Summary
Factor XI (FXI) has structural and mechanistic features that distinguish it from other coagulation proteases. A relatively recent addition to vertebrate plasma coagulation, FXI is a homodimer, with each subunit containing four apple domains and a protease domain. The apple domains form a disk structure with binding sites for platelets, high molecular weight kininogen, and the substrate factor IX (FIX). FXI is converted to the active protease FXIa by cleavage of the Arg369−Ile370 bond on each subunit. This converts the catalytic domains to the active forms, and unmasks exosites on the apple domains required for FIX binding. FXI activation by factor XIIa or thrombin proceeds through an intermediate with only one activated submit (1/2-FXIa). 1/2-FXIa activates FIX in a similar manner to FXIa. While the importance of the homodimeric structure of FXI is not certain, it may represent a strategy for binding to FIX and a platelet surface simultaneously.
Keywords: dimer, factor XI activation, factor XI structure
Introduction
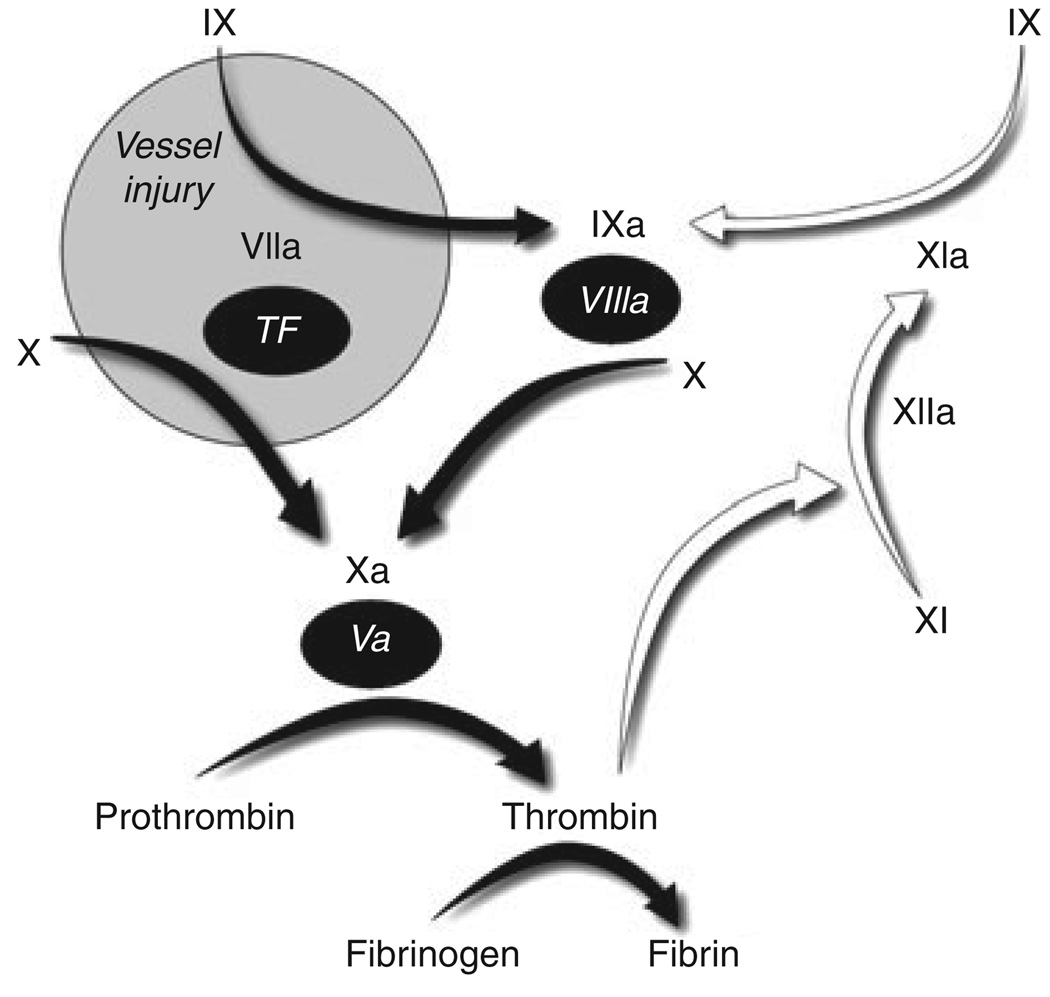
Factor XI (FXI) is the zymogen of a plasma protease (FXIa) that contributes to fibrin formation and stability by activating factor IX (FIX) [1]. Initially described as a component of a mechanism (contact activation) that initiates coagulation when blood is exposed to a surface, FXIa appears to be required for maintaining clot integrity in tissues with high fibrinolytic activity [2]. Figure 1 depicts the major protease-substrate interactions that occur during coagulation. In this scheme, FXI is activated after coagulation has been initiated by the factor VIIa/tissue factor (FVIIa/TF) complex, with FXIa sustaining thrombin generation through FIX activation [3].
