Abstract
Purpose
Alternative CCND1 splicing results in cyclin D1b, which has specialized, pro-tumorigenic functions in prostate not shared by the cyclin D1a (full-length) isoform. Here, the frequency, tumor relevance, and mechanisms controlling cyclin D1b were challenged.
Experimental Design
First, relative expression of both cyclin D1 isoforms was determined in prostate adenocarcinomas. Second, relevance of the androgen axis was determined. Third, minigenes were created to interrogate the role of the G/A870 polymorphism (within the splice site), and findings validated in primary tissue. Fourth, impact of G/A870 on cancer risk was assessed in two large case-control studies.
Results
Cyclin D1b is induced in tumors, and a significant subset expressed this isoform in the absence of detectable cyclin D1a. Accordingly, the isoforms showed non-correlated expression patterns, and hormone status did not alter splicing. While G/A870 was not independently predictive of cancer risk, A870 predisposed for transcript-b production in cells and in normal prostate. The influence of A870 on overall transcript-b levels was relieved in tumors, indicating that aberrations in tumorigenesis likely alter the influence of the polymorphism.
Conclusions
These studies reveal that cyclin D1b is specifically elevated in prostate tumorigenesis. Cyclin D1b expression patterns are distinct from that observed with cyclin D1a. The A870 allele predisposes for transcript-b production in a context-specific manner. While A870 does not independently predict cancer risk, tumor cells can bypass the influence of the polymorphism. These findings have major implications for the analyses of D-cyclin function in the prostate, and provide the foundation for future studies directed at identifying potential modifiers of the G/A870 polymorphism.
Keywords: Cyclin D1a, Cyclin D1b, Prostate Adenocarcinoma, rs603965 Polymorphism, CCND1 Minigene
Introduction
It is increasingly apparent that alternative splicing of the CCND1 transcript results in a protein with divergent functions from full-length cyclin D1 (cyclin D1a). The alternatively spliced transcript, transcript-b, arises from a failure to splice at the exon-4/intron-4 boundary. The presence of a stop codon within intron-4 gives rise to a truncated protein (cyclin D1b) which harbors a unique C-terminus that is devoid of exon-5 encoded sequences (1, 2). Functional studies showed that cyclin D1b has unique activities, distinct from cyclin D1a, with particular cancer significance. In many cell types, cyclin D1a promotes cell cycle progression through activation of cyclin-dependent kinases (CDK) 4 or 6 (3, 4). The principal substrate of cyclin D1a-CDK4/6 complexes is the retinoblastoma tumor suppressor protein (RB), whose anti-proliferative capacity is weakened by cyclin D1a/CDK-mediated phosphorylation (5, 6). Although cyclin D1b binds CDK4, this variant is markedly deficient in inducing RB phosphorylation (7-10). Based on these and other observations which showed that cyclin D1a fails to alter cell cycle progression in RB-deficient cells (11-13), it might be expected that cyclin D1b is ineffective at promoting cellular proliferation and/or tumorigenesis. On the contrary, it was unexpectedly observed that cyclin D1b has enhanced oncogenic capacity compared to cyclin D1a (9, 10, 14-16).
The tumorigenic effects of cyclin D1b have been recently explored, and include unique functions in different tumor types. In mouse fibroblast models, cyclin D1b (but not cyclin D1a) induced in vitro focus formation, in vivo tumor formation (9, 10), and anchorage independence (16). In breast cancer cells, cyclin D1b exclusively induced resistance to therapeutic intervention (17). The importance of differential cyclin D1b function is further underscored by observations that the alternatively spliced transcript has been detected in multiple cancers, including Ewing's sarcoma (18, 19), mantle cell lymphoma (8, 20), esophageal cancer (9), colon cancer (21, 22), B-lymphoid malignancies (7, 23), and breast cancer (45). Given the divergent and decidedly pro-tumorigenic effects of cyclin D1b expression, it is imperative to explore cyclin D1b production and the factors that may regulate its production.
In prostate cancer (PCa) there is a particular interest in discerning the mechanisms that govern cyclin D1b production, as the D-cyclins serve specialized roles that appear to impact tumor progression. Prostate cancer is resistant to most forms of genotoxic stress (24); therefore, disseminated PCa is treated based on the androgen dependence of this tissue (25, 26). Prostate cancer cells require the androgen receptor (AR), a ligand-dependent transcription factor for survival and progression (27, 28). Significantly, AR activation results in mTOR-mediated induction of cyclin D1a, which impinges on CDK4 to initiate cell cycle progression (29). However, accumulated cyclin D1a appears to induce a negative feedback loop wherein cyclin D1 binds and inhibits AR transcriptional activity, thus regulating the strength and duration of AR signaling (reviewed in (14, 27)). The ability of cyclin D1a to regulate AR appears critical, as tumors devoid of cyclin D1a are associated with enhanced AR activity (30). Moreover, cyclin D1a can be sequestered to the cytoplasm in PCa (and other tissues (9, 31-34)), indicating that aberrant regulation of cyclin D1a may be common in PCa (30). The most striking evidence of altered regulation was observed with cyclin D1b, wherein the transcript-b was shown to be elevated in a small sample set of PCa and pre-cancerous PIN (prostatic intraepithelial neoplasia) specimens as compared to matched non-neoplastic controls (35). The consequence of this event is of clinical relevance, as cyclin D1b is compromised in its ability to control AR-dependent signaling (35). Thus, it is hypothesized that cyclin D1b serves multiple, oncogenic functions in PCa.
Despite the importance of cyclin D1b in cancer, few studies have assessed the factors that influence its production, or the relative expression of each isoform in PCa. Here, for the first time, through large-scale analyses we show that PCa specimens express elevated cyclin D1b, thus indicating the importance of delineating the factors that contribute to cyclin D1b production. Moreover, cyclin D1b is specifically and significantly induced as a function of PCa tumorigenesis. Subsequent analyses showed that cyclin D1b showed distinct expression patterns from that of cyclin D1a, which was not elevated in cancer (as compared to non-neoplastic tissue) under the conditions utilized. Unlike observations in breast cancer, hormone status had no detectable impact on transcript-b production. By contrast, the A-allele of the CCND1 G/A870 single nucleotide polymorphism (SNP rs603965) predisposed for transcript-b production in a context specific manner. Introduction of engineered CCND1 minigenes into cyclin D1-deficient cells showed that the A-allele promoted transcript-b and cyclin D1b production, whereas the G-allele promoted transcript-a and cyclin D1a production. These observations were consistent with analyses of non-neoplastic prostate tissue, wherein overall transcript-b levels were higher in patients with an A-allele. However, the requirement of the A-allele for high transcript-b production was relieved in tumor tissue and cell lines. Moreover, analyses of two large population-based studies, revealed that the A-allele is not independently predictive of PCa risk. Together, these data indicate that a subset of PCa induce cyclin D1b, and that the G/A870 polymorphism contributes to transcript-b and cyclin D1b expression. These studies show for the first time that cyclin D1b is elevated in PCa, and demonstrate the relevance of the G/A870 polymorphism on cancer risk and cyclin D1b production.
Materials and Methods
Immunohistochemistry
Human prostate specimens organized as a tissue microarray (cohort 1) were previously described (36). A second human prostate tissue microarray (cohort 2) was purchased (PR801; US Biomax, Ijamsville, MD). Immunostaining was performed after de-paraffinization/re-hydration and microwave-induced antigen retrieval with citrate buffer (DAKO, Carpinteria, CA). Serial sections were blocked with goat serum (10%), followed by incubation (1 hr) with the following antibodies: cyclin D1b [(17), 1:50] or cyclin D1a (Ab-3; Lab Vision, Fremont, CA). After washing with TBS, slides were incubated with a cytokeratin antibody (AE1/AE3; DAKO, Carpinteria, CA). Cyclin D1 was detected using an anti-rabbit-HRP–conjugated antibody (EnVision-Plus; DAKO, Carpinteria, CA), followed by incubation with Tyramide-Cy5 (TSA System; Perkin-Elmer, Waltham, MA). Cytokeratin was visualized with an anti-mouse-Alexa 488-conjugated antibody (Invitrogen/Molecular Probes, Eugene, OR). Nuclear visualization via 4',6-diamidino-2-phenylindole (DAPI).
