Abstract
Background
Cancer has profound effects on gene expression, including a cell’s glycosylation machinery. Thus, tumors produce glycoproteins that carry oligosaccharides with structures that are markedly different from the same protein produced by a normal cell. A single protein can have many glycosylation sites that greatly amplify the signals they generate as compared to their protein backbones.
Content
We survey clinical tests that target carbohydrate modifications. for diagnosing and treating cancer. Next, we present the biological relevance of glycosylation to disease progression by highlighting the role these structures play in adhesion, signaling and metastasis, and then address current methodological approaches to biomarker discovery that capitalize on selectively capturing tumor-associated glycoforms to enrich and identify disease-related candidate analytes. Finally, we discuss emerging technologies—multiple reaction monitoring and lectin-antibody arrays—as potential tools for biomarker validation studies in pursuit of clinically useful tests.
Summary
The future of carbohydrate-based biomarker studies has arrived. At all stages, from discovery through verification and deployment into clinics, glycosylation should be considered a primary readout or a way of increasing the sensitivity and specificity of protein-based analyses.
Keywords: glycosylation, cancer, proteomics, biomarker
Clinical tests based on cancer-related changes in glycosylation
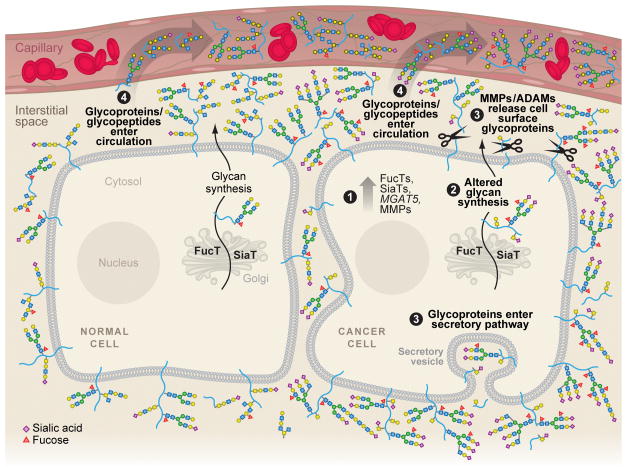
Aberrant glycosylation has been recognized for more than 30 years as a hallmark of cancer (1). However, the complex nature of glycan structure and synthesis has constrained the pace of discoveries relating to their biological significance. Recent advances in carbohydrate chemistry, chemical biology, and mass spectrometric (MS) techniques have opened the door to rapid progress in correlating glycan structure and function (2–10). At the same time, the maturation of proteomics has put cancer biomarker discovery studies at the top of many to-do lists (11). The confluence of these two fields has led many investigators to the same conclusion: Exploiting differences in glycosylation between malignant and healthy tissues likely affords excellent opportunities to identify sensitive and specific cancer biomarkers (12–20). Specifically, as diagrammed in Figure 1, glycosylation machinery appears to be particularly sensitive to malignant transformation; as a result, the saccharide structures that are added to normal cellular proteins change, resulting in neoglycoforms that can be released from the cell through conventional secretory pathways, or as the result of enhanced proteinase activity. It is possible that a portion of these alternatively glycosylated molecules reach the bloodstream. As such, they could serve as early sentinels that enable cancer detection.
Figure 1. Malignant cells release glycoproteins carrying disease-related carbohydrate epitopes into the interstitial space, where they can reach the circulation.
Cancer is associated with major changes in the glycan biosynthetic machinery, including (1) upregulation of fucosyltransferases (FucTs), sialyltransferases (SiaTs), and the MGAT5 gene product, which is involved in the elaboration of highly branched N-linked glycans. Disease-relevant proteinases such as MMPs and ADAM family members are also upregulated. Changes in the expression of glycosyltransferases result in altered glycan assembly, which occurs in the endoplasmic reticulum and Golgi (2). Accordingly, the glycoprotein products of tumor cells carry aberrant carbohydrate structures as compared to their normal counterparts. Typical changes include increased levels of fucose (red triangle), sialic acid (purple diamond), the addition of polylactosamine units (repeating sequences of galactose [yellow circle] and N-acetylglucosamine [blue square]), and higher-ordered branching of N-linked glycans. O-linked glycans are also affected in cancer, typically carrying incomplete or prematurely truncated structures relative to those found on normal cells. After secretion or proteolytic cleavage, glycosylated molecules and/or their cleavage products are released into the interstitial space (3), where they can enter the circulation (4). Since glycoproteins and mucins usually carry many carbohydrate chains, the signals they produce are highly amplified as compared to proteins, making them attractive candidates as biomarkers.
Intriguingly, many of the oldest and mostwidely used clinical cancer biomarker tests detect glycoproteins. These include carcinoembryonic antigen (CEA), commonly used as a marker of colorectal cancer, CA-125, frequently employed to diagnose ovarian cancer, and prostate-specific antigen (PSA) (14, 21, 22). All of these diagnostic markers are glycoproteins, and the glycans that they present change during oncogenesis (18, 23, 24). Capitalizing on alterations in cancer-related protein glycoforms affords the possibility of increasing diagnostic sensitivity and specificity. The separation of serum PSA glycoforms by lectins -proteins that specifically bind glycans according to their structural epitopes -highlights this point. The Maackia amurensis agglutinin lectin can distinguish (p < 0.001) between blood samples from individuals with benign prostatic hypertrophy and prostate cancer patients (18), which standard PSA tests fail to do (25).
Tellingly, several other clinically useful cancer biomarkers directly detect glycan epitopes. CA 19-9 binds to sialyl Lewisa (sLea) (Siaα2–3Galβ1–3(Fucα1–4)GlcNAc) (26), a terminal epitope that imparts unique biological functions (see next section for details). Although serum CA 19-9 concentrations are the most commonly used biomarker for diagnosing pancreatic cancer, monitoring treatment efficacy, and detecting recurrence, the current test lacks sensitivity and specificity (27). In this context, it is important to consider that CA 19-9 and related epitopes exhibit genetic variation within the population. For example, the Secretor (FUT2) and Le (FUT3) genes control the expression of enzymes in the synthetic pathway that governs expression of critical antigenic saccharide residues and, thus, an individual’s presentation of Le blood group structures (28). Accordingly, depending on their genetic makeup, many populations have high background concentrations of these epitopes present as normal components of serum and other secreted glycoproteins (29, 30). Several groups have proposed including analysis of an individual’s Secretor and Le blood group status to improve the diagnostic sensitivity and specificity of serum CA19-9 tests (28, 31). Indeed, one study assessed serum CA 19-9 concentrations in 500 healthy individuals and categorized the donors into six groups based on FUT2 and FUT3 genotype (28). They observed a six-fold difference in the upper reference limit (the concentrations of antigen above which disease is suspected) between the groups with the highest and lowest endogenous CA 19-9 production, suggesting the importance of including this information in the analysis.
