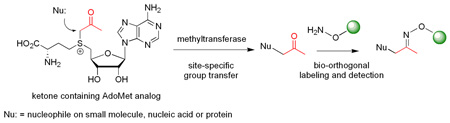
Abstract
S-Adenosylmethionine (AdoMet or SAM)-dependent methyltransferases belong to a large and diverse family of group-transfer enzymes that perform vital biological functions on a host of substrates. Despite the progress in genomics, structural proteomics and computational biology, functional annotation of methyltransferases remains a challenge. Herein, we report the synthesis and activity of a new AdoMet analog functionalized with a ketone group. Using catechol O-methyltransferase (COMT, EC 2.1.1.6) and thiopurine S-methyltransferase (TPMT, EC 2.1.1.67) as model enzymes, this robust and readily accessible analog displays kinetic parameters that are comparable to AdoMet and exhibits multiple turnovers with enzyme. More importantly, this AdoMet surrogate displays the same substrate specificity as the natural methyl donor. Incorporation of the ketone group allows for subsequent modification via bio-orthogonal labeling strategies and sensitive detection of the tagged ketone products. Hence, this AdoMet analog expands the toolbox available to interrogate the biochemical functions of methyltransferases.
Elucidation of the biochemical activity and specificity of enzymes is an essential step towards understanding their physiological roles. Despite the progress in genomics, structural proteomics and computational biology, functional annotation of enzymes is often limited to the family level, i.e., the precise substrate specificity for a given enzyme remains largely undefined. These challenges are exemplified by the large and diverse family of S-adenosylmethionine (AdoMet or SAM)-dependent methyltransferases (MTases); more than 150 different members have been cataloged by the Enzyme Commission and the numbers are still rapidly increasing. These enzymes conduct their activities on a host of substrates, including small molecules, DNA, RNA and proteins.1 Of the five known MTase structural folds, only the Class IV SPOUT family of RNA MTases and the Class V SET-domain containing family of protein lysine MTases exhibit generally definable targets.1a, b The other classes (I–III) do not correlate to the chemical nature of the substrates which themselves are distinguished by chemical diversity and include amines and guanidines (N), alcohols and carboxylic acids (O), thiols and sulfides (S), alkenes and even alkanes (C).1 As such, the structures of many putative MTases have been determined but their substrates and functions remains elusive.2
Progress in functional analyses of MTases is hampered by the inert properties and small size of the methyl group, making it difficult to directly tag or selectively bind methylation products. The last decade has witnessed the development of chemical approaches for the selective labeling of biomolecules that undergo enzyme-catalyzed group transfer, including phosphorylation, acylation and glycosylation.3 These approaches typically involve enzymatic introduction of traceable tags using permissive substrate analogs and subsequent recognition of labeled products with bio-orthogonal chemistry.3 For instance, AdoMet analogs substituted with activated alkyl groups in place of the methyl group served as alkyl donors for nucleic acid MTases, and the unnatural groups were transferred in a sequence-specific manner.4 When functionalized with an aliphatic amine, the tagged products were visualized by amine reactive fluorescent reporter groups. In a different approach, aziridine containing adenosyl analogs were used as surrogates of AdoMet. To mark the resulting bi-substrate adducts, the purine is substituted with either a fluorescent group or bear an azide group for subsequent modification by Staudinger ligation.5
Herein, we report the synthesis and activity of an AdoMet analog functionalized with a ketone group (keto-AdoMet, Scheme 1), which serves as an attractive probe due to the absence of ketone groups in nucleic acids and proteins and its orthogonal reactivity with hydroxylamines and hydrazides.6 The ketone group reacts with these agents rapidly and quantitatively in aqueous solutions and the resulting oximes and hydrazones are stable, thus, these versatile chemistries are widely used for biological labeling studies.6
Scheme 1.
