altered motility, contributes to enhanced overall invasive activity. The current data support this conclusion, as downregulation of CTGF promotes invasion of 3DCI gels, while overexpression retards collagen invasion. 3DCI collagen culture also leads to upregulation of the collagenolytic proteinase MT1-MMP that plays a key role in collagen invasion by ovarian cancer cells 6, 7, 15. In addition to its function as an interstitial collagenase, MT1-MMP also promotes activation of proMMP-2, and it has been demonstrated that MMP-2 and other MMPs can efficiently cleave CTGF 51, 52. Thus, 3DCI provides a potential dual mechanism for loss of CTGF function through both downregulation of expression and post-translational processing of CTGF via MMP-2. Ovarian cancer cells adhere preferentially to CTGF/collagen surfaces relative to either component alone. A similar function of CTGF was previously reported, as it was shown to enhance adhesion of chondrocytes to fibronectin 53. Based on blocking antibody studies, α3β1 and α6β1 integrins function as receptors for CTGF adhesion in ovarian carcinoma cells. Several studies have supported a role for CTGF:αVβ3 integrin interaction in modulation of adhesion in rat activated hepatic stellate cells 54 and breast carcinoma cells 43. The current data demonstrate a very modest effect of blocking αVβ3 integrin on ovarian carcinoma cell adhesion to CTGF, and in addition, only 3 of the 46 tested human EOC specimens expressed αV integrin. While these findings may reflect the specific behavior of the cell lines and human tumor samples used in this study, the current data suggest that CTGF interaction with αVβ3 integrin may not play a major role in ovarian carcinoma metastasis.
Interestingly, culturing cells in a three-dimensional environment, be it natural or synthetic matrix components or MCA culture, resulted in downregulation of CTGF in ovarian carcinoma cells. Similar results were obtained using the invasive breast carcinoma cell line MDA-MB231, rat cortical neurons, HT1080 fibrosarcoma, and HUVEC. These data support the hypothesis that compliant surfaces lead to downregulation of CTGF in many cell types. Several mechanisms of regulation of CTGF expression have been previously reported.). Expression of CTGF in the normal ovary follows hormonal fluctuations in the menstrual cycle. CTGF is negatively regulated by the follicle-stimulating hormone (FSH) and human chorionic gonadotropin (hCG) 55, 56 but stimulated by estradiol 57. Mechanical changes in cytoarchitecture have also been shown to modulate CTGF levels. For example, disruption of microtubules in renal fibroblasts leads to upregulation of CTGF through alterations of the cytoskeleton leading to activation of RhoA via Src-FAK, while disintegration of the actin skeleton with latrunculin B and increased levels of G-actin resulted in the reduction of CTGF expression 58-61. Interestingly, cell culture on rigid surfaces such as collagen coated plates as well as mechanical strain caused upregulation of CTGF in smooth muscle cells and renal fibroblasts 62-64. In ovarian carcinoma cells, 10% uniaxial mechanical strain produced no effect on CTGF expression (data not shown). Finally, epigenetic downregulation of CTGF in ovarian carcinoma has been reported 23. Our data suggest that epigenetic silencing does not contribute to CTGF downregulation in EOC cultured on 3DCI, because addition of the methyltransferase inhibitor 5-aza-2’-deoxycytidine did not restore CTGF expression under these conditions (data not shown).
In human cancers, CTGF has been associated both positively and negatively with tumor invasion and metastasis. In lung adenocarcinoma, overexpression of CTGF correlated with lower invasive and metastatic ability, while reduced expression of CTGF was associated with disease progression, metastasis, and shorter survival 65. However, colorectal cancer patients with high level of CTGF had higher survival rate compared to those with low CTGF level, and reduction of CTGF led to increased metastasis 66. Our data showing that loss of CTGF expression promotes invasion, together with real time RT-PCR data showing lower CTGF expression in ovarian tumors relative to normal ovarian epithelium, suggest that loss of CTGF expression may be one of many factors that contribute to ovarian cancer metastatic dissemination.