The prevalence of glycoprotein and glycan cancer biomarkers can be traced to de novo or altered glycan expression by transformed cells (see Figure 1 and next section). Detection of these changes at the cytological (e.g., bladder cancer) and histological levels by lectins and glyco-reactive antibodies guides disease diagnoses and enables more accurate prognoses. The lectins Helix pomatia agglutinin (HPA), which detects α-GalNAc ±β1–4Gal, and Ulex europaeus I agglutinin (UEA 1), which recognizes Fucα1–2Galβ, are used to assess breast cancer biopsy specimens (32–34). HPA is also part of a panel of markers for histological characterization of gastric cancer specimens (35). In both cases, HPA expression is correlated with increased lymph node metastases, and expression of saccharide sequences that react with either HPA or UEA 1 is related to decreased survival (32–35). Interestingly, many of the most informative tests assess expression of Le antigens. For example, CA 19-9 and other anti-sLea, -Lex, -sLex, and -Ley antibodies are used in the evaluation of biopsy specimens from breast, bladder, colorectal, esophageal and non small cell lung carcinoma (36–43). In all instances, Le antigen expression is correlated with increased metastasis, advanced stage of disease, and reduced survival time.
Thus, it is clear that many of the clinical tests currently used to diagnose and manage the treatment of cancer exploit changes in glycosylation that accompany the disease process. In this context, it is interesting to consider the fact that very few biomarker discovery strategies are designed to focus on this class of post-translational modifications. One reason may be that the complexity and heterogeneity of carbohydrate structures carried by glycoproteins have made it difficult to understand their functions. As discussed in the next section, recent progress has been made in this important area.
Biological significance of altered glycosylation in cancer
Cancer-related changes in glycosylation reflect interesting and disease-specific alterations in glycan biosynthetic pathways. These include variations in the expression of glycosyltransferases, enzymes that add activated donor monosaccharides in specific stereochemistries to acceptors, i.e., growing carbohydrate chains. Expression of enzymes and their corresponding substrates can be downregulated, as is observed with Core 3 O-carbohydrate structures and laminin-binding glycans in gastric and colorectal tumor biopsies or prostate and breast cancer cell lines, respectively (44, 45). Reduction in these glycoforms appears to be related to increased invasive/metastatic potential of carcinoma cells (46). Alternatively, glycan structures can be upregulated in tumors as compared to normal cells. For example, β1,6 N-acetylglucosaminyltransferase V (MGAT5), which is involved in branching of N-linked glycans, is overexpressed in numerous cancers (47); the product of this enzyme, which the Phaseolus vulgaris leukoagglutinating lectin detects, is associated with metastasis and decreased survival time in colorectal cancer (48). The upregulation of cell surface expression of specific monosaccharides is also observed in cancer; prominent examples include increased sialylation and fucosylation. Regarding sialic acid, the sialyltransferases ST3Gal I, which adds N-acetylneuraminic acid (NeuAc) to O-linked galactose (Gal) moieties, and ST6GalNAc I, which adds sialic acid to the Tn antigen (GalNAcα1-O-Ser/Thr), are both upregulated in breast cancer and are associated with aggressive disease and poor prognosis (49–51). Another sialyltransferase, ST6Gal I, is also reported to be overexpressed in many malignant tumors (52). Regarding fucose, expression of several relevant biosynthetic pathways correlates with cancer. These include the GDP-mannose 4,6-dehydratase, GDP-fucose transporter, and fucosyl transferases FucT-IV and FucTVII, all of which are upregulated to varying degrees in hepatocellular carcinomas as compared to normal tissues (reviewed in (53). Similarly, increases in the expression of both FucT-IV and the sialyltransferase ST3Gal II are observed in human colorectal cancer biopsy specimens as compared to adjacent noncancerous tissue (54).
Members of the matrix metalloproteinase (MMP) and ADAM (a disintegrin and metalloproteinase) families are another important class of molecules that are upregulated in malignant tissues (55, 56). These degradative enzymes cleave proteins and glycoproteins from the cell surface and the adjacent extracellular matrix (ECM), often altering cellular behavior and stromal signals, respectively. In aggregate, these changes are thought to facilitate the growth and metastasis of tumor cells. Forexample, “ectodomain shedding” releases and activates plasma membrane-associated molecules such as epidermal growth factor (EGF). These alterations afford several clinical applications; e.g., high MMP expression correlates with adverse outcomes in breast, gastric, pancreatic, and prostate cancers (57, 58). Their glycan-containing products, which have important biological activities, can also be exploited for diagnostic purposes. Thus, tumor-associated enhanced proteinase activity increases the likelihood that molecules carrying these neoglycans will be identified in the surrounding tissue compartment, where they may eventually enter the circulation (Figure 1 and following sections).
Cancer-related changes in glycan biosynthesis and proteinase activity have enormous biological ramifications, many of which advance tumor formation and disease progression. For instance, some carbohydrate modifications seem to act as “molecular switches” affecting glycan conformation and thus lectin binding. In this context, cancer-related substitutions, including sialylation and core fucosylation, govern myriad functions (e.g., serum clearance and hepatic uptake) (59). Additionally, upregulation of MMPs/ADAMs and presentation of cancer-related glycans generally decrease cell-cell and cell-ECM adhesion and promote migration (56, 60). Regulation of integrin function appears to play a key role in these processes. Integrins are heterodimeric adhesion molecules that mediate cell interactions with ECM substrates such as fibronectin and laminin, and with cell surface proteins such as vascular cell adhesion molecule (61). As a group, integrins are highly N-glycosylated, with some dimers, e.g., α5β1 and α3β1, possessing a total of 26 potential N-linked glycosylation sites (62).
Numerous aspects of integrin biology—including expression, heterodimerization, ligand binding, and downstream signaling—are modulated by glycosylation (63). A minimal glycan repertoire allows cell surface expression of these heterodimers; further elaborations to glycan structure refine integrin function. For instance, increased integrin sialylation, mediated by the glycosyltransferase ST6Gal I, leads to integrin activation, increased adhesion to ECM substrates, and augmented migration and invasion of colon cancer cell lines in vitro (52, 64). Similarly, in vivo, ST6Gal I expression by spontaneously derived mouse mammary tumors modulates integrin signaling and tumor differentiation (65). In addition to sialylation, integrins are also modified by the glycosyltransferase MGAT5 and, as a result, carry highly branched structures (47). Spontaneous mammary tumors, which developed in an MGAT5-deficient mouse model, exhibited decreased growth, metastasis and signaling downstream of the focal adhesion kinase, evidence that the observed effects are mediated, in part, by loss of integrin-associated N-linked branching (47).
The highly branched MGAT5 glycan products are often further elaborated with the addition of polylactosamine chains, polymeric repeating units of galactose and N-acetylglucosamine (i.e., lactosamines). Galectins, a family of endogenous lectins, bind these epitopes. Galectins can multimerize through homotypic interactions, forming a lattice-type network that incorporates glycosylated ligands and other galectin molecules (66). This architecture stabilizes bound cell surface molecules, preventing lateral movements and endocytosis. Thus, this phenomenon can affect many processes. MGAT5 regulates tumor motility, in part, through galectin-3 binding to newly presented lactosamine sequences, and the consequences include changes in integrin signaling (67). MGAT5 also modifies N-linked glycans on receptor tyrosine kinases such as EGF receptor (EGFR) and transforming growth factor β receptors I and II (TβRI and II). Upon polylactosaminylation, they become ligands for galectin-3, which leads to lattice formation and receptor entrapment. The lattice helpsthe cell surface receptors resist removal by constitutive endocytosis, and thus increases receptor signaling (68). EGFR is highly N-glycosylated while TβRI and TβRII carry minimal modifications. Accordingly, cells display a strong MGAT5-dependent response when ligands bind to the former receptor, and less so when the latter receptors become engaged (68). Interestingly, MGAT5 also plays a role in epithelial to mesenchymal transitions (68), an early stage of oncogenesis that is correlated with metastasis (69). This effect might be mediated, in part, through MGAT5 influences on integrin and growth factor receptor signaling.