AQUA system analysis
Immunohistochemical sections were quantified using the unbiased automated image acquisition system AQUA/PM2000 Imaging Platform (HistoRx, New Haven, CT) as previously described (37). Briefly, immunostained sections were scanned to detect FITC/Alexa 488, Cy5, and DAPI. The cyclin D1a or cyclin D1b AQUA score (average signal intensity) was determined within epithelial compartments based on cytokeratin masking. Analysis of the cyclin D1 isoforms was performed and statistical significance (p<0.05) was determined using Mann-Whitney U.
PCa risk associations
The Multiethnic Cohort (MEC) is a large prospective study, approved by the Institutional Review Boards at the University of Hawaii and University of Southern California, and is comprised of five racial/ethnic groups as previously described (38). Incident cancers and stage of disease were identified (up to April 1, 2002) using SEER cancer registries covering Hawaii and California. Controls were men without PCa prior to entry into the study and were matched to cases by age and ethnicity. This study consisted of 2,302 cases and 2,277 controls. Genotyping was performed by the 5’-nuclease TaqMan allelic discrimination assay (Applied Biosystems; Foster City, CA). Blinded duplicate samples (10%) were included to assess genotyping reproducibility.
The Australian Risk Factors for Prostate Cancer Study (RFPCS) is a population-based case-control study including 829 cases and 739 controls predominantly of European descent and was previously described (39). Eligible cases (diagnosed and histopathologically confirmed) were notified to the State Cancer Registry during 1994 to 1997. Cases were excluded if age at diagnosis was ≥ 70 years or if the tumor Gleason score <5. Controls were randomly selected from the State Electoral Rolls (compulsory to vote in Australia) and matched to cases by 5-year age group and city of residence. Informed consent was obtained from all study participants and approved by the Human Research Ethics Committee of the Cancer council of Victoria (HREC 9500). Genotyping was performed in duplicate using the Sequenom MassARRAY™ Compact System (Sequenom, San Diego, CA) with mass spectrometry (MALDI-TOF) and 5’-nuclease TaqMan allelic discrimination assay. Genotyping was performed in a blinded manner and discordance was validated using amplicon specific amplification and denaturing gradient gel electrophoresis (DGGE). Allele frequency estimates and tests of deviation from Hardy–Weinberg (H-W) equilibrium were carried out using standard procedures based on asymptotic likelihood theory. Tests for association between genotypes and case-control status were performed under dominant, recessive, co-dominant, and additive models. Case-control analyses were conducted using unconditional logistic regression and ORs and 95% confidence intervals (CI) were estimated for each genotype (relative to GG). All tests were two-sided and 5% level was used as a threshold for statistical significance.
Cell culture and treatments
The androgen-dependent (LNCaP, LAPC4) and castration resistant (PC3, 22Rv1, DU145) PCa cell lines were maintained as described (35). The breast cancer (MCF-7) cell line was maintained as described (17). PCR-RFLP analysis of PCa cell lines was previously described (35, 40). To assess AR activity, LNCaP cells were plated on poly-L-lysine in steroid-free conditions for 48 hrs in 5% CDT (HyClone; Logan, UT), and subsequently treated for 24 hrs with dihydrotestosterone (DHT, 1 nM). Treatment (1 uM for 24 hr) with the anti-androgen, Casodex (AstraZeneca Pharmaceuticals; Wilmington, DE) or the mTOR inhibitor, RAD001 (Everolimus, LC Laboratories; Woburn, MA, 10 uM for 4 hr) was performed as indicated.
CCND1 minigenes
The G/A870 region of CCND1 was amplified from LNCaP (GA-genotype) genomic DNA using the Expand Long Template PCR System (Roche; Indianapolis, IN). The amplified fragment (3297bp containing intron-4) and pcDNA3.1 (with BGH polyadenylation signal) containing the coding sequence (exons 1-5) for human cyclin D1 (tagged with EGFP and HA) were digested with Bsu36I (internal exon-4 site) and EcoRV (introduced site after 6bp of CCND1 3’UTR), purified, ligated, and screened using BsrGI (internal intron-4 site) to verify insertion of intron-4 containing sequences. Positive clones were sequenced to identify those that harbor either the G- or A-allele. Transfection of the minigene constructs was performed using the standard calcium phosphate method. Immunoblotting was performed with the following antibodies: GFP (sc-9996, Santa Cruz Biotechnology; Santa Cruz, CA), cyclin D1a, and cyclin D1b.
Gene expression analysis
For PCa cell lines, RNA was extracted using TRIzol, and cDNA generated with SuperScript II (Invitrogen; Carlsbad, CA). RT-PCR was performed using GoTaq (Promega; Madison, WI) with the following primers: KLK3/PSA [prostate specific antigen, described previously (41)], transcript-a, transcript-b, and GAPDH (Supplementary Fig. S1). GAPDH and KLK3/PSA were amplified at 26 cycles. Transcript-b and transcript-a were amplified at 32 and 27 cycles, respectively. Products were resolved on agarose (2%) and visualized with ethidium bromide. Quantitative PCR (qPCR) was performed using a StepOne machine (Applied Biosystems). Taqman assays (Applied Biosystems) were used to determine KLK3/PSA (Hs00426859_g1) and GAPDH (Hs00266705_g1). Transcript-a, transcript-b, and GAPDH (primers described above) were determined by qPCR using Express SYBR (Invitrogen). Relative expression was determined as described (42). Human prostate tissue was obtained from the University of Cincinnati and Thomas Jefferson University in accordance with Institutional Review Board guidelines. OCT-embedded tissues were incubated in RNAlater-ICE (Ambion; Austin, TX). After OCT removal, tissue was minced and homogenized in TRIzol to obtain RNA (and DNA for PCR-RFLP as above). Subsequently, cDNA was generated using SuperScript VILO (Invitrogen; Carlsbad, CA). Cyclin D1a, cyclin D1b, and GAPDH (primers described above), expression was determined by RT-PCR in the presence of a [α-32P]-dCTP (PerkinElmer; Waltham, MA), which allows accurate quantification over a large dynamic range (43, 44). Products were separated on a 6% PAGE gel and quantified with a Typhoon 9400 PhosphorImager (GE Healthcare/Amersham; Piscataway, NJ). Total transcript-b expression was determined relative to GAPDH.