Supplementary Material
Acknowledgments
We thank Dr. Min-Liang Kuo for providing a CTGF overexpressing plasmid, Dr. Jonathan Jones for providing HT1080 cells, as well as Dr. Z.Xie and Cassandra Shum for providing dissected rat cortical neurons.
Research support by 2005-2006 and 2007-2008 Penny Severns Breast, Cervical and Ovarian Cancer Fund grant from the Illinois Department of Public Health (to M.V.B.), 2006 Ovarian Cancer Research Foundation Program of Excellence award (to M.V.B.), and National Cancer Institute Research Grants RO1 CA86984 (to M.S.S.) and CA109545 (M.S.S).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA: a Cancer Journal for Clinicians. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Cannistra SA. Cancer of the ovary. New England Journal of Medicine. 1993;329:1550–9. doi: 10.1056/NEJM199311183292108. [see comment][erratum appears in N Engl J Med 1994 Feb 10;330(6):448] [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey W, Amlot PL. Collagen production by human mesothelial cells in vitro. J Pathol. 1983;139:337–47. doi: 10.1002/path.1711390309. [DOI] [PubMed] [Google Scholar]
- 6.Barbolina MV, Adley BP, Ariztia EV, Liu Y, Stack MS. Microenvironmental Regulation of Membrane Type 1 Matrix Metalloproteinase Activity in Ovarian Carcinoma Cells via Collagen-induced EGR1 Expression. J Biol Chem. 2007;282:4924–31. doi: 10.1074/jbc.M608428200. [DOI] [PubMed] [Google Scholar]
- 7.Ellerbroek SM, Wu YI, Overall CM, Stack MS. Functional interplay between type I collagen and cell surface matrix metalloproteinase activity. J Biol Chem. 2001;276:24833–42. doi: 10.1074/jbc.M005631200. [DOI] [PubMed] [Google Scholar]
- 8.Kenny HA, Krausz T, Yamada SD, Lengyel E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. International Journal of Cancer. 2007;121:1463–72. doi: 10.1002/ijc.22874. [DOI] [PubMed] [Google Scholar]
- 9.Yamada KM, Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell. 2007;130:601–10. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 10.Zietarska M, Maugard CM, Filali-Mouhim A, Alam-Fahmy M, Tonin PN, Provencher DM, Mes-Masson AM. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC) Molecular Carcinogenesis. 2007;46:872–85. doi: 10.1002/mc.20315. [DOI] [PubMed] [Google Scholar]
- 11.Harvey W, Amlot PL. Collagen production by human mesothelial cells in vitro. Journal of Pathology. 1983;139:337–47. doi: 10.1002/path.1711390309. [DOI] [PubMed] [Google Scholar]
- 12.Zhu GG, Risteli J, Puistola U, Kauppila A, Risteli L. Progressive ovarian carcinoma induces synthesis of type I and type III procollagens in the tumor tissue and peritoneal cavity. Cancer Res. 1993;53:5028–32. [PubMed] [Google Scholar]
- 13.Barbolina M, Adley BP, Kelly DL, Fought AJ, Scholtens DM, Shea LD, Stack MS. Motility-related actinin alpha-4 is associated with advanced and metastatic ovarian carcinoma. Lab Invest. 2008;88:602–14. doi: 10.1038/labinvest.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbolina MV, Adley BP, Shea LD, Stack MS. Wilms tumor gene protein 1 is associated with ovarian cancer metastasis and modulates cell invasion. Cancer. 2008;112:1632–41. doi: 10.1002/cncr.23341. [DOI] [PubMed] [Google Scholar]
- 15.Ellerbroek SM, Fishman DA, Kearns AS, Bafetti LM, Stack MS. Ovarian carcinoma regulation of matrix metalloproteinase-2 and membrane type 1 matrix metalloproteinase through beta1 integrin. Cancer Res. 1999;59:1635–41. [PubMed] [Google Scholar]
- 16.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–10. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Barbolina MV, Adley BP, Shea LD, Stack MS. Wilms’ Tumor Gene Protein 1 Is Associated with Ovarian Cancer Metastasis and Modulates Cell Invasion. Cancer. 2008 doi: 10.1002/cncr.23341. in press. [DOI] [PubMed] [Google Scholar]