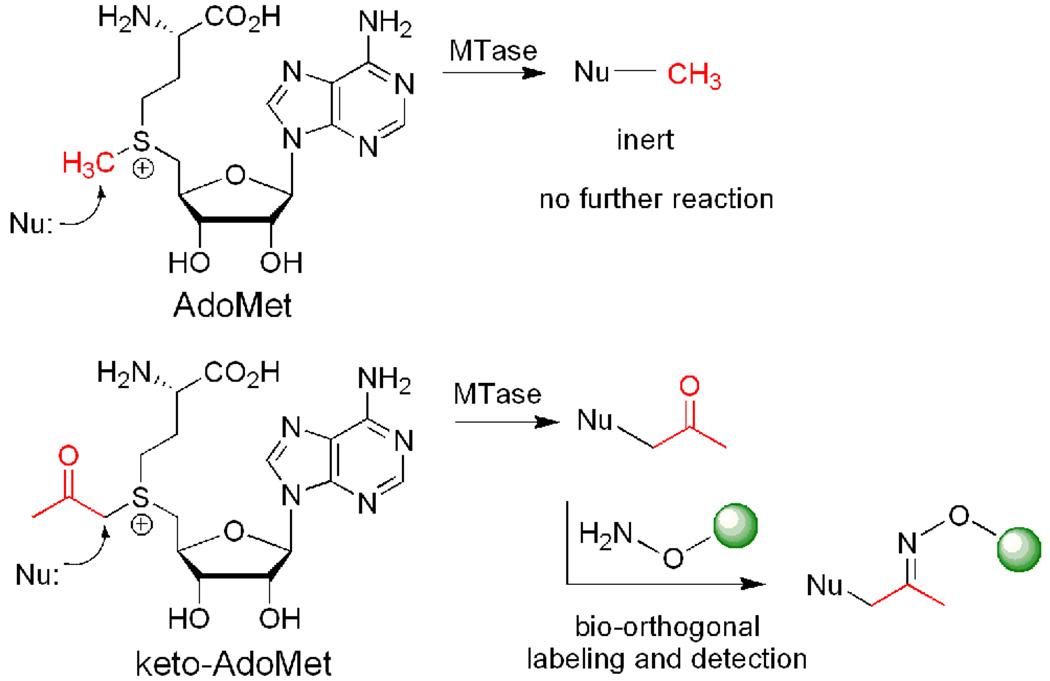
Enzyme-catalyzed transmethylation from AdoMet, transfer of ketone group from keto-AdoMet, and the detection of ketone containing products via selective derivatization.
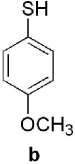
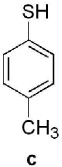
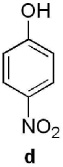
Keto-AdoMet was synthesized in a single step from S-adenosylhomocysteine (AdoHcy) and bromoacetone in a manner analogous to the preparation of other AdoMet analogs.4 The activity and specificity of keto-AdoMet was first assessed in vitro using Class I MTases catechol O-methyltransferase (COMT, EC 2.1.1.6) from porcine liver and human thiopurine S-methyltransferase (TPMT, EC 2.1.1.67). Our results show the ketone group is transferred from keto-AdoMet to the nucleophilic substrates for both MTases in a time and enzyme concentration dependent manner (See SI 3 and 4.1). Detailed studies were carried out with TPMT, an enzyme which exhibits substrate promiscuity in the methylation of a broad spectrum of aromatic thiols, including thiopurine drugs.7 Despite its role in drug metabolism, the endogenous substrates of TPMT, if any, remain unknown. As summarized in Table 1, the substrate specificity of AdoMet and keto-AdoMet mirror each other. For example, three known substrates (aromatic thiols a–c) are alkylated by keto-AdoMet. Conversely, the non-substrate (4-nitrophenol, d) is not alkylated by either AdoMet or keto-AdoMet.
Table 1.