Results
Cyclin D1b is specifically elevated in prostate cancer
Despite the emerging knowledge that cyclin D1b has enhanced oncogenic functions, little is known about the frequency of cyclin D1b production and the mechanisms that regulate alternative CCND1 splicing. Previous studies detected induction of transcript-b in a small subset of PIN and PCa specimens; these findings were of significance, as cyclin D1b engages in prostate-specific activities that are proposed to enhance tumorigenesis (35). Thus, it was imperative to determine the status of cyclin D1b and define the factor(s) that regulate its production in PCa. To address this, a cyclin D1b-specific antibody was used that has been extensively characterized by immunoblot and immunohistochemistry (IHC) approaches in multiple studies (8, 17, 19, 35). The cyclin D1b-specific antibody is directed against an epitope not present in cyclin D1a, and a recent study further validated the specificity of the cyclin D1b antisera using both fluorescence-based immunocytochemistry and IHC (45). Although the cyclin D1b antisera is slightly more sensitive than the cyclin D1a antisera, conditions identical to those used in the present study could detect endogenous protein, as well as discern tumor-associated induction of either isoform in other tumor types (17, 45). To further confirm specificity, several studies were performed. First, U2OS cells (which express low levels of cyclin D1 (17)) were transfected with expression vectors encoding individual GFP-tagged cyclin D1 isoforms. As shown in Figure 1A, immunocytochemistry revealed no cross-reactivity with the cyclin D1 isoform-specific antisera. Similar results were observed in SAOS2 cells (which express low levels of cyclin D1 (17)) transduced with adenovirus encoding either cyclin D1 isoform (Supplementary Fig. S2A), as detected by AQUA System (HistoRx) analysis. Second, immunodetection of endogenous cyclin D1b in prostate cancer (LAPC-4) cells was depleted by hormone ablation (Supplementary Figure S2B), which blocks CCND1 mRNA translation. Third, immunofluorescence (Supplementary Figure S2C) and immunohistochemistry (using cells treated in vitro, clotted, embedded, and subsequently processed for immunohistochemistry, Supplementary Figure S2D) showed identical profiles when detecting endogenous cyclin D1b in prostate cancer cells in the presence of androgen or after androgen ablation. Combined, these studies validated the specificity of the cyclin D1b antibody.
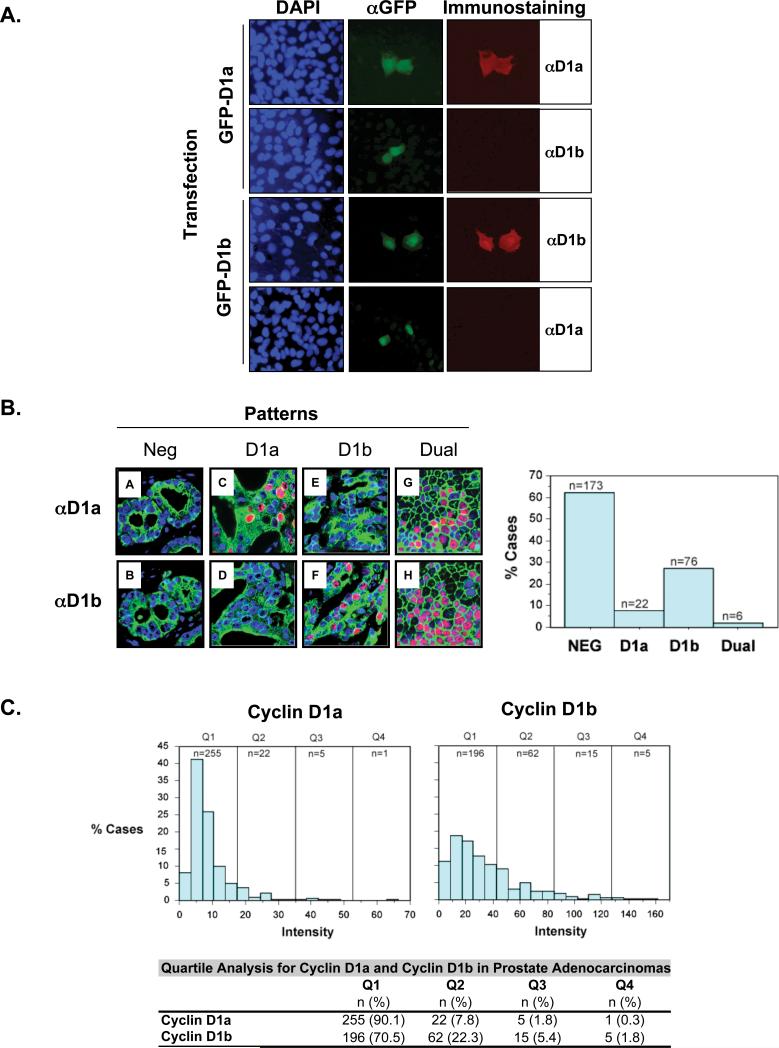
Figure 1. The cyclin D1b isoform is frequently expressed in prostate cancer.
A). U2OS cells were transfected with expression plasmids encoding GFP-cyclin D1a (upper panels) or GFP–cyclin D1b (lower panels) then immunostained with GFP-specific antisera (green, middle column) or isoform-specific cyclin D1 antibodies, as indicated (red, right column). Nuclei are indicated by DAPI (blue, left column). Representative images were captured with equal exposure times. B). Cyclin D1a and cyclin D1b isoforms were detected in serial sections from a large PCa tissue microarray using a fluorescence-based tissue biomarker platform. Representative images (left) and quantification (right) from the tumor dataset with antibodies specific to cyclin D1a (upper panels) or cyclin D1b (lower panels). Cyclin D1a and cyclin D1b are indicated by red fluorescence, cytokeratin (epithelial marker) is indicated by green fluorescence, and nuclei (DAPI) are indicated by blue fluorescence. Staining patterns are indicated: Negative (A, B), cyclin D1a-positive (C, D), cyclin D1b-positive (E, F), and cyclin D1a/D1b-positive “dual-positive” (G, H). C). Frequency distribution analysis of cyclin D1a (left) and cyclin D1b (right) in adenocarcinomas. Data are presented as percent cases vs. staining intensity and quartiles (Q1-4) are indicated by vertical line. The n-number for each quartile is indicated.
Relative cyclin D1a and cyclin D1b expression was subsequently assessed in serial sections using isoform-specific antisera in a cohort of human prostate specimens (1,626 cores) containing non-neoplastic, PIN lesions, and adenocarcinomas (36). The fluorescent signal was determined using a tissue biomarker platform (AQUA System, HistoRx) that allows unbiased quantification of signal intensity in tissue microarrays. Quantification of cyclin D1a or cyclin D1b was performed in cells of epithelial origin, as achieved through co-immunostaining for epithelial cytokeratins. Representative staining of parallel samples (left) and overall quantification (right) are shown in Figure 1B, wherein it was apparent that distinct patterns emerged with regard to each isoform. Consistent with previous reports (9, 30, 31), a large fraction (62.5%) of tumors had low or undetectable expression of either cyclin D1 isoform. Notably, identical conditions were used in a recent study which found significant cyclin D1a elevation in breast cancer (45), thus confirming that the conditions used are capable of detecting differences in cyclin D1a levels. These data indicate that neither protein is necessarily required for tumor maintenance in the prostate, and are congruent with a previous report which suggested that loss of cyclin D1a may facilitate AR activity (30). A second cohort showed cytoplasmic localization of cyclin D1a (data not shown), whereas nuclear cyclin D1a was detected either in isolation (7.94%) or in conjunction (2.16%) with cyclin D1b. Strikingly, a subset (27.4%) of tumors appeared to express cyclin D1b but showed minimal immunoreactivity against cyclin D1a. The differential staining patterns for cyclin D1a and cyclin D1b provide additional evidence that the isoform-specific antibodies recognize distinct epitopes, and provide the first evidence that the two cyclin D1 isoforms show distinct expression profiles in PCa. This concept was substantiated by quartile analysis, wherein comparison of frequency and intensity of immunostaining for each isoform within the adenocarcinomas was quantified (Fig. 1C). As shown, little to no cyclin D1a expression was observed in the majority of cases (Quartile-1 “Q1” =90.1%). By contrast, cyclin D1b expression showed a larger range of intensity (Q1=70.5%, Q2=22.3%, Q3=5.4%, Q4=1.8%). Together, these data demonstrate that cyclin D1a and cyclin D1b show distinct profiles in PCa, and that a significant subset of tumors express cyclin D1b.