Metastasis is a critical event in the progression of tumor development. This multistep process begins when individual primary tumor cells breach the basement membrane and access the local vasculature, where they interact with endothelial cells, platelets and leukocytes (70, 71). Exit from the circulation is accomplished by extravasation, which requires a series of steps. This shear stress-dependent process begins with rolling on endothelial cells, which leads to tethering and integrin-dependent stable adhesion. It is thought that extravasation occurs at receptive sites primed by circulating tumor-derived factors. Successful metastasis requires coordinated interactions between the newly engrafted tumor cell, the “seed,” and the “soil” at sites of metastasis (70, 71).
The first step of extravasation, rolling of cells along the endothelium, is regulated by glycosylation. Specifically, rolling is mediated by adhesion under shear stress of the selectin family of endogenous lectins to their carbohydrate ligands on cell surface glycoproteins. The selectin family consists of three members that exhibit differential expression patterns. Endothelial cells display E-selectin and P-selectin, platelets express P-selectin, and leukocytes present L-selectin (72, 73). Carbohydrate ligands for selectins are modified Le blood group antigens, i.e., contain fucosylated and sialylated epitopes. These structures, detectable by antibodies such as CA19-9 (26), are abundant in malignant tissues, where their expression is strongly correlated with metastasis and poor disease outcome (see first section above). It is thought that tethering and rolling of tumor cells occurs through selectin-mediated interactions between malignant cells and the endothelium (via E-and P-selectin). However, interactions between malignant cells and circulating platelets and leukocytes (via P-and L-selectins, respectively) are also thought to participate in tumor cell extravasation. One model envisions that circulating aggregates of tumor cells, platelets, and/or leukocytes form microvascular occlusions that arrest the cells, providing a possible means of egress from the vessel (70, 73). Thus, although expression patterns vary by virtue of location and carbohydrate selectivity, each selectin family member is poised to influence tumor metastasis.
Evidence is accumulating that the clinical correlation observed between tumor expression of selectin ligands and metastasis (see Section 1) is a reflection of a causal relationship (74). Mice lacking L-selectin, P-selectin, or both molecules, exhibit greater resistance to the metastasis of colon carcinoma cells (75, S76). E-selectin is also implicated in tumor metastasis. Notably, one study followed dissemination of melanoma tumor cells, either presenting or lacking E-selectin ligands, in a mouse model in which E-selection was overexpressed in the liver. Tumor cell expression of E-selectin ligands was required for metastasis to the liver of transgenic but not wild-type mice (S77). Thus, the pattern of E-selectin expression governs metastasis, a finding that has interesting implications for the pathogenesis of human cancers. Additionally, cancer-related protein scaffolds have been identified as selectin-ligand carriers, andthus as participants in the extravasation process. For instance, CEA, the glycoprotein commonly used as a marker for colorectal cancer (first section above), and CD44, a highly glycosylated cell surface adhesion protein, can serve as E-and L-selectin ligands (74).
In summary, the examples in this section illustrate the biological importance of glycosylation to tumor development and disease progression. As biomarker discovery becomes more sophisticated, it is clearthat using glycobiology to inform experimental design will lead to more sensitive and specific diagnostic tests (S78, S79). In this context, cancer-related changes in glycosylation can guide biomarker studies. As discussed in the next section, many laboratories have already begun exploring approaches to exploit the latest technological and biological breakthroughs in these areas.
Current approaches to glycan-based biomarker discovery
The challenge of biomarker discovery is to identify the disease-related “needle” in the “haystack” of normally expressed proteins. Although several accessible human fluids, including urine and saliva, are available for biomarker studies, the subject of the majority of biomarker discovery approaches to date—and the one with the highest order of complexity—is processed blood (as plasma or serum). In this context, the normal plasma or serum proteome/glycome is the “haystack,” which creates a myriad of background issues. In this regard, it is noteworthy that 82 of the top 100 most abundant plasma proteins are glycoproteins (Supplemental Table 1) (S80). However, the majority of these endogenous molecules carry basic glycan motifs lacking typical cancer-related structures, including fucosylation, sialylation, more complex N-linked branching and polylactosaminylation. Thus, judicious selection of lectins or antibodies that recognize carbohydrate epitopes for capture of cancer-related glycoforms enables separation of potential biomarker targets. The list of potentially useful lectins and antibodies shown in Table 1 demonstrates the versatility of these tools for identifying structural differences.
Table 1.
Specificities of lectins and carbohydrate-reactive antibodies
| Detection reagent Lectin | General reactivity | Lectin or Antibody | Carbohydrate structural motif | References |
|---|---|---|---|---|
| αβ-D-galactosyl-specific | ||||
| Agaricus bisporus (ABA) | Galβ1–3GalNAc-serine/threonine | S120 | ||
| Allomyrina dichotoma (AlloA) | NeuAcα2–3Galβ1–4GlcNAc | S121 | ||
| Artocarpus integrifolia (Jacalin) | Galβ1–3GalNAc, Galα1–6Gal | S122 | ||
| Erythrina christagalli (ECA) | Galβ1–4GlcNAc | S123 | ||
| Arachis hypogaea (peanut, PNA) | Galβ1–3GalNAc | S124 | ||
| D-N-acetylgalactosaminyl-specific | ||||
| Dolichos biflorus (DBA) | GalNAcα1–3GalNAc | S125 | ||
| GalNAcα1–3Gal | S126 | |||
| Glycine max (soybean, SBA) | GalNAcα1–3GalNAc | S126 | ||
| GalNAcα/β1–3/4Gal | ||||
| Helix pomatia (HPA) | α-GalNAc, GalNAcβ1–4Gal | S125, S126 | ||
| Phaseolus lunatus (lima bean, LBA) | GalNAcα1–3(Fucα1–2)Galβ | S127 | ||
| GalNAcα1–2Galβ | ||||
| Wisteria floribunda (WFA) | GalNAcα1–3GalNAc | S125 | ||
| GalNAcβ1–3/4Gal | S126 | |||
| Vicia villosa (VVA) | GalNAc-Ser | S128 | ||
| D-N-acetylglucosamine-specific | ||||
| Datura stramonium (DSA) | (GlcNAcβ1–4)2–4, Galβ1–4GlcNAc | S129 | ||
| Griffonia simplifilica II (GS-II) | GlcNAcα1–4Galβ1–4GlcNAc | S130 | ||
| Lycopersicon esculentum (Tomato) | (GlcNAcβ1–4)1–4 | S131 | ||
| Triticum vulgare (wheat germ, WGA) | (GlcNAcβ1–4)2–5, Neu5Ac | S132, S133 | ||
| Manβ1–4GlcNAcβ1–4GlacNAc | S134 | |||
| D-mannosyl-specific | ||||
| Canavalia ensiformis (Con A) | branched N-linked hexa-saccharide | S135 | ||
| Galanthus nivalis (GNA) | Manα1–3Man | S136 | ||
| Lens culinaris (LCA) | Fucα1–6GlcNAc-N-Asn containing N-linked oligosaccharides | S135 | ||