Specificity of TPMT with AdoMet and keto-AdoMet.
| Substrate | Kinetic Parameters | 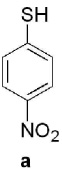 |
 |
 |
 |
|---|---|---|---|---|---|
| Alkyl Donor | |||||
| AdoMet | Km: 6.7 ± 0.7 µM kcat: 13.6 ± 0.4 min−1 |
known | known | known | non- substrate |
| keto-AdoMet | Km: 17.9 ± 1.5 µM kcat: 0.13 ± 0.01 min−1 |
yes | yes | yes | no |
Subsequent kinetic analysis using 4-nitrobenzenethiol (a) as the alkyl accepting substrate indicates that keto-AdoMet is a respectable substrate of TPMT (Table 1); the calculated kcat/Km value (120 M−1 s−1) is approximately 300-fold lower than the value measured with AdoMet. Competition assays conducted at varied ratios of AdoMet to keto-AdoMet indicate these substrates bind to the same site on TPMT (see SI 4.3).
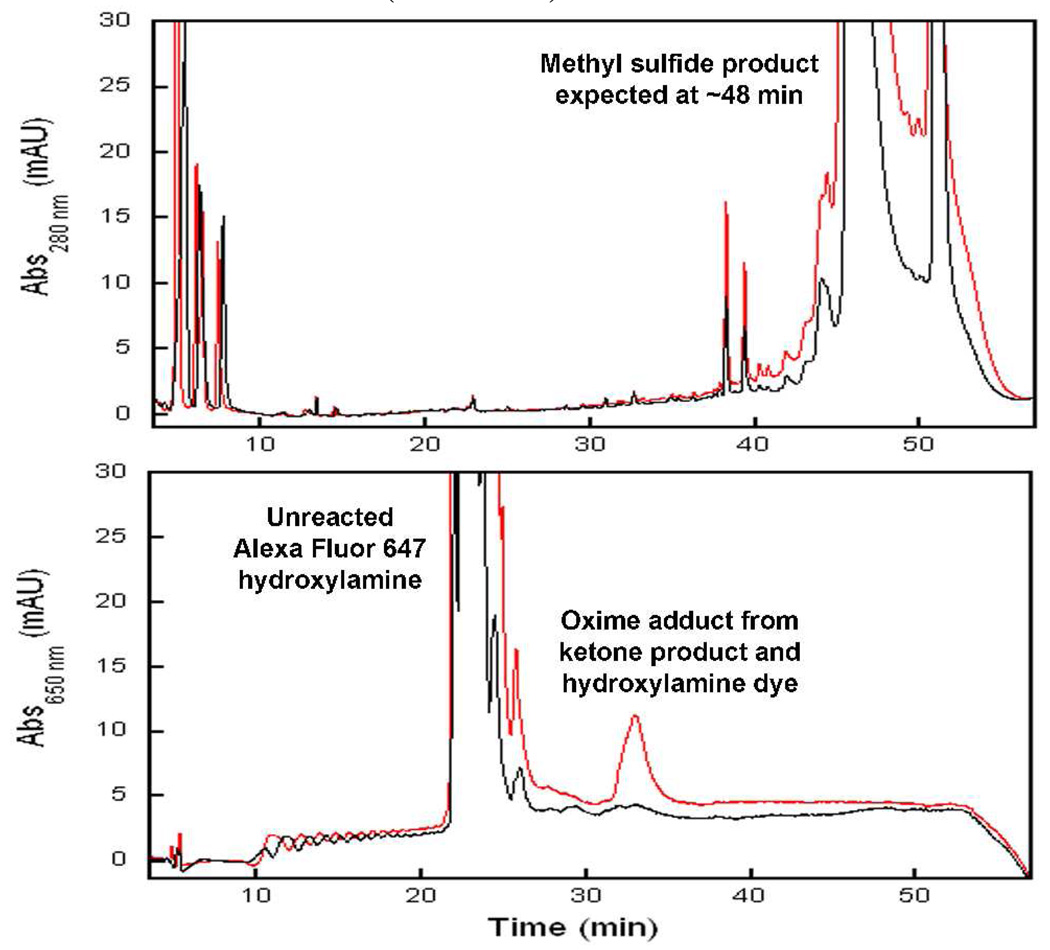
Labeling of the substrate using the analog was demonstrated ex vivo with human erythrocyte lysate which contains TPMT.8 HPLC analysis of the reaction containing AdoMet and 4-nitrobenzenethiol indicates the methyl sulfide product is obscured by the broad peaks eluting between 44–54 min resulting from heme and other metabolites (Figure 1, top), even though a fairly large amount of the product (1.1 nmoles) was loaded onto the column. This observation further underscores the challenges inherent in the identification of unknown products; under standard analytical conditions, reaction products may not be detectable against the background of cellular components. In comparison, the TPMT product was readily detected using keto-AdoMet as alkyl donor and derivatization with Alexa Fluor 647 hydroxylamine (Figure 1, bottom). At 650 nm, the complex background associated with the erythrocyte lysate, including the strong absorptions from heme, is absent. Other than the excess hydroxylamine dye, the oxime product is the single prominent peak visible at this wavelength and due to the large extinction coefficient at 650 nm (203,000 M−1 cm−1), 52 pmoles of the ketone product (1/20th of the methylation reaction) was sufficient to provide a strong signal.9
Figure 1.