Since the above data are suggestive that cyclin D1b may play a significant role in PCa, comparison of relative expression between the two variants was performed using non-neoplastic and tumor tissue. As shown, no significant induction in cyclin D1a levels were observed in PIN or adenocarcinomas as compared to non-neoplastic tissue (Fig. 2A, left panel). Interestingly, recent reports have shown that cyclin D1a levels can be reduced in PCa specimens (30) or xenograft models (46), and this event correlates with enhanced AR activity. By contrast, cyclin D1b levels (right panel) were elevated 1.2-fold in tumors as compared to non-neoplastic tissue (p<0.004). Interestingly, no induction of cyclin D1b protein was noted in PIN lesions, indicating that cyclin D1b maybe specifically associated with tumorigenesis. To expand these findings, parallel analyses were performed using a second independent cohort of non-neoplastic and PCa tissue, wherein cyclin D1b status was elevated 1.5-fold in tumors compared to non-neoplastic epithelia (p<0.004) (Fig. 2B). The observed increase in cyclin D1b in tumors but not matched non-neoplastic tissue was also seen by immunoblot analysis from a radical prostatectomy specimen (Supplementary Fig. S2E). Combined, these data provide evidence that different from cyclin D1a expression profiles, elevated cyclin D1b is observed in a significant fraction of tumors.
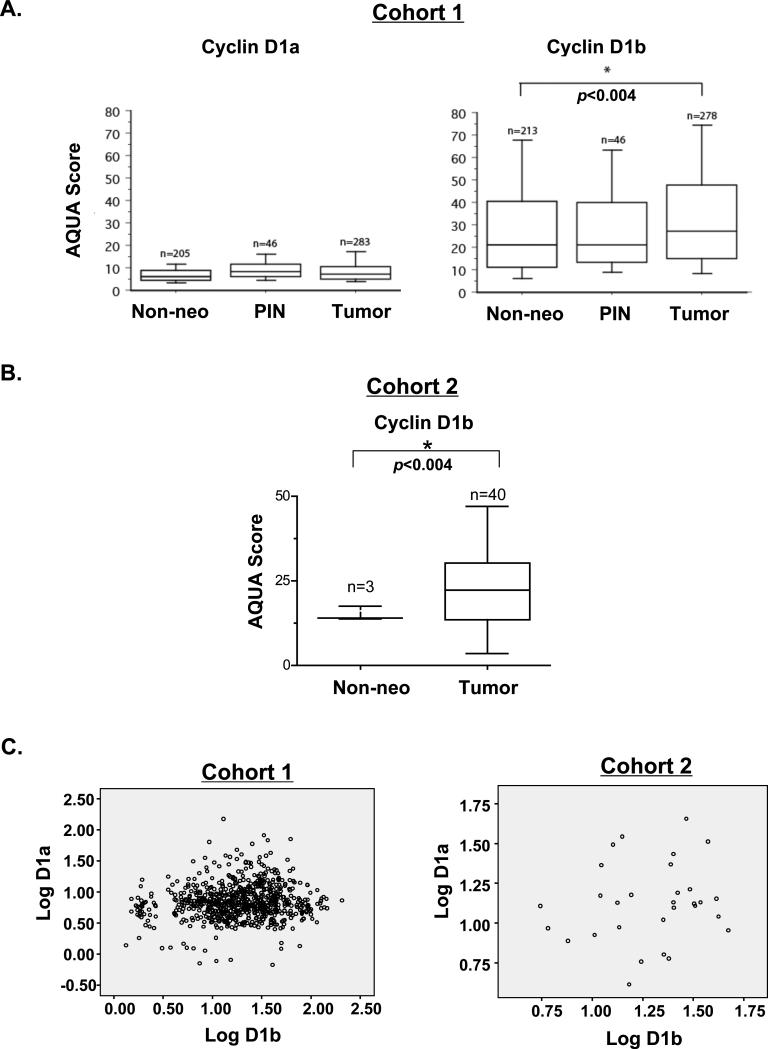
Figure 2. Cyclin D1b is uniquely associated with tumorigenesis.
A). Immunostaining for non-neoplastic, PIN, and adenocarcinoma specimens obtained in Figure 1 was assessed to determine the association of cyclin D1a (left) and cyclin D1b (right) with tumorigenesis. Immunostaining data (AQUA Scores) are shown, and the n-number is indicated. Statistical significance (p<0.05) was determined by Mann-Whitney U analysis and is indicated by *. B). Parallel analysis was performed on a second tissue array containing non-neoplastic and tumor tissue. The n-number is indicated. Statistical significance (p<0.05) was determined by Mann-Whitney U analysis and is indicated by *. C). The relationship between cyclin D1b and cyclin D1a were determined by Pearson correlation for cohort-1 (left) and cohort-2 (right). Data is presented as log transformed AQUA scores from individual tumor specimens for cyclin D1b and cyclin D1a. The Pearson correlations (r) for cohort 1 and cohort 2 were 0.106 and 0.153, respectively.
Cyclin D1b expression patterns are distinct from cyclin D1a
The above data demonstrate that cyclin D1b is induced in PCa; however, the mechanism(s) by which production of this isoform is enhanced remains unknown. Furthermore, it is likely that the mechanism(s) involve various aspects of transcription, translation, and stability. The notion that divergent pathways exist to regulate cyclin D1 isoform levels is consistent with the observation that human PCa can produce differential cyclin D1 isoform expression patterns (Fig. 1). Despite these complexities, up-regulation of cyclin D1b may be a reflection of overall cyclin D1 isoform levels. To address this possibility, Pearson-correlations were determined between cyclin D1b and cyclin D1a. As shown in Figure 2C, no relationship was observed between cyclin D1b and cyclin D1a in tumor specimens in either cohort-1 (left, r=0.016) or cohort-2 (right, r=0.153). Overall, these data indicate that the protein expression patterns of cyclin D1b are distinct from that of cyclin D1a and that disparate mechanism(s) may exist in PCa to regulate each isoform.
Alternative splicing occurs independent of AR activity in PCa cell model systems
Given the observation that cyclin D1b production is associated with tumorigenesis (Fig. 2A) and that cyclin D1b (unlike cyclin D1a) has a diminished capacity to negatively regulate AR function (35), the relationship between alternative splicing of cyclin D1 and AR activity was examined. These studies are imperative, as AR is the dominant signaling pathway that drives PCa progression, therapeutic failure (27, 28, 47), and androgen-dependent splicing of other factors (e.g., clusterin (48)). Analyses of relative transcript expression in asynchronous cells (cultured in the presence of complete serum) revealed that transcript-b levels were slightly higher in AR-positive (LNCaP, LAPC4, 22Rv1) cells as compared to AR-negative (PC3, DU145) PCa cells and an AR-positive (MCF-7) breast cancer cell line (Supplementary Figure S3A). To directly assess the impact of AR activity on transcript-b levels, LNCaP cells were cultured in the absence of steroid hormones prior to stimulation with DHT. Representative transcript analyses are shown in Supplementary Figure S3B and quantification provided in Figure 3A. AR activation was verified through induction of KLK3/PSA, a well-characterized AR target gene (Figure 3A and Supplemental Figure S3B lanes 1 and 2, upper panel). Unlike PSA (Figure 3A, top panel), transcript-a levels were unchanged by the presence or absence of androgen (middle panel) consistent with the ability of the AR-signaling axis to regulate cyclin D1a levels through post-transcriptional mechanisms involving mTOR (29). Likewise, transcript-b levels were indistinguishable between androgen-deprived and -stimulated conditions (Figure 3A, bottom panel), and similar results were observed in parallel experiments wherein an AR antagonist resulted in attenuated KLK3/PSA expression but no alteration in either cyclin D1 transcript. These data suggest that in PCa cell lines, production of transcript-b occurs independently of hormone or AR activity.
Figure 3. Transcript-b levels are refractory to androgen and AR activity.