Fig. 1.
Major protease-mediated reactions in mammalian coagulation. Black Roman numerals represent plasma protease zymogens, with activated proteases indicated by a lower case ‘a’. Cofactors are represented by numerals in black ovals. White arrows indicate reactions involving FXI/XIa. Coagulation is initiated at an injury site by binding of plasma factor (F) VIIa to tissue factor (TF), which is present on cells underlying blood vessel endothelium. FVIIa/TF activates FX to initiate thrombin generation and fibrin formation, and also FIX to sustain coagulation through activation of FX. In some situations, FIX activation by FXIa is also required. FXI may be activated by more than one protease, but probably is most important during consolidation of coagulation rather than its initial phase.
There are distinct structural differences between FXI and the vitamin K-dependent coagulation protease zymogens, factors II (prothrombin) VII, IX and X [1]. Human FXI is a dimer composed of identical 80 kDa subunits [4]; a unique configuration among coagulation proteases. Starting at the N-terminus, each FXI subunit consists of four apple domains (A1–A4) and a catalytic domain (CD) [4]. In mammals, apple domains are found only in FXI and its homolog prekallikrein (PK) [5]. FXI also lacks the N-terminal calcium binding Gla domain that is characteristic of vitamin K-dependent proteases. It is difficult, therefore, to extrapolate from work with the vitamin K-dependent coagulation enzymes to structure-function relationships for FXI. Here we consider the FXI/XIa structure as it relates to the evolutionary history of the protein, its mechanism of activation, and the manner in which it interacts with FIX.
The natural history of factor XI
Availability of complete genome sequences for multiple species has facilitated analysis of the evolution of the vertebrate blood coagulation mechanism. Teleosts (jawed boney fish) have homologs for the vitamin K-dependent coagulation proteases found in mammals [6], but lack FXI and FXII [5]. The apple domains of FXI and PK are members of the PAN domain family, with homology to the N-termini of hepatoctye growth factor and plasminogen. A gene for a trypsin-like protease with four apple domains that is clearly ancestral to FXI and PK first appears in amphibians [5], and is also found in the chicken and duck-billed platypus (a monotreme – egg-laying mammal), indicating its spread to all major terrestrial vertebrate groups. The function of this protease is not known, but it is unlikely that it would activate FIX. In the opossum and all placental mammals, the locus for this gene is occupied by separate genes for FXI and PK [5]. Thus, FXI and PK are products of a duplication of a predecessor gene that occurred after monotremes diverged from the lineage that gave rise to marsupial and placental mammals, making FXI the newest member of the vertebrate plasma coagulation mechanism. Interestingly, the gene for FXII, the precursor of a protease (FXIIa) that activates FXI and PK, also makes its debut in amphibians through a duplication of the gene for hepatocyte growth factor activator [5], suggesting that FXIIa may also be an activator of the FXI/PK predecessor.