| Pisum sativum (PSA) | Fucα1–6GlcNAc-N-Asn containing N-linked oligosaccharides | S135 | ||
| α-L-Fucosyl-specific | ||||
| Aleuria aurantia (AAL) | Fucα1–6/3GlcNAc | S137 | ||
| Anguilla anguilla (AAA) | α-L-Fuc | S138 | ||
| Lotus tetragonolobus (LCA) | Fucα1–2Galβ1–4(Fucα1–3)GlcNAc | S139 | ||
| Ulex europaeus I (UEA I) | Fucα1–2Galβ | S140 | ||
| Neu5Ac-specific | ||||
| Cancer antennarius (CAA) | 4/9-O-Ac-Neu5Ac | S141 | ||
| Maackia amurensis II (MAA II) | Neu5Acα2–3Galβ1–4Glc/GlcNAc | S142 | ||
| Sambucus nigra (SNA) | Neu5Acα2–6Gal/GalNAc | S143 | ||
| Lectins with complex specificities | ||||
| Cicer arietinum (CPA) | complex structure, binding inhibited by human IgM, fetuin | S144 | ||
| Euonymus europaeus (EEA) | Galα1–3(Fucα1–2)Galβ1–3/4GlcNAc | S145 | ||
| Phaseolus vulgaris erythroagglutinating (PHA-E) | N-linked bi-antennary | S146 | ||
| Phaseolus vulgaris leukoagglutinating (PHA-L) | N-linked tri/tetra-antennary | S146 | ||
| Antibody | ||||
| Lewis and ABH blood group antigens | Anti blood group O (H) | (Fuca1–2)Galb1–3/4GlcNAc | S147 | |
| Anti blood group A | GalNAca1–3(Fuca1–2)Gal | S147 | ||
| Anti-blood group B | Gala1–3(Fuca1–2)Gal | S148 | ||
| Anti-Lewis a (Lea) | Galb1–3(Fuca1–4)GlcNAc | S149 | ||
| Anti-sialylated Lewis a (sLea) | NeuAca2–3Galb1–3(Fuca1–4)GlcNAc | S150 | ||
| Anti-Lewis b (Leb) | Fuca1–2Galb1–3(Fuca1–4)GlcNAc | S151 | ||
| Anti-Lewis x (Lex, CD15) | Galb1–4(Fuca1–3)GlcNAc | S152 | ||
| Anti-sialylated Lewis x (sLex, CSLEX-1) | NeuAca2–3Galb1–4(Fuca1–3)GlcNAc | S153 | ||
| Anti-Lewis y (Ley) | Fuca1–2Galb1–4(Fuca1–3)GlcNAc | S154 | ||
| HECA-452 | NeuAca2–3Galb1–4(Fuca1–3)GlcNAc, | S153 | ||
| NeuAca2–3Galb1–4(Fuca1–3) (SO3–6)GlcNAc, | ||||
| NeuAca2–3(SO3–6)Galb1–4(Fuca1–3)GlcNAc, | ||||
| MECA-79 | NeuAca2–3(SO3–6)Galb1–4(Fuca1–3)(SO3–6)GlcNAc complex structure containing SO3–6GlcNAc | S155 | ||
Given that affinity selection is the hallmark of workflows that are designed to capture glycan-containing molecules, itis possible to analyze unfractionated samples. However, many investigators commonly include immunoaffinity depletion of the most abundant proteins, which enables deeper coverage of the remaining lower-abundance proteome where biomarkers typically reside. Multiple platforms have been employed for this purpose, including ProteomeLab IgY (Beckman Coulter, Fullerton, CA; www.beckmancoulter.com) Seppro IgY and SuperMix (Sigma-Aldrich, Saint Louis, MO; www.sigmaaldrich.com) (S81)and the Multiple Affinity Removal System (MARS; Agilent Technologies, Santa Clara, CA; www.agilent.com) (S81–S83).
Current glycan-based biomarker approaches typically involve one or more enrichment steps, proteolytic digestion, and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. Enrichment of carbohydrate-containing species is performed either at the glycoprotein or glycopeptide level, with distinct rationales for each strategy. Major advantages of performing affinity selection at the glycoprotein level include the increased likelihood of obtaining sequence information from multiple peptides, which in turn facilitates glycoprotein identification (S84). Disadvantages of this approach, which are overcome by selecting at the glycopeptide level, include a higher probability of nonspecific protein protein interactions, a particular concern since many lectins are hydrophobic molecules (S85).
As compared to their O-linked counterparts, N-linked glycopeptides are more commonly analyzed in glycan-based biomarker workflows, as they are amenable to peptide N-glycosidase F (PNGase F) digestion, which hydrolyzes the amide bond of the asparagine residue to which the glycan is attached (S86). This reaction results in a 1-dalton molecular mass increase in the deglycosylated form of the peptide. Additional certainty is gained by requiring that thismass shift be observed in the context of an N-linked consensus sequence (NXT/S, where X is any residue except proline). Since PNGase F is an asparagine deamidase, it is also possible to enzymatically incorporate oxygen-18 at the site of glycan release (S87). The development of equivalent enzymes for the release of O-linked structures, and identification of their sites of attachment have, yet to be achieved. In this regard, it may be possible to engineer a proteinase that can hydrolyze the GalNAc-Ser/Thr linkage.
Lectin-and antibody-based affinity selection
A variety of protocols have been developed for lectin capture of glycoproteins and glycopeptides. These include using single lectins (S82, S84, S88), as well as mixtures, e.g., concanavalin A, wheat germ agglutinin, and Artocarpus integrifolia (jacalin). The latter approach has been dubbed multi-lectin affinity chromatography (S89–S92). Less commonly, carbohydrate-reactive antibodies such as those recognizing Le blood group antigens (Table 1) are used for enrichment (S93). Additional lectin-capture steps and dimensions of fractionation (e.g., hydrophilic interaction, ion exchange, reverse-phase chromatography) have been used in series to achieve greater protein and peptide coverage (S82, S84, S87, S89–S91, S93). Elution methods include displacement by specific mono-or disaccharide solutions (S88, S89, S94), or by high-salt and/or acidic conditions (S84, S93, S95). Numerous workflows employ commercially available agarose-conjugated lectins, in either a centrifugal, gravity flow, or low-pressure HPLC format (S84, S89, S92, S93). Magnetic bead technology has also been used (S95, S96). When desirable, a higher degree of reproducibility and speed is achieved by employing lectins conjugated to HPLC-compatible matrices (S83, S88), which enable high-pressure/high-flow-rate chromatography.
In general, glycoprotein/glycopeptide enrichment methods identify fewer proteins than in-depth protein analyses. For instance, in four studies involving lectin or antibody glycoprotein/glycopeptide capture and reverse-phase LC/MS/MS analyses, 25 to 150 glycoproteins were observed (S84, S89, S93, S94). As expected, the inclusion of additional separation steps often increases the number of proteins identified. Including a third dimension of separation and/or depletion of abundant proteins enabled identification of 50 to 250 glycoproteins (S82, S87, S89, S94). Interestingly, narrow-specificity lectins that recognize disease-related carbohydrate structures capture fewer plasma proteins than do those with broader specificities, making them attractive affinity selectors for isolating cancer-related glycosylated biomarkers. Jung et al. observed that concanavalin A, which has broad reactivity for the mannose cores typically found in N-linked glycans, captured ~30% of the plasma proteome. In contrast, a panel of lectins with narrower specificities bound a much smaller fraction (S84). Two fucose-specific lectins, Aleuria aurantia lectin (AAL) and Lens culinaris agglutinin (LCA), bound ~4% and 8% of plasma proteins, respectively; Lycopersicon esculentum lectin (LEL), which targets polylactosamine, and HPA (see first section above) selected ≤0.1% of the proteins contained in plasma.