A). To assess AR activity on cyclin D1 transcript-a and transcript-b levels, LNCaP cells were cultured in either in the absence of steroid hormones (charcoal dextran treated serum, CDT) and stimulated 24 hrs with (or not) with 1 nM DHT, as indicated, or in complete serum (5% FBS) supplemented where indicated with 1 uM Casodex. Real time PCR for relative KLK3/PSA (upper panel), transcript-a (middle panel), transcript-b (lower panel) are shown. Representative images are shown in Supplemental Figure S3. B). To evaluate the contribution of mTOR-mediated post-transcriptional regulation of cyclin D1b, LAPC4 cells were treated with 10 uM RAD001 for 4 hrs. Cyclin D1b levels were determined by immunoblot analysis using the cyclin D1b-specific antibody with lamin (control). Representative immunoblot is shown.
Since these data suggested that cyclin D1b production, like cyclin D1a, may be regulated in an mTOR-dependent manner, cyclin D1b protein levels were monitored in LAPC-4 cells treated with an mTOR inhibitor (RAD001). As shown in Figure 3B and Supplemental Figures S2C, and S2D, cyclin D1b protein levels were suppressed by RAD001 treatment, indicating that cyclin D1b is also regulated by mTOR. Combined, these data indicate that both cyclin D1 transcripts are regulated independently of androgen and AR transcriptional activity, and that cyclin D1b is also modulated through translational mechanisms involving mTOR. The observation that both cyclin D1 transcripts are refractory to androgen is distinct from breast cancer, wherein estrogen activates cyclin D1 expression (49, 50). In the context of PCa, estrogen treatment (which activates the gain-of-function AR mutant found in LNCaP cells (41, 51)) had no effect on the abundance of transcript-b expression (Supplementary Fig. S3C). Collectively, these data indicate that altered AR transcriptional activity cannot account for the tumor-associated induction of cyclin D1b in PCa.
The G/A870 polymorphism is not independently predictive of PCa risk
The data above indicate that cyclin D1b production must be attributed to factors independent of hormone or overall cyclin D1a levels. It has been long hypothesized that the G/A870 polymorphism could contribute to cyclin D1b production due to its location in a conserved splice donor site (15). This polymorphism is of interest, as the A-allele has been shown to be tentatively associated with increased risk for many cancers (15, 52). Surprisingly, this provocative hypothesis has not been tested, and the relevance of the polymorphism for PCa not well delineated. To address these issues, the contribution of the G/A870 polymorphism to PCa risk was analyzed using two large population-based case-control studies from the Multiethnic Cohort (MEC) study and the Australian Risk Factors for Prostate Cancer Study. A combined analysis among 3,131 invasive cases and 3,016 controls revealed minimal differences in the overall distribution of genotypes between cases [34.9% (GG), 48.2% (GA), 16.9% (AA)] and controls [35.9% (GG), 46.5% (GA), and 17.6% (AA)]. Moreover, no significant association between the G/A870 polymorphism and PCa risk overall or in any ethnic population was observed (Table 1). Combined, these data indicate that the CCND1 polymorphism is not independently predictive of PCa risk.
Association between CCND1 G/A870 (rs603965) genotype and prostate cancer risk
| Genotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | Effect per allele | |||||||
| Ethnicity | n | Freq. A-allele | n | OR | n | OR (95% CI)a | n | OR (95% CI)a | OR (95% CI)a | P Trend |
| African Americans | ||||||||||
| Controls | 647 | 0.232 | 374 | 246 | 27 | |||||
| Cases | 675 | 0.235 | 387 | 1.0 | 258 | 1.01 (0.81-1.27) | 30 | 1.05 (0.61-1.80) | 1.02 (0.84-1.23) | 0.85 |
| Latinos | ||||||||||
| Controls | 646 | 0.424 | 214 | 315 | 117 | |||||
| Cases | 643 | 0.427 | 212 | 1.0 | 313 | 1.01 (0.79-1.29) | 118 | 1.02 (0.74-1.41) | 1.01 (0.86-1.18) | 0.89 |
| Japanese | ||||||||||
| Controls | 467 | 0.485 | 126 | 229 | 112 | |||||
| Cases | 457 | 0.480 | 121 | 1.0 | 233 | 1.08 (0.79-1.47) | 103 | 0.96 (0.67-1.39) | 0.98 (0.81-1.18) | 0.86 |
| Native Hawaiians | ||||||||||
| Controls | 68 | 0.588 | 12 | 32 | 24 | |||||
| Cases | 71 | 0.521 | 14 | 1.0 | 40 | 1.10 (0.44-2.71) | 17 | 0.55 (0.20-1.51) | 0.71 (0.43-1.18) | 0.18 |
| European Americans | ||||||||||
| Controls | 449 | 0.451 | 134 | 225 | 90 | |||||
| Cases | 456 | 0.478 | 117 | 1.0 | 242 | 1.24 (0.91-1.68) | 97 | 1.23 (0.84-1.80) | 1.12 (0.93-1.35) | 0.24 |
| Australians | ||||||||||
| Controls | 739 | 0.456 | 225 | 354 | 160 | |||||
| Cases | 829 | 0.455 | 241 | 1.0 | 422 | 1.12 (0.89-1.41) | 166 | 0.98 (0.74-1.30) | 1.00 (0.87-1.15) | 0.99 |
| All Groups | ||||||||||
| Controls | 3,016 | 1,085 | 1,401 | 530 | ||||||
| Cases | 3,131 | 1,092 | 1.0 | 1,508 | 1.00 (0.86-1.17) | 531 | 1.08 (0.96-1.21) | 1.01 (0.94-1.09) | 0.73 | |
Odds ratios (OR) and 95% confidence intervals (95% CI) estimated using unconditional logistic regression adjusted for age, race/country where necessary.
Minigene analyses reveal a role for the A-allele in cyclin D1b production
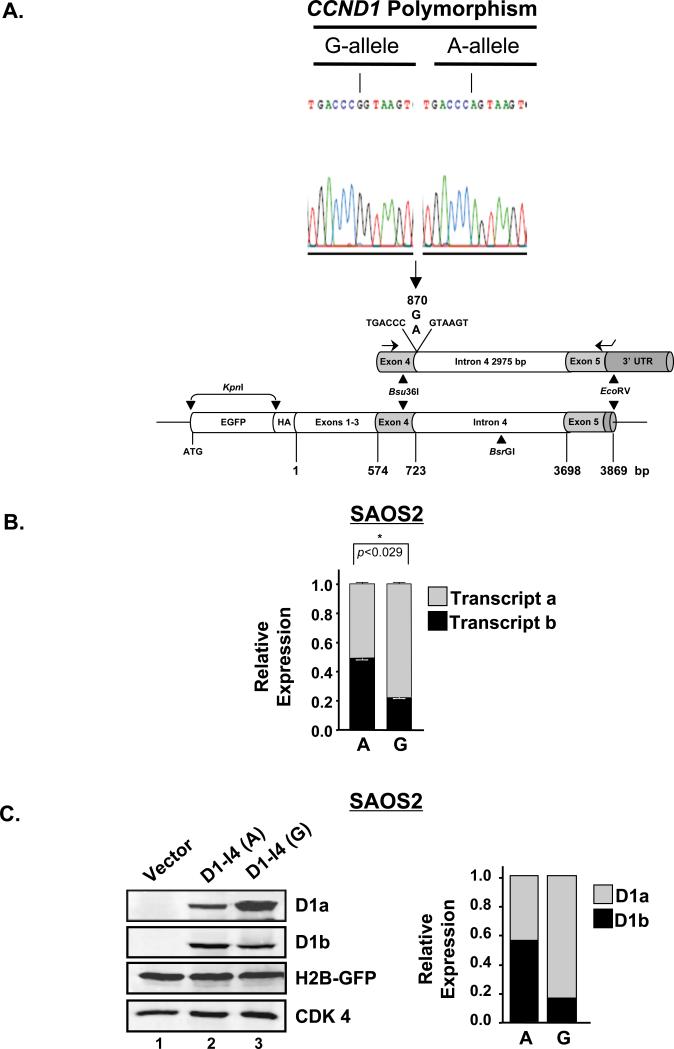
The observation that the G/A870 polymorphism is not independently predictive of PCa risk was surprising, since it has been hypothesized that the A-allele supports transcript-b production (14, 15, 53), and that the resultant cyclin D1b protein harbors oncogenic activity (9, 10, 16, 17). To assess the contribution of the G/A870 polymorphism on transcript-b production, CCND1 minigenes were generated. For these studies, the intron-4 sequence containing either the G- or A-allele was cloned and inserted into the full-length, EGFP-tagged cyclin D1 cDNA (Fig. 4A). Using these minigenes, the ability of G/A870 polymorphism to control transcript-b expression was assessed by mRNA analysis (Fig. 4B) in SAOS2 cells that express low levels of both cyclin D1 isoforms. Transfection of the A-allele containing minigene resulted in 2.2-fold enhanced transcript-b expression when compared to the wild-type G-allele minigene (p<0.029). These data are consistent with published observations suggesting that the A-allele correlates with transcript-b expression (54-57). To assess the impact on isoform production, protein analysis was performed (Fig. 4C, left panel). For these studies, co-transfection with H2B-GFP was used as a control for transfection efficiency and evaluated by immunoblot. As shown, vector transfected SAOS2 cells (lane 1) were devoid of detectable cyclin D1a (upper blot) and D1b (middle blot), consistent with previous reports (10, 12, 17). Introduction of the CCND1 minigenes resulted in a reciprocal relationship. As shown, a significant increase in cyclin D1a was observed with the G-allele minigene (lanes 2 and 3, upper blot) as compared to the A-allele equivalent. By contrast, the A-allele minigene resulted in an approximate 3-fold increase in cyclin D1b (lanes 2 and 3, upper-middle blot) as compared to the G-allele equivalent. Similar results were obtained in MCF-7 cells (Supplementary Fig. S4). These results suggest that the CCND1 minigenes are a viable means to assess CCND1 splicing, regulation, and demonstrate that the A-allele can confer a selective advantage for cyclin D1b production.