- 18.Berrier AL, Yamada KM. Cell-matrix adhesion. Journal of Cellular Physiology. 2007;213:565–73. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 19.Lock JG, Wehrle-Haller B, Stromblad S. Cell-matrix adhesion complexes: master control machinery of cell migration. Seminars in Cancer Biology. 2008;18:65–76. doi: 10.1016/j.semcancer.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Brigstock DR. The CCN family: a new stimulus package. Journal of Endocrinology. 2003;178:169–75. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 21.Lau LF, Lam SC. The CCN family of angiogenic regulators: the integrin connection. Experimental Cell Research. 1999;248:44–57. doi: 10.1006/excr.1999.4456. [DOI] [PubMed] [Google Scholar]
- 22.Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–4. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- 23.Kikuchi R, Tsuda H, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, Inazawa J, Imoto I. Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Research. 2007;67:7095–105. doi: 10.1158/0008-5472.CAN-06-4567. [DOI] [PubMed] [Google Scholar]
- 24.Moser TL, Pizzo SV, Bafetti LM, Fishman DA, Stack MS. Evidence for preferential adhesion of ovarian epithelial carcinoma cells to type I collagen mediated by the alpha2beta1 integrin. Int J Cancer. 1996;67:695–701. doi: 10.1002/(SICI)1097-0215(19960904)67:5<695::AID-IJC18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Liao D, Scannevin RH, Huganir R. Activation of silent synapses by rapid activity-dependent synaptic recruitment of AMPA receptors. Journal of Neuroscience. 2001;21:6008–17. doi: 10.1523/JNEUROSCI.21-16-06008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller GM. In vitro differentiation of embryonic stem cells. Current Opinion in Cell Biology. 1995;7:862–9. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–4. [PubMed] [Google Scholar]
- 28.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 29.Batrakova EV, Kelly DL, Li S, Li Y, Yang Z, Xiao L, Alakhova DY, Sherman S, Alakhov VY, Kabanov AV. Alteration of genomic responses to doxorubicin and prevention of MDR in breast cancer cells by a polymer excipient: pluronic P85. Molecular Pharmaceutics. 2006;3:113–23. doi: 10.1021/mp050050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan CY, Salabat MR, Ding XZ, Kelly DL, Talamonti MS, Bell RH, Jr, Adrian TE. Identification and in silico characterization of a novel gene: TPA induced trans-membrane protein. Biochemical & Biophysical Research Communications. 2005;329:755–64. doi: 10.1016/j.bbrc.2005.02.037. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Team. RDC. R Foundation for Statistical Computing Vienna, Austria. 2005. R: A language and environment for statistical computing. [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 35.Zirn B, Hartmann O, Samans B, Krause M, Wittmann S, Mertens F, Graf N, Eilers M, Gessler M. Expression profiling of Wilms tumors reveals new candidate genes for different clinical parameters. International Journal of Cancer. 2006;118:1954–62. doi: 10.1002/ijc.21564. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Barbolina MV, Adley BP, Kelly DL, Fought AJ, Scholtens D, Shea LD, Stack MS. Motility Realted Actinin Alpha-4 Is Associated with Advanced and Metastatic Ovarian Carcinoma. Lab Invest. 2008 doi: 10.1038/labinvest.2008.25. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbolina MV, Stack MS. Membrane type 1-matrix metalloproteinase: Substrate diversity in pericellular proteolysis. Semin Cell Dev Biol. 2007 doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munshi HG, Stack MS. Reciprocal interactions between adhesion receptor signaling and MMP regulation. Cancer & Metastasis Reviews. 2006;25:45–56. doi: 10.1007/s10555-006-7888-7. [DOI] [PubMed] [Google Scholar]