Top: HPLC trace of the erythrocyte lysate reaction containing 4-nitrobenzenethiol in the presence (red) and absence (black) of AdoMet. Bottom: Chromatogram at 650 nm after reactions with (red) and without (black) keto-AdoMet followed by derivatization with Alexa Fluor 647 hydroxylamine.
One of the goals is to identify unknown substrates of MTases. Toward this end, we attempted to identify endogenous substrates of TPMT. Labeled products were not observed from red blood cell lysate treated with keto-AdoMet (see SI 5.1). Since active TPMT was present, endogenous thiol substrates might already be converted into their methylated forms. Another possibility is that no endogenous substrates exist. It has been reported that human subjects with little TPMT activity due to genetic background display no observable phenotype except when undergoing thiopurine treatment.7e Unexpectedly, we found that an aliphatic thiol (4-methoxybenzylthiol) was labeled by keto-AdoMet. Subsequently, TPMT-catalyzed methylation by AdoMet was confirmed. Since benzylthiols were not previously considered as substrates of TPMT, this aliphatic thiol was intended as a negative control. This serendipitous finding highlights the utility of keto-AdoMet to identify undescribed MTase activities.
A general concern regarding substrate analogs, in particular those with intrinsically reactive aziridines or sulfoniums, is whether the analog modifies and inactivates enzymes.4,5 Using mass spectrometry, no modification was detected for either TPMT or AdoHcy nucleosidase, even in the presence of excess keto-AdoMet. More importantly, the activities of these enzymes were not affected. An additional advantage of keto-AdoMet as a labeling reagent is that multiple turnovers were observed with various substrates (e.g., at least 14 for 4-nitrobenzenethiol). We further report that the analog is stable for over two months when stored under mildly acidic conditions and at either −20 or −80 °C.
In summary, keto-AdoMet, a readily accessible analog of AdoMet, expands the toolbox available to interrogate the biochemical functions of MTases. When coupled with specific MTases, the analog may further serve as a site-specific labeling reagent for bioconjugation and structural diversification.4,5
Supplementary Material
Acknowledgments
This work was supported by the Herman Frasch Foundation (541-HF02 to Z.S.Z.), NIH (1R01AI058146 to Z.S.Z. and 1F31GM073559 to J.F.A.) and the American Heart Association (09PRE2300071 to T.Z.) We thank the reviewers for their constructive comments. This is contribution number 943 from the Barnett Institute.
Footnotes
Supporting Information Available: Full experimental details, synthesis and characterization of keto-AdoMet. This material is free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Schubert HL, Blumenthal RM, Cheng X. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Klimasauskas S, Weinhold E. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [DOI] [PubMed] [Google Scholar]; (c) Cheng X, Blumenthal R. S-Adenosylmethionine-dependent methyltransferases. Singapore: World Scientific; 1999. p. 400. [Google Scholar]; (d) Clarke SG, Tamanoi F. Protein methyltransferases. Amsterdam: Academic Press; 2006. p. 570. [Google Scholar]
- 2. For two examples, see Grana M, Haouz A, Buschiazzo A, Miras I, Wehenkel A, Bondet V, Shepard W, Schaeffer F, Cole ST, Alzari PM. Protein Sci. 2007;16:1896–1904. doi: 10.1110/ps.072982707. Lim K, Zhang H, Tempczyk A, Bonander N, Toedt J, Howard A, Eisenstein E, Herzberg O. Proteins. 2001;45:397–407. doi: 10.1002/prot.10004.