Several studies have applied glycan-based enrichment strategies to cancer biomarker discovery. In general, these studies tend to be small-scale proof-of-principle endeavors. It is noteworthy that, on the whole, they have succeeded in identifying target biomarker candidates for further validation. The published literature suggests that focusing the selection process on cancer-relevant glycans increases the likelihood of identifying potential disease targets. For example, in one study, approximately 10% of the proteins that bound to HPA and LEL were increased in abundance by at least threefold in samples obtained from breast cancer patients as compared to control subjects (S84). Comunale et al. used AAL to enhance recovery of fucosylated glycoproteins from pooled serum samples obtained from three study groups: patients with liver cirrhosis ± hepatocellular carcinoma and healthy control subjects. Approximately 50 glycoproteins tracked with the hepatocellular carcinomadiagnosis, and a subset were validated in a cohort of 332 patients using an antibody-lectin sandwich array (discussed below). From these studies, fucosylated hemopexin emerged as a potential biomarker with a specificity and sensitivity of 92% (S82).
Chemical-based methods
Chemical biology presents numerous approaches to enrich glycosylated molecules by exploiting glycan structures and manipulating their biosynthetic pathways (S97). Two similar approaches, involving hydrazide or boronic acid chemistry, capitalize on the cis-diols present in monosaccharides. Zhang et al. pioneered the use of hydrazide chemistry for purification by directly coupling glycosylated proteins to a solid support (S98). In this scheme, carbohydrate cis-diols are oxidized into aldehydes by sodium periodate treatment, then covalently coupled through hydrazide chemistry to functionalized beads. Bound glycoproteins are trypsin-digested in situ, andnon-glycosylated/unbound peptides are removed by washing. Then, covalently coupled N-linked glycoproteins are released by PNGase F treatment and analyzed by LC/MS/MS. Using this approach the authors identified 57 serum glycoproteins. The same laboratory published a revised version of this method that captures glycopeptides rather than intact glycoproteins (S99). This modified procedure markedly improved glycopeptide recovery to >90%. In the aggregate, ~300 unique glycosylation sites were observed in serum samples and tissue lysates. A similar method was used by Sun et al., who identified 302 proteins from an ovarian cancer cell line with 91% selectivity for N-linked glycopeptides in the captured fractions (S100). A complementary approach was devised by Sparbier et al., who employed boronic acid-functionalized beads to covalently capture (through formation of heterocyclic boronic acid diesters) glycoproteins followed by elution with acid (S101). This method was recently applied to the analysis of serum samples; 113 proteins were identified, of which 74 were known or proposed glycoproteins (S102). Other captured molecules included proteins known to associate with glycoproteins, suggesting that this method may also be useful for characterizing complexes that contain glycosylated molecules. Building upon data derived from these methods, Aebersold’s group developed a searchable online catalog for depositing data regarding N-linked glycosylation sites and glycopeptide sequences (www.UniPep.org). This repository is an important companion to sequence-based algorithms that predict N-linked glycosylation sites, and as such is a valuable resource for plasma-and serum-based studies (S103). Finally, hijacking glycan biosynthetic pathways for the incorporation of monosaccharides bearing bio-orthogonal functional handles affords an alternative approach to chemically labeling glycoconjugates (S104).
Glycan-focused methods
Whereas the above-described biomarker approaches strive to identify disease-related glycoproteins, another method targets the glycans themselves as cancer biomarkers. This approach involves the enzymatic or chemical release of N-and O-linked glycans from biological sources (e.g., cancer cell lines, tumors or plasma/serum), followed by MS-based characterization and identification of cancer-related glycoforms. Studies using these techniques identified distinct glycan subsets that correlated with malignant as compared to non-transformed cell lines, or that were preferentially expressed by cancer cell lines derived from specific tissues (S105, S106). Li et al. used gel electrophoresis to separate proteins from medium that was conditioned by cancer cell lines or serum from individuals with ovarian cancer. Glycoprotein-containing bands, which were identified by virtue of their reactivity with Pro-Q Emerald 300, were isolated, and the attached carbohydrates were characterized by MS (S107). The results yielded information about glycan structures that were unique to the disease state. Interestingly, investigators used specific serum glycoforms to distinguish individuals with prostate or ovarian cancer from those without disease (S108, S109). A study by Kirmiz et al. demonstrated a successful progression of inquiry, which began with analysis of glycans released from a panel of breast cancer cell lines (S110). The study next moved into a mouse mammary tumor model, wherecancer-related glycans were observed in serum at various time points over a 6-week course of disease progression. Finally, a small number of serum samples were analyzed from healthy control subjects and patients with breast cancer. Remarkably, the molecular ions corresponding to glycans observed in breast cancer cell lines and serum samples of mice with mammary tumors were also present in patient sera.
Strategies to exploit cancer-related glycobiology for developing diagnostic tests
The use of glycobiology to inform biomarker studies holds clear promise for the rapid identification of disease markers that enable risk prediction as well as diagnosis, prognosis and monitoring response to therapy. If recent trends prevail, it is highly likely that future biomarker discovery efforts will focus on developing panels of disease sentinels, rather than pursuing single molecules. This change in strategy has been driven by a better understanding of the inherent complexity of human biology, which has in turn highlighted the importance of patient-specific therapies. In this context, approaches that allow the simultaneous analysis of numerous markers, or multiplexing, are highly advantageous.
At present, two relatively new technology platforms are particularly suited for the analysis of cancer-related neoglycans. The first is multiple reaction monitoring (MRM), an MS-based approach. MRM achieves high analytical specificity and sensitivity by selecting predetermined precursor molecular ions for collision-induced dissociation and monitoring the appearance of several diagnostic product ions. When identified in combination, the precursor and product ions confirm the presence of the analyte of interest. Importantly, addition of isotopically labeled internal standards allows absolute quantification. Numerous precursor ions can be monitored in one experiment, particularly when using “scheduled MRMs,” which incorporate prior knowledge of peptide elution times into the LC-MS/MS program (S111, S112). Recently, a large National Cancer Institute-sponsored inter-laboratory study showed that an MRM approach can quantify target proteins in a background of unfractionated human plasma with highly robust and reproducible results, supporting the notion that this technology is suitable for biomarker discovery (S113).
The immediate challenge is to increase the sensitivity of these assays to reach the depth of coverage that will be required to identify molecules in the range of abundances in which disease-specific biomarkers likely circulate. In this regard, the stable isotope standards and capture by anti-peptide (or anti-carbohydrate) antibodies (SISCAPA) approach is designed to enhance the sensitivity of MRM assays. First reported by Anderson and colleagues (S114), this technique uses antibodies to capture target analytes, followed by elution in a small volume for simultaneous enrichment and concentration. When coupled to an MRM readout, SISCAPA increases the limit of detection by up to 3 orders of magnitude over the original assay (S115). Thus, a SISCAPA-MRM approach allows detection by MS of biomolecules present in blood at or near concentrations typical of disease biomarkers (e.g., ng/L to μg/L), with a sensitivity approaching that of clinical immunoassays (S115, S116). Interestingly, Aebersold and colleagues reached a similar level of plasma glycoprotein detection and quantification by employing hydrazide-mediated capture of carbohydrate-containing molecules followed by release of N-linked glycopeptides and detection by scheduled MRM (S112).