Factor XI structure
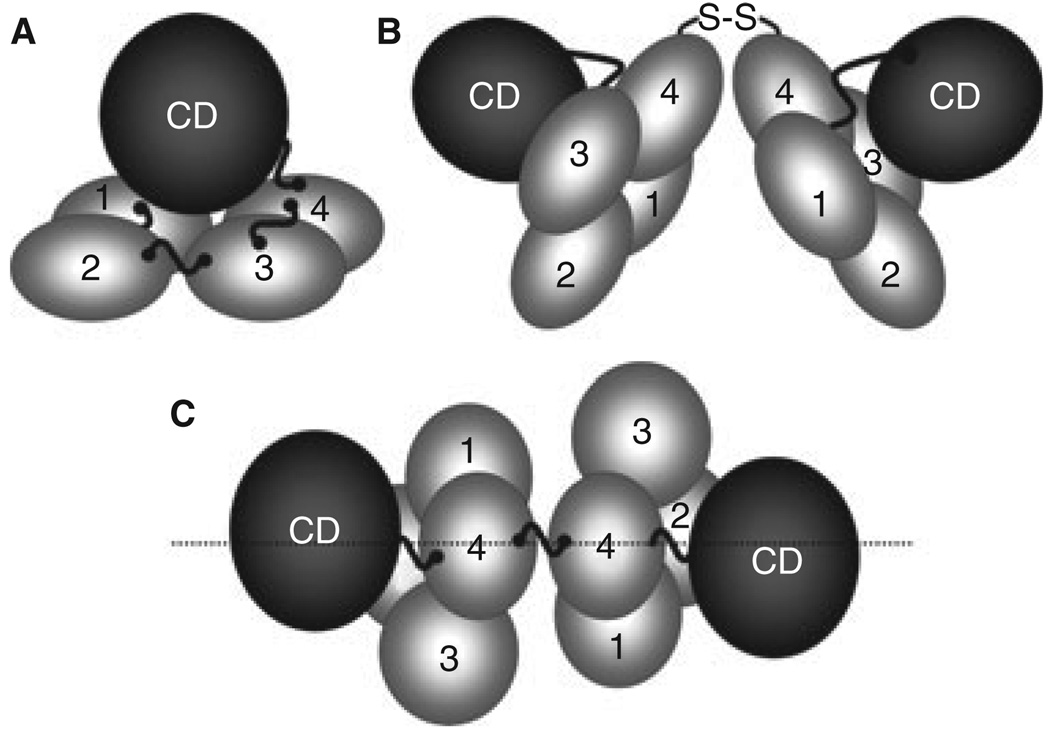
The availability of a structure for zymogen FXI has greatly facilitated analyses of structure–function relationships for this unusual protease [4]. Each apple domain contains seven β-strands that form an anti-parallel sheet, with an α-helix attached to the concave side of the sheet by disulfide bonds. The apple domains of FXI are packed into a 60 × 60 × 20 Å planar structure, with the N-terminal A1 domain making contact with A4 (Fig. 2A). The catalytic domain rests on the apple domains in a ‘cup and saucer’ arrangement [4]. It is likely that PK has a similar structure [7]. Unlike PK, however, FXI is a homodimer (Fig. 2B). The site of contact between FXI subunits is within the A4 domain [4], and NMR studies [8] and site-directed mutagenesis [9] confirm that electrostatic and hydrophobic interactions are responsible for the tight association between FXI subunits, with Leu284, Ile290 and Tyr329 forming the hydrophobic interface. In most species, Cys321 forms an interchain disulfide bond between the subunits of the dimer, but is not required to maintain the dimeric conformation [4].
Fig. 2.
Factor XI Structure. (A) A factor (F) XI subunit contains four apple domains (1–4) that form a planar structure on which the catalytic domain (CD) rests. High molecular weight kininogen likely binds to the underside of the apple domain disc. Cleavage at Arg369-Ile370 between A4 and the CD generates FXIa. (B) FXI is a dimer of two identical subunits, with the A4 domains forming the interface. (C) In this view of the dimer from above the A4 domains, the positions of the two A3 domains on opposite sides of the longitudinal plane of the molecule can be seen.
The apple domains contain binding sites for a number of ligands, including the FXIa substrate FIX [3]. As FIX does not bind to the zymogen, FXI [10], a change must occur on conversion to FXIa that exposes one or more FIX binding sites. Consistent with this, a study using rotary shadow EM indicates that FXI and FXIa have difference shapes [8]. Amino acids 183–191 in the FXIa A3 domain are required for activation of FIX, and probably are part of a FIX binding site [11]. In FXI, these residues are obscured by the catalytic domain [4] consistent with the notion that a conformational change in FXI is required to expose the FIX binding site on FXIa. The A3 domain of FXI also binds to platelet glycoprotein 1bα (GP1bα) [12]. In the FXI structure, the A3 domains are positioned on opposite sides of a major plane of the molecule (Fig. 2C) [4]. This arrangement may allow a FXI subunit to bind to a platelet through one A3 domain, while binding to IX through the other.
Both FXI and PK circulate in plasma in complex with high molecular weight kininogen (HK). The A2 domains of both proteins contain a major binding site for HK, with contributions from A1 and A4 [13]. Hooley et al. [7] proposed that HK binds to FXI and PK via a charged channel that traverses these domains on the underside of the apple domain disc (the surface opposite the catalytic domain).