Figure 4. The A-allele predisposes for cyclin D1b production using CCND1 minigenes.
A). Sequence analysis (top) and cloning strategy (bottom) surrounding the CCND1 polymorphism (G/A870) from two independent constructs showing the G- and A-allele of human cyclin D1. Briefly, a 3.3kb genomic PCR fragment containing either the G- or A-allele and the entire intron-4 was digested with Bsu36I and EcoRV and sub-cloned into pcDNA3.1 containing a EGFP- and HA-tagged human cyclin D1. B). Quantitative PCR analyses (n=4) of transcript-a and transcript–b expression in SAOS2 cells (express low levels of cyclin D1) transfected with the A- and G-allele containing CCND1 minigenes [D1-I4 (A) or D1-I4 (G)]. Statistical significance (p<0.05) was determined by Mann-Whitney U analysis and is indicated by *. C). Left: Representative immunoblots for cyclin D1a and cyclin D1b from SAOS2 cells transfected with the A-allele (lane 2) or G-allele (lane 3) CCND1 minigenes and a plasmid encoding H2B-GFP (used as a loading/transfection control). Right: Quantification of cyclin D1a and cyclin D1b immunoblots, as a function of allele, after correcting for transfection efficiency (transfected H2B-GFP) and loading (endogenous CDK 4).
AA-genotype predicts transcript-b expression in non-neoplastic but not PCa tissue
Given the propensity for the A-allele minigene to confer cyclin D1b production in a heterologous system, the influence of the G/A870 polymorphism on cyclin D1b expression in human prostate specimens was determined. First, the CCND1 genotype was determined by PCR-RFLP from non-neoplastic and tumor specimens after prostatectomy (data not shown). The genotype distribution in non-neoplastic specimens was 50% (GG), 30% (GA), and 20% (AA) and a similar distribution of 45.5% (GG), 31.8% (GA), and 22.7% (AA) was observed in tumor specimens. These data are consistent with analyses in Table 1, which show that the A-allele is not over-represented in PCa.
The relevance of the G/A870 polymorphism on transcript–a and transcript–b expression was subsequently determined using quantitative RT-PCR from available prostate specimens. As depicted in Figure 5A, analyses of overall transcript levels as a function of genotype in non-neoplastic tissue revealed a significant (2.3-fold, p<0.014) increase in overall transcript-b levels in tissues with the AA-genotype (as compared to GG). These data are consistent with the minigene analyses, and suggest that A870 can influence the alternative splicing event and transcript-b production. Parallel analyses in a subset of tissue with sufficient material to also assess transcript-a levels suggest that the AA-genotype may not significantly alter the transcript-b/transcript-a ratios in tissue, but analyses of a larger data set will be needed to more fully address this complex question (Supplementary Fig. S5A). Finally, overall transcript-b levels were analyzed in tumor tissue from radical prostatectomy (Fig. 5B). In these tissues, the AA-genotype may influence the expression of cyclin D1b isoforms, but does not predict overall transcript-b levels (p<0.371). Together, these data reflect a context-specific influence of the A870 polymorphism, which can predispose for transcript-b expression; however, additional events may circumvent or cooperate with A870 to induce cyclin D1b production.
Figure 5. The AA-genotype correlates with overall transcript-b levels in a context-dependent manner.
Transcripts-b levels relative to total mRNA (GAPDH) were determined by quantitative RT-PCR (described in the Materials and Methods) and plotted as a function of genotype for: A). non-neoplastic and B). tumor tissue. The n-number is indicated. Statistical significance (p<0.05) was determined by Mann-Whitney U analysis and is indicated by *.
Discussion
It has been previously demonstrated that the cyclin D1b splice variant harbors differential oncogenic activities and likely serves specialized functions in the prostate that promote development and/or progression of cancer (14, 15, 35). Despite these observations, little is known concerning the factors that control cyclin D1b production. The present study is the first to examine expression profiles for each cyclin D1 isoform and to address the mechanisms of cyclin D1b production in PCa. Consistent with our previous report, a large number of prostatic adenocarcinomas show mis-localization or are devoid of detectable cyclin D1a (30). However, a significant fraction of tumors express cyclin D1b, which showed distinct expression profiles not correlated to detected cyclin D1a. Notably, cyclin D1b was significantly enhanced in PCa as compared to non-neoplastic tissue. Thus, the cyclin D1b isoform appears to be specifically induced in a significant fraction of prostate cancers. Functional studies demonstrated that transcript-b levels are refractory to androgen or AR activation, but the G/A870 polymorphism is a critical effector of cyclin D1b production. Subsequent analyses in human specimens showed that while the A-allele predicts for enhanced overall transcript-b levels in prostate tissue, G/A870 is not independently predictive of PCa risk. Parallel studies revealed that the requirement of the A-allele for enhanced transcript-b expression is relieved in tumor tissue. Together, these data demonstrate that the cyclin D1b isoform is predominantly associated with tumorigenesis in PCa, and implicate both the polymorphism and modifiers thereof as effectors of the cyclin D1b oncogenic variant.
Only recently have the cyclin D1 isoforms been examined in human disease, and current observations suggest that cyclin D1b regulation is under stringent but tissue-specific control. First, induction of both isoforms has been documented in primary breast carcinomas (17, 45). These observations are distinct from what was observed in PCa, wherein no significant induction in cyclin D1a was observed in neoplastic disease. In breast cancer models, anti-estrogen therapies enhanced cyclin D1b production, while cyclin D1a was reduced (17). In contrast, neither cyclin D1 transcript in the current study was influenced by anti-androgen therapy, suggesting that unique distinctions exist between these two hormone-dependent cancers. Second, like the current study, the majority of primary esophageal carcinomas expressed cyclin D1b (9). Subsequent studies revealed esophageal tumor-derived mutations that increased the oncogenic capacity of cyclin D1a in a manner synonymous to cyclin D1b (58). Finally, increased cyclin D1b (not cyclin D1a) has been associated with histologic grade in non-small cell lung carcinoma patients (31). Importantly, cyclin D1b correlated with poor survival and was found to be an independent risk factor for lung cancer development. These collective studies (summarized in Supplementary Fig. S6) highlight the importance of examining cyclin D1b levels and demonstrate the need to discern the contributing isoform in tumorigenesis. Moreover, previous studies examining cyclin D1 used reagents that can potentially detect either isoform (13, 59-61). Nonetheless, it is apparent from the present findings that cyclin D1b induction is characteristic of a subset of PCa, and ongoing studies will address the impact of this induction on clinical outcome.