- 3.(a) Sawa M, Hsu TL, Itoh T, Sugiyama M, Hanson SR, Vogt PK, Wong CH. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12371–12376. doi: 10.1073/pnas.0605418103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Saghatelian A, Cravatt BF. Nat. Chem. Biol. 2005;1:130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]; (c) Laughlin ST, Bertozzi CR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Foley TL, Burkart MD. Curr. Opin. Chem. Biol. 2007;11:12–19. doi: 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]; (e) Allen JJ, Lazerwith SE, Shokat KM. J. Am. Chem. Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Heal WP, Wickramasinghe SR, Leatherbarrow RJ, Tate EW. Org. Biomol. Chem. 2008;6:2308–2315. doi: 10.1039/b803258k. [DOI] [PubMed] [Google Scholar]
- 4.(a) Lukinavicius G, Lapiene V, Stasevskij Z, Dalhoff C, Weinhold E, Klimasauskas S. J. Am. Chem. Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]; (b) Dalhoff C, Lukinavicius G, Klimasauskas S, Weinhold E. Nat. Chem. Biol. 2006;2:31–32. doi: 10.1038/nchembio754. [DOI] [PubMed] [Google Scholar]
- 5.(a) Osborne T, Roska RL, Rajski SR, Thompson PR. J. Am. Chem. Soc. 2008;130:4574–4575. doi: 10.1021/ja077104v. [DOI] [PubMed] [Google Scholar]; (b) Pignot M, Siethoff C, Linscheid M, Weinhold E. Angew. Chem. Int. Ed. 1998;37:2888–2891. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2888::AID-ANIE2888>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]; (c) Comstock LR, Rajski SR. J. Am. Chem. Soc. 2005;127:14136–14137. doi: 10.1021/ja054128y. [DOI] [PubMed] [Google Scholar]; (d) Zhang C, Weller RL, Thorson JS, Rajski SR. J. Am. Chem. Soc. 2006;128:2760–2761. doi: 10.1021/ja056231t. [DOI] [PubMed] [Google Scholar]
- 6.(a) Mahal LK, Yarema KJ, Bertozzi CR. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]; (b) Cornish VM, Hahn KM, Schultz PG. J. Am. Chem. Soc. 1996;118:8150–8151. [Google Scholar]; (c) Dirksen A, Hackeng TM, Dawson PE. Angew. Chem. Int. Ed. Engl. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]; (d) Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew. Chem. Int. Ed. Engl. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 7.(a) Wu H, Horton JR, Battaile K, Allali-Hassani A, Martin F, Zeng H, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Cheng X. Proteins. 2007;67:198–208. doi: 10.1002/prot.21272. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ames MM, Selassie CD, Woodson LC, Van Loon JA, Hansch C, Weinshilboum RM. J. Med. Chem. 1986;29:354–358. doi: 10.1021/jm00153a009. [DOI] [PubMed] [Google Scholar]; (c) Krynetski EY, Krynetskaia NF, Yanishevski Y, Evans WE. Mol. Pharmacol. 1995;47:1141–1147. [PubMed] [Google Scholar]; (d) Cannon LM, Butler FN, Wan W, Zhou ZS. Anal. Biochem. 2002;308:358–363. doi: 10.1016/s0003-2697(02)00267-1. [DOI] [PubMed] [Google Scholar]; (e) Woodson LC, Weinshilboum RM. Biochem. Pharmacol. 1983;32:819–826. doi: 10.1016/0006-2952(83)90582-8. [DOI] [PubMed] [Google Scholar]
- 8.Khalil MN, Erb N, Khalil PN, Escherich G, Janka-Schaub GE. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;821:105–111. doi: 10.1016/j.jchromb.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Anderson GP, Nerurkar NL. J. Immunol. Methods. 2002;271:17–24. doi: 10.1016/s0022-1759(02)00327-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.