Factor XI activation
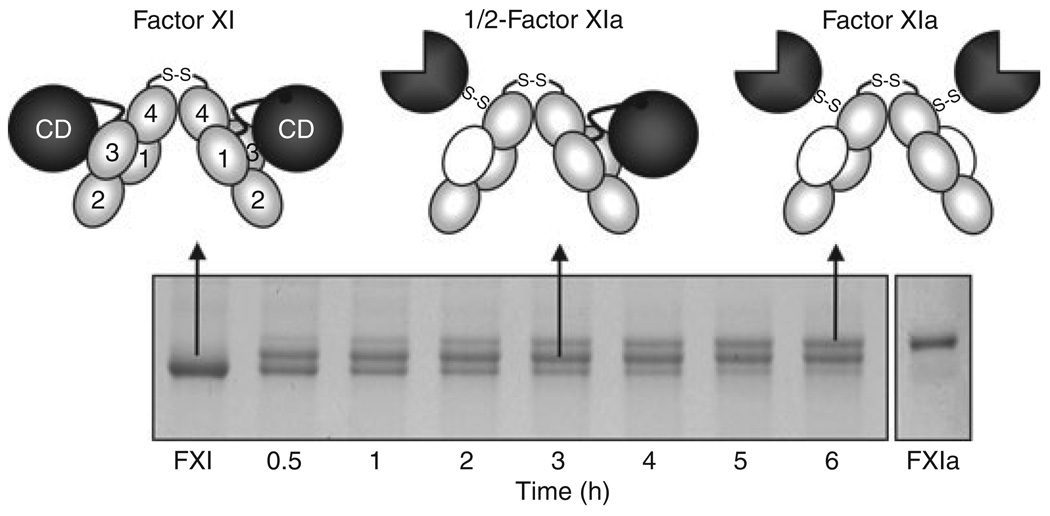
FXI subunits are activated by cleavage of the Arg369–Ile370 bond, which resides between A4 and the CD. While the nature of the physiologic activator remains controversial, both FXIIa and thrombin can make this cleavage (Fig. 1) [2,3]. Until recently, FXI activation was treated as a simple process, with both Arg369–Ile370 bonds on the homodimer cleaved to form FXIa. However, there is no obvious reason why a form of FXIa with only one activated subunit could not form. As a consequence of conformational changes that accompany activation, FXI migrates slightly faster than FXIa on SDS–polyacrylamide gels. During factor XI activation by thrombin (Fig. 3) or FXIIa (not shown), a species that migrates between FXI and FXIa is clearly evident. Conversion of FXI to FXIa appears to proceed through this intermediate. Characterization of this species showed it to be a form of FXI with only one activated subunit (1/2-FXIa) [14]. 1/2-FXIa can be detected in plasma induced to undergo contact activation. As it is likely that very small amounts of FXI are actually activated in plasma during coagulation, 1/2-FXIa may turn out to be a major species of activated FXI.
Fig. 3.
Factor XI Activation. Each factor (F) XI subunit is activated by cleavage between Arg369 and Ile370. FXI migrates slightly faster than FXIa on SDS–PAGE. Activation of FXI by α-thrombin (shown) or FXIIa proceeds through an intermediate in which only one dimer subunit is cleaved (1/2-FXIa). Subsequent conversion of 1/2-FXIa to fully activated FXIa appears to be a slower process. In the schematic diagrams at the top of the figure, gray ovals indicate apple (A) domains, black circles unactivated catalytic domains, and three-quarter circles activate catalytic domains. Note that with activation, an exosite for factor IX binding is exposed on the A3 domain (indicated by white ovals).