Based on these observations, it is imperative to define the mechanism of cyclin D1b production. In addition to alternative splicing, it is likely that multiple mechanisms exist to regulate cyclin D1b production, including: gene activation (17), translational control (29), transcriptional elongation (19), and mRNA stability (62). In breast cancer, both cyclin D1 isoforms are regulated at the transcriptional level through the actions of the estrogen receptor (49, 50). This is in marked contrast to PCa cells, wherein both transcripts were unchanged by manipulation of the AR pathway. By contrast, mTOR-mediated enhancement of translation appears to be required for cyclin D1b production, which has been established for cyclin D1a (29). Thus, current therapies directed at ablation of AR activity are not likely to alter alternative splicing, but may assist in preventing translation of transcript-b.
Based on these findings, critical questions were addressed with regard to the induction of transcript-b and resultant cyclin D1b protein production. The data herein show that the CCND1 G/A870 polymorphism is a potent effector of this event. Using novel CCND1 minigenes that harbor a single base change (G- or A-allele), it was evident that the A-allele predisposes for cyclin D1b production in cells. To our knowledge, this is the first report to demonstrate that the A-allele, in isolation, influences cyclin D1b production. However, a noted caveat is that the minigenes lack most of the 3’UTR and natural polyadenylation site which could potentially contribute to transcript stability (62) which could potentially influence the relative levels of cyclin D1a and cyclin D1b. Nonetheless, analyses of non-neoplastic prostate tissue showed that transcript-b levels were enhanced in tissue with the AA-genotype, consistent with the minigene analyses and additional tumor types (54-57). However, functional studies demonstrate that the G-allele can also produce cyclin D1b, supporting the hypothesis that the A-allele is not universally required for transcript-b or cyclin D1b production. Future studies will be directed at factors that influence the A-allele effect, and on the importance of the transcript b:a ratio. Collectively, these data indicate that the A-allele predisposes for transcript-b and cyclin D1b production in PCa, but that additional factors may influence the alternative splicing of cyclin D1.
An association between the A-allele and increased PCa risk (63, 64) has been suggested in prior studies of limited size (<300 cases and <300 controls) and composition. Consistent with the supposition that the A-allele is not sufficient for transcript-b production, two population-based PCa studies (>3,000 cases and >3,000 controls) determined no independent risk association. These analyses had 80% power to detect effects as low as 1.11 and 1.17 for the A-allele, assuming a minor allelic frequency (MAF) of 40% (which is the average MAF in the multiethnic sample), and a log-additive or dominant effect on risk, respectively. However, assessment of luteinizing hormone (LH) levels in the controls with the AA-genotype were modestly elevated (12%, p=0.02) compared to controls with the G-allele, while other hormones (testosterone, estradiol, SHBG, DHEAS, androstenedione, androstanediol glucuronide, and prolactin) were not significantly different (data not shown). Nonetheless, the consistent lack of a significant association of the polymorphism to PCa risk, in all populations, further supports that this variant is not an independent marker.
Intriguingly, despite the relevance of the A-allele for overall transcript-b production in non-neoplastic prostate tissue, this requirement was alleviated in tumor tissue. These observations suggest a provocative hypothesis that tumor-specific alterations may bypass or synergize with the G/A870 polymorphism to enhance total transcript-b production. While the data herein clearly indicate the importance of the polymorphism, a number of additional factors have been suggested to influence cyclin D1b production. First, recent observations in Ewing's sarcoma cell lines and tumors suggests that chromosomal alterations between the Ewing's sarcoma oncogene and the ets family transcription factor (FLI1) result in up-regulation of cyclin D1b through alteration of transcript elongation (18, 19). Interestingly, chromosomal translocations of other ets family transcription factors are frequently observed in PCa (e.g., TMPRSS2:ERG (65, 66)). Therefore, it will be of importance to characterize the role of these fusions on cyclin D1b production. Second, loss of Brahma (Brm), the core ATPase of the SWI/SNF-chromatin-remodeling complex, significantly increased transcript-b expression in colorectal cells (67). Moreover, Brm is frequently reduced or lost in human PCa, and correlates with enhanced cellular proliferation (68). Finally, recent observations show that AR is alternatively spliced, thus indicating that aberrant splicing of several critical effectors of AR activity and cellular proliferation occur in PCa (69-71). It will be intriguing whether the mechanisms that underlie the alternative splicing events are linked.
In summary, the data herein demonstrate that cyclin D1b is specifically enhanced in PCa, thus showing a distinct expression profile from the cyclin D1a isoform. Accordingly, expression of the two isoforms is likely differentially regulated, and unlike breast cancer, the transcription and splicing of CCND1 (a or b) is independent of hormone. Rather, the A870 polymorphic allele was identified as a significant effector of cyclin D1b production, but additional events in prostate tumorigenesis may alleviate the influence of the A-allele. Together, these studies highlight a unique mechanism of cyclin D1b regulation, thereby providing the basis for future studies directed at identifying modifiers of the G/A870 polymorphism and the oncogenic consequence of cyclin D1b in PCa.
Clinical and Translational Relevance.
This study has marked clinical and translational implications: First, these data reveal for the first time that cyclin D1b is specifically induced in prostate cancer. As cyclin D1b has distinct functions from cyclin D1a, this has important implications for interpretation of biomarker and functional analyses. Second, previous studies tentatively linked the G/A870 polymorphism to cancer risk and cyclin D1b, but until now these postulates remained untested. Here, case-control studies showed that G/A870 is not independently predictive of risk. Third, although not sufficient for risk, functional studies identified A870 as a critical modulator of cyclin D1b in model systems and human prostate specimens. The A870 requirement is bypassed in cancer, indicating that tumor-specific events circumvent or cooperate with A870. Together, these findings identify cyclin D1b markedly enhanced in prostate cancer and reveal the impact of G/A870 for cyclin D1b production and cancer risk.
Supplementary Material
Acknowledgements
We thank Ankur Sharma, Nicholas A. Olshavsky, and Matthew J. Schiewer for critical reading of the manuscript and the K.E.K. laboratory for ongoing discussions. We thank professors: John L. Hopper and Dallas R. English, and Assistant Professor Melissa C. Southey, in the design and conduct of the Australian RFPCS study.
Support: This work was supported by: ACS-IRG114958 (C.E.S.C), NIH-CA099996 (K.E.K.), NHMRC-276408 (R.L.S) with The Cancer Institute NSW, Australian Cancer Research Foundation and the RT Hall Trust. The RFPCS study was supported by the National Health and Medical Research Council, Australia (209057, 251533, 450104), VicHealth, The Cancer Council Victoria, The Whitten Foundation, and Tattersall's. The Multiethnic Cohort Study was supported by the NCI-CA63464, –CA54281, and Department of Health and Human Services (Contract Nos. N01-PC-35139, -35137).