Second, other technologies are being developed that do not require the use of the sophisticated instruments and computer software programs that mass spectrometry entails. For example, lectin-antibody sandwich arrays can be used at every stage in the biomarker discovery and verification pipeline. These assays differentiate disease-associated glycosylation, offer a high level of sensitivity and specificity, and are easily multiplexed. Similar in design to an ELISA, the sandwich assay, in separate steps, detectsthe protein portion of the analyte (via antibodies) and the attached glycan (via lectins and carbohydrate-specific antibodies). Chen et al. used this method to identify cancer-related changes in glycosylation on MUC1 and CEA in sera from individuals with pancreatic cancer (S117). In a subsequent study, sandwich arrays were used to monitor changes in glycosylation of mucins expressed by human pancreatic cancer cell lines in response to inflammatory stimuli (S118).
Conclusions: The sweet future of cancer biomarkers
In a recent editorial, a group of distinguished glycobiologists dubbed this the “Second Golden Age of Glycomics” due to the increasing evidence that disease-related changes in glycosylation will provide the basis for improved cancer biomarkers (S79). As shown in Fig. 1, we think that the products of glycosylation, which are often altered in the setting of cancer, are particularly interesting targets that offer several major advantages as compared to proteins. First, the biology allows for the rational design of discovery efforts. For example, changes in the glycosylation machinery can be identified from microarray data and translated in structural terms. In this regard, the glycosyltransferases upregulated in the cancer cell as compared to its normal counterpart point to specific disease-related aberrations in oligosaccharide structures.
In turn, these observations provide a compelling rationale for designing antibody-and lectin-based separation strategies for their isolation in the discovery phase of biomarker-type projects. Second, one protein can carry many copies of an altered glycan, which may also be added to other scaffolds. Thus, there is an important amplification effect, which could enable the detection of many fewer abnormal cells than would otherwise be possible. Finally, glycosylation acts to shield the peptide backbone from proteolytic degradation. Thus, in theory glycan-based biomarkers are likely to be more stable in a variety of disease settings than unmodified proteins, which are often more labile.
In this context, it is interesting to note that 109 diagnostic tests currently approved by the Food and Drug Administration measure protein concentration alone, while 18 additional assays include assessment of post-translational modifications. Of the latter subset, six assays detect carbohydrate structures (S119), namely—Lens culinaris agglutinin-reactive alpha-fetoprotein, glycated forms of albumin, bone-specific alkaline phosphatase, and LDL; CA 19-9; carbohydrate-deficienttransferrin. Notably, while the currently approved clinical tests monitor single biomolecules in isolation, future diagnostics will include multiplexed analyses of numerous proteins with or without their post-translational modificationsor exclusively glycans. In parallel with the shift toward biology-driven discovery efforts, technological improvements in MS platforms and lectin-antibody array methodologies are opening the door to a new generation of approaches for verification and clinical deployment. In total, the confluence of these changes will strongly affect the way cancer is diagnosed and disease progression/remission is monitored in the context of patient-specific therapies.
Supplementary Material
Acknowledgments
This work was supported by the Clinical Proteomic Technologies for Cancer initiative, 5U24CA126477-04.
Nonstandard abbreviations
- MS
mass spectrometry
- CEA
carcinoembryonic antigen
- PSA
prostate-specific antigen
- HPA
Helix pomatia agglutinin
- MGAT5
β1,6 N-acetylglucosaminyltransferase V
- MMP
matrix metalloproteinase
- ECM
extracellular matrix
- EGF
epidermal growth factor
- MRM
multiple reaction monitoring
- SISCAPA
stable isotope standards and capture by anti-peptide (or anti-carbohydrate) antibodies
Human genes discussed
- FUT2
fucosyltransferase 2 (secretor status included)
- FUT3
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
- MGAT5
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase
References
- 1.Meezan E, Wu HC, Black PH, Robbins PW. Comparative studies on the carbohydrate-containing membrane components of normal and virus-transformed mouse fibroblasts. II. Separation of glycoproteins and glycopeptides by sephadex chromatography. Biochemistry. 1969;8:2518–24. doi: 10.1021/bi00834a039. [DOI] [PubMed] [Google Scholar]
- 2.Backstrom M, Thomsson KA, Karlsson H, Hansson GC. Sensitive liquid chromatography-electrospray mass spectrometry allows for the analysis of the O-glycosylation of immunoprecipitated proteins from cells or tissues: application to MUC1 glycosylation incancer. J Proteome Res. 2009;8:538–45. doi: 10.1021/pr800713h. [DOI] [PubMed] [Google Scholar]
- 3.Barkauskas DA, An HJ, Kronewitter SR, de Leoz ML, Chew HK, de Vere White RW, et al. Detecting glycan cancer biomarkers in serum samples using MALDI FT-ICR mass spectrometry data. Bioinformatics. 2009;25:251–7. doi: 10.1093/bioinformatics/btn610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. Automated N-glycopeptide identification using a combination of single-and tandem-MS. J Proteome Res. 2007;6:3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- 5.Harvey DJ. Structural determination of N-linked glycans by matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. Proteomics. 2005;5:1774–86. doi: 10.1002/pmic.200401248. [DOI] [PubMed] [Google Scholar]
- 6.Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci U S A. 2009;106:12–7. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.North SJ, Hitchen PG, Haslam SM, Dell A. Mass spectrometry in the analysis of N-linked and O-linked glycans. Curr Opin Struct Biol. 2009 doi: 10.1016/j.sbi.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376:1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Tang H, Mechref Y, Novotny MV. Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics. 2005;21 (Suppl 1):i431–9. doi: 10.1093/bioinformatics/bti1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werz DB, Seeberger PH. Carbohydrates as the next frontier in pharmaceutical research. Chemistry. 2005;11:3194–206. doi: 10.1002/chem.200500025. [DOI] [PubMed] [Google Scholar]
- 11.Wong SC, Chan CM, Ma BB, Lam MY, Choi GC, Au TC, et al. Advanced proteomic technologies for cancer biomarker discovery. Expert Rev Proteomics. 2009;6:123–34. doi: 10.1586/epr.09.1. [DOI] [PubMed] [Google Scholar]