Factor XIa activation of factor IX
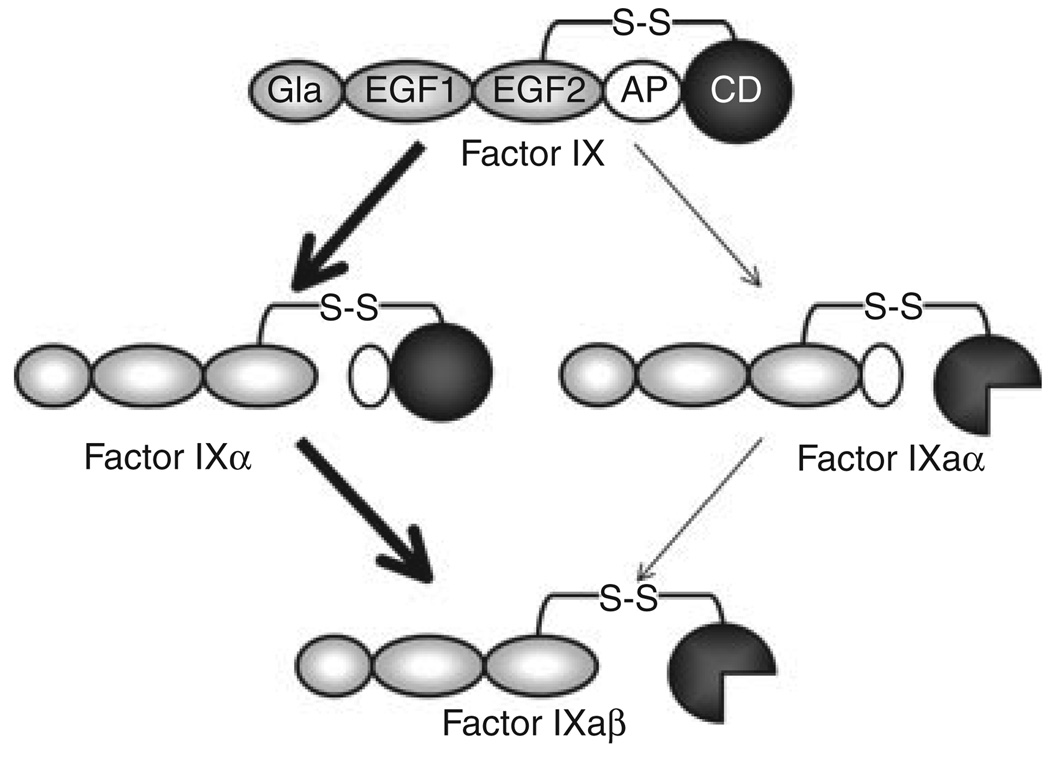
FIX is the natural substrate of FXIa. A diagram of the steps in FIX activation are shown in Fig. 4. FIX is cleaved at two bonds (Arg145–Ala146 and Arg180–Val181) to form the protease FIXaβ. During FIX activation by FVIIa/TF, the Arg145–Ala146 is cleaved to form the intermediate FIXα [3]. FIXα subsequently rebinds to FVIIa/TF for cleavage after Arg180. In contrast, FIX activation by FXIa is not associated with appreciable accumulation of an intermediate [14]. The recent availability of 1/2-FXIa and other novel FXIa species with single active sites allowed us to address one hypothesis that could explain the absence of FIX intermediate accumulation; that the two catalytic domains of FXIa each cleave one bond on a single FIX molecule simultaneously [8,14]. We found that 1/2-FXIa activated FIX similarly to FXIa, indicating the protease does not require two active sites [14]. This was confirmed by subsequent work with monomeric FXIa [9]. It turns out that FIX activation by FXIa follows the same order of sequential bond cleavage as activation by FVIIa/TF. The lack of intermediate accumulation with FXIa, then, indicates either that the second cleavage after Arg180 is faster than cleavage after Arg145, or that the protease cleaves the two FIX bonds sequentially without releasing FIXα. Studies are currently underway to investigate these possibilities.
Fig. 4.
Factor IX Activation. Factor (F) IX is converted to FIXaβ by cleavage after Arg145 and Arg180, releasing the activation peptide (white oval) that connects the catalytic domain (CD) to the non-catalytic light chain (Gla and EGF domains). FVIIa/TF initially cleaves FIX after Arg145 to form the intermediate FIXα, with subsequent cleavage after Arg180 forming FIXaβ (bold arrows). The active catalytic domain of FIXaβ is indicated by the three-quarter circle. During this reaction, FIXα accumulates prior to formation of FIXaβ. FXIa also cleaves FIX initially after Arg145, however, no FIXα accumulates, suggesting that subsequent cleavage after Arg180 is relatively rapid. Activation of FIX by initial cleavage after Arg180 (through FIXaα) is a minor reaction (thin arrows).
Why is factor XI a dimer?
The finding that FXIa requires only one active site to activate FIX leaves open the question of why the protein is a dimer. Early observations that some FXI mutations cause FXI deficiency by interfering with secretion from cells [2] could be interpreted as indicating that the dimeric conformation is required for intracellular processing. This seems unlikely, however, given the recent success in expressing monomeric FXI [9]. Wu et al. [9], showed that FXI monomers are activated poorly, and proposed a mechanism in which FXIIa or thrombin binds to one FXI subunit while activating the other (a trans-activating mechanism). If this is the case, FXI must be considerably different in its mechanism of activation than its homolog PK, which is a monomer.