- 40.Burleson KM, Casey RC, Skubitz KM, Pambuccian SE, Oegema TR, Jr, Skubitz AP. Ovarian carcinoma ascites spheroids adhere to extracellular matrix components and mesothelial cell monolayers. Gynecologic Oncology. 2004;93:170–81. doi: 10.1016/j.ygyno.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 41.Burleson KM, Hansen LK, Skubitz AP. Ovarian carcinoma spheroids disaggregate on type I collagen and invade live human mesothelial cell monolayers. Clinical & Experimental Metastasis. 2004;21:685–97. doi: 10.1007/s10585-004-5768-5. [DOI] [PubMed] [Google Scholar]
- 42.Ball DK, Rachfal AW, Kemper SA, Brigstock DR. The heparin-binding 10 kDa fragment of connective tissue growth factor (CTGF) containing module 4 alone stimulates cell adhesion. Journal of Endocrinology. 2003;176:R1–7. doi: 10.1677/joe.0.176r001. [DOI] [PubMed] [Google Scholar]
- 43.Chen PS, Wang MY, Wu SN, Su JL, Hong CC, Chuang SE, Chen MW, Hua KT, Wu YL, Cha ST, Babu MS, Chen CN, et al. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. Journal of Cell Science. 2007;120:2053–65. doi: 10.1242/jcs.03460. [DOI] [PubMed] [Google Scholar]
- 44.Gao R, Brigstock DR. A novel integrin alpha5beta1 binding domain in module 4 of connective tissue growth factor (CCN2/CTGF) promotes adhesion and migration of activated pancreatic stellate cells. Gut. 2006;55:856–62. doi: 10.1136/gut.2005.079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heng EC, Huang Y, Black SA, Jr, Trackman PC. CCN2, connective tissue growth factor, stimulates collagen deposition by gingival fibroblasts via module 3 and alpha6- and beta1 integrins. Journal of Cellular Biochemistry. 2006;98:409–20. doi: 10.1002/jcb.20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jedsadayanmata A, Chen CC, Kireeva ML, Lau LF, Lam SC. Activation-dependent adhesion of human platelets to Cyr61 and Fisp12/mouse connective tissue growth factor is mediated through integrin alpha(IIb)beta(3) Journal of Biological Chemistry. 1999;274:24321–7. doi: 10.1074/jbc.274.34.24321. [DOI] [PubMed] [Google Scholar]
- 47.Tammela J, Odunsi K. Gene expression and prognostic significance in ovarian cancer. Minerva Ginecologica. 2004;56:495–502. [PubMed] [Google Scholar]
- 48.Bidard FC, Pierga JY, Vincent-Salomon A, Poupon MF. A “class action” against the microenvironment: do cancer cells cooperate in metastasis? Cancer & Metastasis Reviews. 2008;27:5–10. doi: 10.1007/s10555-007-9103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Le Clainche C, Carlier M-F. Regulation of Actin Assembly Associated With Protrusion and Adhesion in Cell Migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 50.Ellerbroek SM, Fishman DA, Kearns AS, Bafetti LM, Stack MS. Ovarian carcinoma regulation of matrix metalloproteinase-2 and membrane type 1 matrix metalloproteinase through beta1 integrin. Cancer Research. 1999;59:1635–41. [PubMed] [Google Scholar]
- 51.Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Identification of candidate angiogenic inhibitors processed by matrix metalloproteinase 2 (MMP-2) in cell-based proteomic screens: disruption of vascular endothelial growth factor (VEGF)/heparin affin regulatory peptide (pleiotrophin) and VEGF/Connective tissue growth factor angiogenic inhibitory complexes by MMP-2 proteolysis. Molecular & Cellular Biology. 2007;27:8454–65. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto G, Inoki I, Fujii Y, Aoki T, Ikeda E, Okada Y. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. Journal of Biological Chemistry. 2002;277:36288–95. doi: 10.1074/jbc.M201674200. [DOI] [PubMed] [Google Scholar]