- 12.Barrabes S, Pages-Pons L, Radcliffe CM, Tabares G, Fort E, Royle L, et al. Glycosylation of serum ribonuclease 1 indicates a major endothelial origin and reveals an increase in core fucosylation in pancreatic cancer. Glycobiology. 2007;17:388–400. doi: 10.1093/glycob/cwm002. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmanov U, Jiang N, Smith CR, Soosaipillai A, Diamandis EP. Differential N-glycosylation of kallikrein 6 derived from ovarian cancer cells or the central nervous system. Mol Cell Proteomics. 2009;8:791–8. doi: 10.1074/mcp.M800516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meany DL, Zhang Z, Sokoll LJ, Zhang H, Chan DW. Glycoproteomics for prostate cancer detection: changes in serum PSA glycosylation patterns. J Proteome Res. 2009;8:613–9. doi: 10.1021/pr8007539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misonou Y, Shida K, Korekane H, Seki Y, Noura S, Ohue M, Miyamoto Y. Comprehensive clinico-glycomic study of 16 colorectal cancer specimens: elucidation of aberrant glycosylation and itsmechanistic causes in colorectal cancer cells. J Proteome Res. 2009;8:2990–3005. doi: 10.1021/pr900092r. [DOI] [PubMed] [Google Scholar]
- 16.Mizuochi T, Nishimura R, Derappe C, Taniguchi T, Hamamoto T, Mochizuki M, Kobata A. Structures of the asparagine-linked sugar chains of human chorionic gonadotropin produced in choriocarcinoma. Appearance of triantennary sugar chains and unique biantennary sugar chains. J Biol Chem. 1983;258:14126–9. [PubMed] [Google Scholar]
- 17.Moore A, Medarova Z, Potthast A, Dai G. In vivo targeting of underglycosylated MUC-1 tumor antigen using a multimodal imaging probe. Cancer Res. 2004;64:1821–7. doi: 10.1158/0008-5472.can-03-3230. [DOI] [PubMed] [Google Scholar]
- 18.Ohyama C, Hosono M, Nitta K, Oh-eda M, Yoshikawa K, Habuchi T, et al. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14:671–9. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- 19.Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13:457–70. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 20.Taylor AD, Hancock WS, Hincapie M, Taniguchi N, Hanash SM. Towards an integrated proteomic and glycomic approach to finding cancer biomarkers. Genome Med. 2009;1:57. doi: 10.1186/gm57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 22.Moss EL, Hollingworth J, Reynolds TM. The role of CA125 in clinical practice. J Clin Pathol. 2005;58:308–12. doi: 10.1136/jcp.2004.018077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jankovic MM, Milutinovic BS. Glycoforms of CA125 antigen as a possible cancer marker. Cancer Biomark. 2008;4:35–42. doi: 10.3233/cbm-2008-4104. [DOI] [PubMed] [Google Scholar]
- 24.van Gisbergen KP, Aarnoudse CA, Meijer GA, Geijtenbeek TB, van Kooyk Y. Dendritic cells recognize tumor-specific glycosylation of carcinoembryonic antigen on colorectal cancer cells through dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin. Cancer Res. 2005;65:5935–44. doi: 10.1158/0008-5472.CAN-04-4140. [DOI] [PubMed] [Google Scholar]
- 25.Vashi AR, Wojno KJ, Henricks W, England BA, Vessella RL, Lange PH, et al. Determination of the “reflex range” and appropriate cutpoints for percent free prostate-specific antigen in 413 men referred for prostatic evaluation using the AxSYM system. Urology. 1997;49:19–27. doi: 10.1016/S0090-4295(96)00511-0. [DOI] [PubMed] [Google Scholar]
- 26.Magnani JL, Nilsson B, Brockhaus M, Zopf D, Steplewski Z, Koprowski H, Ginsburg V. A monoclonal antibody-defined antigen associated with gastrointestinal cancer is a ganglioside containing sialylated lacto-N-fucopentaose II. J Biol Chem. 1982;257:14365–9. [PubMed] [Google Scholar]
- 27.Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20:362–71. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 28.Vestergaard EM, Hein HO, Meyer H, Grunnet N, Jorgensen J, Wolf H, Orntoft TF. Reference values and biological variation for tumor marker CA 19–9 in serum for different Lewis and secretor genotypes and evaluation of secretor and Lewis genotyping in a Caucasian population. Clin Chem. 1999;45:54–61. [PubMed] [Google Scholar]
- 29.Prakobphol A, Boren T, Ma W, Zhixiang P, Fisher SJ. Highly glycosylated human salivary molecules present oligosaccharides that mediate adhesion of leukocytes and Helicobacter pylori. Biochemistry. 2005;44:2216–24. doi: 10.1021/bi0480180. [DOI] [PubMed] [Google Scholar]
- 30.Serpa J, Almeida R, Oliveira C, Silva FS, Silva E, Reis C, et al. Lewis enzyme (alpha1–3/4 fucosyltransferase) polymorphisms do not explain the Lewis phenotype in the gastric mucosa of a Portuguese population. J Hum Genet. 2003;48:183–9. doi: 10.1007/s10038-003-0007-5. [DOI] [PubMed] [Google Scholar]
- 31.Narimatsu H, Iwasaki H, Nakayama F, Ikehara Y, Kudo T, Nishihara S, et al. Lewis and secretor gene dosages affect CA19–9 and DU-PAN-2 serum levels in normal individuals and colorectal cancer patients. Cancer Res. 1998;58:512–8. [PubMed] [Google Scholar]
- 32.Brooks SA, Leathem AJ. Prediction of lymph node involvement in breast cancer by detection of altered glycosylation in the primary tumour. Lancet. 1991;338:71–4. doi: 10.1016/0140-6736(91)90071-v. [DOI] [PubMed] [Google Scholar]
- 33.Fenlon S, Ellis IO, Bell J, Todd JH, Elston CW, Blamey RW. Helix pomatia and Ulex europeus lectin binding in human breast carcinoma. J Pathol. 1987;152:169–76. doi: 10.1002/path.1711520305. [DOI] [PubMed] [Google Scholar]
- 34.Fukutomi T, Itabashi M, Tsugane S, Yamamoto H, Nanasawa T, Hirota T. Prognostic contributions of Helix pomatia and carcinoembryonic antigen staining using histochemical techniques in breast carcinomas. Jpn J Clin Oncol. 1989;19:127–34. [PubMed] [Google Scholar]
- 35.Kakeji Y, Maehara Y, Tsujitani S, Baba H, Ohno S, Watanabe A, Sugimachi K. Helix pomatia agglutinin binding activity and lymph node metastasis in patients with gastric cancer. Semin Surg Oncol. 1994;10:130–4. doi: 10.1002/ssu.2980100214. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich MG, Hellstern A, Hautmann SH, Graefen M, Conrad S, Huland E, Huland H. Clinical use of urinary markers for the detection and prognosis of bladder carcinoma: a comparison of immunocytology with monoclonal antibodies against Lewis X and 486p3/12 with the BTA STAT and NMP22 tests. J Urol. 2002;168:470–4. doi: 10.1016/s0022-5347(05)64660-5. [DOI] [PubMed] [Google Scholar]
- 37.Nakagoe T, Fukushima K, Nanashima A, Sawai T, Tsuji T, Jibiki M, et al. Expression of Lewis(a), sialyl Lewis(a), Lewis(x) and sialyl Lewis(x) antigens as prognostic factors in patients with colorectal cancer. Can J Gastroenterol. 2000;14:753–60. doi: 10.1155/2000/149851. [DOI] [PubMed] [Google Scholar]
- 38.Narita T, Funahashi H, Satoh Y, Watanabe T, Sakamoto J, Takagi H. Association of expression of blood group-related carbohydrate antigens with prognosis in breast cancer. Cancer. 1993;71:3044–53. doi: 10.1002/1097-0142(19930515)71:10<3044::aid-cncr2820711026>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa J, Sano A, Inoue H, Koide S. Expression of Lewis-related antigen and prognosis in stage I non-small cell lung cancer. Ann Thorac Surg. 1995;59:412–5. doi: 10.1016/0003-4975(94)00866-6. [DOI] [PubMed] [Google Scholar]
- 40.Ohyama C. Glycosylation in bladder cancer. Int J Clin Oncol. 2008;13:308–13. doi: 10.1007/s10147-008-0809-8. [DOI] [PubMed] [Google Scholar]