The identification of 1/2-FXIa raises a possibility that is yet to be demonstrated experimentally, but which is consistent with what we know about FXI biochemistry. FXI, but not FXIa, binds to platelet GP1bα [12]. It is conceivable that 1/2-FXIa could bind GP1bα through the A3 domain of its zymogen subunit, leaving the activated subunit free to bind FIX. Most coagulation enzyme–substrate interactions take place on cell or platelet membranes, facilitated by the membrane binding properties of the Gla domains of protease and substrate. As FXI lacks a Gla domain, the dimeric structure may represent an alternative solution to the problem of tethering a prothrombotic molecule to a cellular surface in flowing blood in the vicinity of a wound.
Footnotes
Disclosure of Conflict of Interests
The authors state that they have no conflicts of interest.
References
- 1.Furie B, Furie BC. Molecular basis of blood coagulation. In: Hoffman H, Benze EJ, Shattil SJ, Furie B, Silberstein LE, McGlave P, Heslop H, editors. Hematology, Basic Principles and Practice. 5th edn. Churchill: Livingstone-Elsevier; 2009. pp. 1819–1836. [Google Scholar]
- 2.Seligsohn U. Factor XI in haemostasis and thrombosis: past, present and future. Thromb Haemost. 2007;98:84–89. [PubMed] [Google Scholar]
- 3.Smith SB, Gailani D. Update on the physiology and pathology of factor IX activation by factorXIa. Expert Rev Hematol. 2008;1:87–98. doi: 10.1586/17474086.1.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006;13:557–558. doi: 10.1038/nsmb1095. [DOI] [PubMed] [Google Scholar]
- 5.Ponczek MB, Gailani D, Doolittle RF. Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost. 2008;6:1876–1883. doi: 10.1111/j.1538-7836.2008.03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Y, Doolittle RF. The evolution of vertebrate blood coagulation as viewed from a comparison of puffer fish and sea squirt genomes. Proc Natl Acad Sci USA. 2003;100:7527–7532. doi: 10.1073/pnas.0932632100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooley E, McEwan PA, Emsley J. Molecular modeling of the prekallikrein structure provides insights into high-molecular-weight kininogen binding and zymogen activation. J Thromb Haemost. 2007;5:2461–2466. doi: 10.1111/j.1538-7836.2007.02792.x. [DOI] [PubMed] [Google Scholar]
- 8.Samuel D, Cheng H, Riley P, Canutescu A, Nagaswami C, Weisel J, Bu Z, Walsh PN, Roder H. Solution structure of the A4 domain of factor XI sheds light on the mechanism of zymogen activation. Proc Natl Acad Sci USA. 2007;104:15693–15698. doi: 10.1073/pnas.0703080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu W, Sinha D, Shikov S, Yip C, Walz T, Billings P, Lear J, Walsh P. Factor XI homodimer structure is essential for normal proteolytic activation by factor XIIa, thrombin, and factor XIa. J Biol Chem. 2008;283:18655–18664. doi: 10.1074/jbc.M802275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktimur A, Gabrie lM, Gailani D, Toomey J. The factor IX gamma-carboxyglutamic acid (Gla) domain is involved in interactions between factor IX and factor XIa. J Biol Chem. 2003;278:7981–7987. doi: 10.1074/jbc.M212748200. [DOI] [PubMed] [Google Scholar]
- 11.Sun M-F, Zhao M, Gailani D. Identification of amino acids in the factor XI apple 3 domain required for activation of factor IX. J Biol Chem. 1999;274:36373–36378. doi: 10.1074/jbc.274.51.36373. [DOI] [PubMed] [Google Scholar]
- 12.Baglia F, Gailani D, López J, Walsh P. Identification of a binding site for glycoprotein Ib-alpha in the apple 3 domain of factor XI. J Biol Chem. 2004;279:45470–45476. doi: 10.1074/jbc.M406727200. [DOI] [PubMed] [Google Scholar]
- 13.Renné T, Gailani D, Meijers J, Müller-Esterl W. Characterization of the H-kininogen-binding site on factor XI: a comparison of factor XI and plasma prekallikrein. J Biol Chem. 2002;277:4892–4899. doi: 10.1074/jbc.M105221200. [DOI] [PubMed] [Google Scholar]
- 14.Smith S, Verhamme I, Sun M-F, Bock P, Gailani D. Characterization of novel forms of coagulation factor XIa: independence of factor XIa subunits in factor IX activation. J Biol Chem. 2008;283:6696–6705. doi: 10.1074/jbc.M707234200. [DOI] [PMC free article] [PubMed] [Google Scholar]