- 53.Hoshijima M, Hattori T, Inoue M, Araki D, Hanagata H, Miyauchi A, Takigawa M. CT domain of CCN2/CTGF directly interacts with fibronectin and enhances cell adhesion of chondrocytes through integrin alpha5beta1. FEBS Letters. 2006;580:1376–82. doi: 10.1016/j.febslet.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 54.Gao R, Brigstock DR. Connective tissue growth factor (CCN2) induces adhesion of rat activated hepatic stellate cells by binding of its C-terminal domain to integrin alpha(v)beta(3) and heparan sulfate proteoglycan. Journal of Biological Chemistry. 2004;279:8848–55. doi: 10.1074/jbc.M313204200. [DOI] [PubMed] [Google Scholar]
- 55.Duncan WC, Hillier SG, Gay E, Bell J, Fraser HM. Connective tissue growth factor expression in the human corpus luteum: paracrine regulation by human chorionic gonadotropin. Journal of Clinical Endocrinology & Metabolism. 2005;90:5366–76. doi: 10.1210/jc.2005-0014. [DOI] [PubMed] [Google Scholar]
- 56.Harlow CR, Hillier SG. Connective tissue growth factor in the ovarian paracrine system. Molecular & Cellular Endocrinology. 2002;187:23–7. doi: 10.1016/s0303-7207(01)00702-x. [DOI] [PubMed] [Google Scholar]
- 57.Harlow CR, Bradshaw AC, Rae MT, Shearer KD, Hillier SG. Oestrogen formation and connective tissue growth factor expression in rat granulosa cells. Journal of Endocrinology. 2007;192:41–52. doi: 10.1677/joe.1.06689. [DOI] [PubMed] [Google Scholar]
- 58.Graness A, Cicha I, Goppelt-Struebe M. Contribution of Src-FAK signaling to the induction of connective tissue growth factor in renal fibroblasts. Kidney International. 2006;69:1341–9. doi: 10.1038/sj.ki.5000296. [DOI] [PubMed] [Google Scholar]
- 59.Graness A, Giehl K, Goppelt-Struebe M. Differential involvement of the integrin-linked kinase (ILK) in RhoA-dependent rearrangement of F-actin fibers and induction of connective tissue growth factor (CTGF) Cellular Signalling. 2006;18:433–40. doi: 10.1016/j.cellsig.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 60.Heusinger-Ribeiro J, Eberlein M, Wahab NA, Goppelt-Struebe M. Expression of connective tissue growth factor in human renal fibroblasts: regulatory roles of RhoA and cAMP. Journal of the American Society of Nephrology. 2001;12:1853–61. doi: 10.1681/ASN.V1291853. [DOI] [PubMed] [Google Scholar]
- 61.Ott C, Iwanciw D, Graness A, Giehl K, Goppelt-Struebe M. Modulation of the expression of connective tissue growth factor by alterations of the cytoskeleton. Journal of Biological Chemistry. 2003;278:44305–11. doi: 10.1074/jbc.M309140200. [DOI] [PubMed] [Google Scholar]
- 62.Chaqour B, Goppelt-Struebe M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. FEBS Journal. 2006;273:3639–49. doi: 10.1111/j.1742-4658.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- 63.Schild C, Trueb B. Mechanical stress is required for high-level expression of connective tissue growth factor. Experimental Cell Research. 2002;274:83–91. doi: 10.1006/excr.2001.5458. [DOI] [PubMed] [Google Scholar]
- 64.Schild C, Trueb B. Three members of the connective tissue growth factor family CCN are differentially regulated by mechanical stress. Biochimica et Biophysica Acta. 2004;1691:33–40. doi: 10.1016/j.bbamcr.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Chang CC, Shih JY, Jeng YM, Su JL, Lin BZ, Chen ST, Chau YP, Yang PC, Kuo ML. Connective tissue growth factor and its role in lung adenocarcinoma invasion and metastasis. Journal of the National Cancer Institute. 2004;96:364–75. doi: 10.1093/jnci/djh059. see comment. [DOI] [PubMed] [Google Scholar]
- 66.Lin BR, Chang CC, Che TF, Chen ST, Chen RJ, Yang CY, Jeng YM, Liang JT, Lee PH, Chang KJ, Chau YP, Kuo ML. Connective tissue growth factor inhibits metastasis and acts as an independent prognostic marker in colorectal cancer. Gastroenterology. 2005;128:9–23. doi: 10.1053/j.gastro.2004.10.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