- 41.Pode D, Golijanin D, Sherman Y, Lebensart P, Shapiro A. Immunostaining of Lewis X in cells from voided urine, cytopathology and ultrasound for noninvasive detection of bladder tumors. J Urol. 1998;159:389–92. doi: 10.1016/s0022-5347(01)63928-4. discussion 93. [DOI] [PubMed] [Google Scholar]
- 42.Steplewska-Mazur K, Gabriel A, Zajecki W, Wylezol M, Gluck M. Breast cancer progression and expression of blood group-related tumor-associated antigens. Hybridoma. 2000;19:129–33. doi: 10.1089/02724570050031167. [DOI] [PubMed] [Google Scholar]
- 43.Tauchi K, Kakudo K, Machimura T, Makuuchi H, Mitomi T. Immunohistochemical studies of blood group-related antigens in human superficial esophageal carcinomas. Cancer. 1991;67:3042–50. doi: 10.1002/1097-0142(19910615)67:12<3042::aid-cncr2820671219>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 44.Bao X, Kobayashi M, Hatakeyama S, Angata K, Gullberg D, Nakayama J, et al. Tumor suppressor function of laminin-binding alpha-dystroglycan requires a distinct beta3-N-acetylglucosaminyltransferase. Proc Natl Acad Sci U S A. 2009;106:12109–14. doi: 10.1073/pnas.0904515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwai T, Kudo T, Kawamoto R, Kubota T, Togayachi A, Hiruma T, et al. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc Natl Acad Sci U S A. 2005;102:4572–7. doi: 10.1073/pnas.0407983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SH, Hatakeyama S, Yu SY, Bao X, Ohyama C, Khoo KH, et al. Core3 O-glycan synthase suppresses tumor formation and metastasis of prostate carcinoma PC3 and LNCaP cells through down-regulation of alpha2beta1 integrin complex. J Biol Chem. 2009;284:17157–69. doi: 10.1074/jbc.M109.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–12. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 48.Seelentag WK, Li WP, Schmitz SF, Metzger U, Aeberhard P, Heitz PU, Roth J. Prognostic value of beta1,6-branched oligosaccharides in human colorectal carcinoma. Cancer Res. 1998;58:5559–64. [PubMed] [Google Scholar]
- 49.Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9:1307–11. doi: 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- 50.Leivonen M, Nordling S, Lundin J, von Boguslawski K, Haglund C. STn and prognosis in breast cancer. Oncology. 2001;61:299–305. doi: 10.1159/000055337. [DOI] [PubMed] [Google Scholar]
- 51.Sewell R, Backstrom M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, et al. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. J Biol Chem. 2006;281:3586–94. doi: 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- 52.Chiricolo M, Malagolini N, Bonfiglioli S, Dall’Olio F. Phenotypic changes induced by expression of beta-galactoside alpha2,6 sialyltransferase I in the human colon cancer cell line SW948. Glycobiology. 2006;16:146–54. doi: 10.1093/glycob/cwj045. [DOI] [PubMed] [Google Scholar]
- 53.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143:725–9. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 54.Kudo T, Ikehara Y, Togayachi A, Morozumi K, Watanabe M, Nakamura M, et al. Up-regulation of a set of glycosyltransferase genes in human colorectal cancer. Lab Invest. 1998;78:797–811. [PubMed] [Google Scholar]
- 55.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–8. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–41. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 57.Hojilla CV, Wood GA, Khokha R. Inflammation and breast cancer: metalloproteinases as common effectors of inflammation and extracellular matrix breakdown in breast cancer. Breast Cancer Res. 2008;10:205. doi: 10.1186/bcr1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jezierska A, Motyl T. Matrix metalloproteinase-2 involvement in breast cancer progression: a mini-review. Med Sci Monit. 2009;15:RA32–40. [PubMed] [Google Scholar]
- 59.Andre S, Kozar T, Kojima S, Unverzagt C, Gabius HJ. From structural to functional glycomics: core substitutions as molecular switches for shape and lectin affinity of N-glycans. Biol Chem. 2009;390:557–65. doi: 10.1515/BC.2009.072. [DOI] [PubMed] [Google Scholar]
- 60.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5:526–42. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 61.Gahmberg CG, Fagerholm SC, Nurmi SM, Chavakis T, Marchesan S, Gronholm M. Regulation of integrin activity and signalling. Biochim Biophys Acta. 2009;1790:431–44. doi: 10.1016/j.bbagen.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu J, Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj J. 2004;21:9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 63.Isaji T, Sato Y, Fukuda T, Gu J. N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: identification of the minimal N-glycosylation requirement for alpha5beta1. J Biol Chem. 2009;284:12207–16. doi: 10.1074/jbc.M807920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL. Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 2005;65:4645–52. doi: 10.1158/0008-5472.CAN-04-3117. [DOI] [PubMed] [Google Scholar]
- 65.Hedlund M, Ng E, Varki A, Varki NM. alpha 2–6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 2008;68:388–94. doi: 10.1158/0008-5472.CAN-07-1340. [DOI] [PubMed] [Google Scholar]
- 66.Ahmad N, Gabius HJ, Andre S, Kaltner H, Sabesan S, Roy R, et al. Galectin-3 precipitates as a pentamer with synthetic multivalent carbohydrates and forms heterogeneous cross-linked complexes. J Biol Chem. 2004;279:10841–7. doi: 10.1074/jbc.M312834200. [DOI] [PubMed] [Google Scholar]
- 67.Lagana A, Goetz JG, Cheung P, Raz A, Dennis JW, Nabi IR. Galectin binding to Mgat5-modified N-glycans regulates fibronectin matrix remodeling in tumor cells. Mol Cell Biol. 2006;26:3181–93. doi: 10.1128/MCB.26.8.3181-3193.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, et al. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–4. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 69.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 70.Gout S, Tremblay PL, Huot J. Selectins and selectin ligands in extravasation of cancer cells and organ selectivity of metastasis. Clin Exp Metastasis. 2008;25:335–44. doi: 10.1007/s10585-007-9096-4. [DOI] [PubMed] [Google Scholar]
- 71.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–93. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosen SD. Ligands for L-selectin: homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–56. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 73.Witz IP. The selectin-selectin ligand axis in tumor progression. Cancer Metastasis Rev. 2008;27:19–30. doi: 10.1007/s10555-007-9101-z. [DOI] [PubMed] [Google Scholar]
- 74.Thomas SN, Zhu F, Schnaar RL, Alves CS, Konstantopoulos K. Carcinoembryonic antigen and CD44 variant isoforms cooperate to mediate colon carcinoma cell adhesion to E-and L-selectin in shear flow. J Biol Chem. 2008;283:15647–55. doi: 10.1074/jbc.M800543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borsig L, Wong R, Feramisco J, Nadeau DR, Varki NM, Varki A. Heparin and cancer revisited: mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci U S A. 2001;98:3352–7. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.