Abbreviations
- CDK
Cyclin Dependent Kinase
- RB
Retinoblastoma
- PCa
Prostate Cancer
- PIN
Prostatic Intraepithelial Neoplasia
- AR
Androgen Receptor
- mTOR
mammalian Target of Rapamycin
References
- 1.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11:1005–11. [PubMed] [Google Scholar]
- 2.Hosokawa Y, Tu T, Tahara H, Smith AP, Arnold A. Absence of cyclin D1/PRAD1 point mutations in human breast cancers and parathyroid adenomas and identification of a new cyclin D1 gene polymorphism. Cancer Lett. 1995;93:165–70. doi: 10.1016/0304-3835(95)03805-7. [DOI] [PubMed] [Google Scholar]
- 3.Lee YM, Sicinski P. Targeting cyclins and cyclin-dependent kinases in cancer: lessons from mice, hopes for therapeutic applications in human. Cell Cycle. 2006;5:2110–4. doi: 10.4161/cc.5.18.3218. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 5.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008:18685599. doi: 10.1038/nrc2401. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: where cancer meets the cell cycle. Exp Biol Med (Maywood) 2006;231:1271–81. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- 7.Leveque C, Marsaud V, Renoir JM, Sola B. Alternative cyclin D1 forms a and b have different biological functions in the cell cycle of B lymphocytes. Exp Cell Res. 2007;313:2719–29. doi: 10.1016/j.yexcr.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Marzec M, Kasprzycka M, Lai R, et al. Mantle cell lymphoma cells express predominantly cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108:1744–50. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–61. [PubMed] [Google Scholar]
- 10.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278:30339–47. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 11.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–11. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukas J, Muller H, Bartkova J, et al. DNA tumor virus oncoproteins and retinoblastoma gene mutations share the ability to relieve the cell's requirement for cyclin D1 function in G1. J Cell Biol. 1994;125:625–38. doi: 10.1083/jcb.125.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer. 1994;57:353–61. doi: 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 14.Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:1–12. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–8. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 16.Holley SL, Heighway J, Hoban PR. Induced expression of human CCND1 alternative transcripts in mouse Cyl-1 knockout fibroblasts highlights functional differences. Int J Cancer. 2005;114:364–70. doi: 10.1002/ijc.20750. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Dean JL, Millar EK, et al. Cyclin D1b is aberrantly regulated in response to therapeutic challenge and promotes resistance to estrogen antagonists. Cancer Res. 2008;68:5628–38. doi: 10.1158/0008-5472.CAN-07-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez G, Delattre O, Auboeuf D, Dutertre M. Coupled alteration of transcription and splicing by a single oncogene: boosting the effect on cyclin D1 activity. Cell Cycle. 2008;7:2299–305. doi: 10.4161/cc.6445. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez G, Bittencourt D, Laud K, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci U S A. 2008;105:6004–9. doi: 10.1073/pnas.0710748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shakir R, Ngo N, Naresh KN. Correlation of cyclin D1 transcript levels, transcript type and protein expression with proliferation and histology among mantle cell lymphoma. J Clin Pathol. 2008;61:920–7. doi: 10.1136/jcp.2008.057455. [DOI] [PubMed] [Google Scholar]
- 21.Hong Y, Eu KW, Seow-Choen F, Fook-Chong S, Cheah PY. GG genotype of cyclin D1 G870A polymorphism is associated with increased risk and advanced colorectal cancer in patients in Singapore. Eur J Cancer. 2005;41:1037–44. doi: 10.1016/j.ejca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 22.Bala S, Peltomaki P. CYCLIN D1 as a genetic modifier in hereditary nonpolyposis colorectal cancer. Cancer Res. 2001;61:6042–5. [PubMed] [Google Scholar]
- 23.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25:998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comstock CE, Knudsen KE. The complex role of AR signaling after cytotoxic insult: implications for cell-cycle-based chemotherapeutics. Cell Cycle. 2007;6:1307–13. doi: 10.4161/cc.6.11.4353. [DOI] [PubMed] [Google Scholar]
- 25.Balk SP. Androgen receptor as a target in androgen-independent prostate cancer. Urology. 2002;60:132–8. doi: 10.1016/s0090-4295(02)01593-5. discussion 8-9. [DOI] [PubMed] [Google Scholar]
- 26.Culig Z, Bartsch G. Androgen axis in prostate cancer. J Cell Biochem. 2006;99:373–81. doi: 10.1002/jcb.20898. [DOI] [PubMed] [Google Scholar]
- 27.Balk SP, Knudsen KE. AR, the cell cycle, and prostate cancer. Nucl Recept Signal. 2008;6:1–12. doi: 10.1621/nrs.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapman J, Brinkmann AO. The androgen receptor in prostate cancer. Pathol Res Pract. 1996;192:752–60. doi: 10.1016/S0344-0338(96)80097-5. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Chen SY, Ross KN, Balk SP. Androgens induce prostate cancer cell proliferation through mammalian target of rapamycin activation and post-transcriptional increases in cyclin D proteins. Cancer Res. 2006;66:7783–92. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]
- 30.Comstock CE, Revelo MP, Buncher CR, Knudsen KE. Impact of differential cyclin D1 expression and localisation in prostate cancer. Br J Cancer. 2007;96:970–9. doi: 10.1038/sj.bjc.6603615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, An SJ, Chen ZH, et al. Expression of cyclin D1 splice variants is differentially associated with outcome in non-small cell lung cancer patients. Hum Pathol. 2008:18715616. doi: 10.1016/j.humpath.2008.05.008. PMID. [DOI] [PubMed] [Google Scholar]
- 32.Sumrejkanchanakij P, Tamamori-Adachi M, Matsunaga Y, Eto K, Ikeda MA. Role of cyclin D1 cytoplasmic sequestration in the survival of postmitotic neurons. Oncogene. 2003;22:8723–30. doi: 10.1038/sj.onc.1206870. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto M, Tamakawa S, Yoshie M, Yaginuma Y, Ogawa K. Neoplastic hepatocyte growth associated with cyclin D1 redistribution from the cytoplasm to the nucleus in mouse hepatocarcinogenesis. Mol Carcinog. 2006;45:901–13. doi: 10.1002/mc.20204. [DOI] [PubMed] [Google Scholar]
- 34.Ratschiller D, Heighway J, Gugger M, et al. Cyclin D1 overexpression in bronchial epithelia of patients with lung cancer is associated with smoking and predicts survival. J Clin Oncol. 2003;21:2085–93. doi: 10.1200/JCO.2003.03.103. [DOI] [PubMed] [Google Scholar]
- 35.Burd CJ, Petre CE, Morey LM, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103:2190–5. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henshall SM, Horvath LG, Quinn DI, et al. Zinc-alpha2-glycoprotein expression as a predictor of metastatic prostate cancer following radical prostatectomy. J Natl Cancer Inst. 2006;98:1420–4. doi: 10.1093/jnci/djj378. [DOI] [PubMed] [Google Scholar]
- 37.Rubin MA, Zerkowski MP, Camp RL, et al. Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol. 2004;164:831–40. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolonel LN, Henderson BE, Hankin JH, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–57. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severi G, Giles GG, Southey MC, et al. ELAC2/HPC2 polymorphisms, prostate-specific antigen levels, and prostate cancer. J Natl Cancer Inst. 2003;95:818–24. doi: 10.1093/jnci/95.11.818. [DOI] [PubMed] [Google Scholar]
- 40.Sawa H, Ohshima TA, Ukita H, et al. Alternatively spliced forms of cyclin D1 modulate entry into the cell cycle in an inverse manner. Oncogene. 1998;16:1701–12. doi: 10.1038/sj.onc.1201691. [DOI] [PubMed] [Google Scholar]
- 41.Wetherill YB, Petre CE, Monk KR, Puga A, Knudsen KE. The xenoestrogen bisphenol A induces inappropriate androgen receptor activation and mitogenesis in prostatic adenocarcinoma cells. Mol Cancer Ther. 2002;1:515–24. [PubMed] [Google Scholar]
- 42.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:1–10. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddiqui H, Solomon DA, Gunawardena RW, Wang Y, Knudsen ES. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol Cell Biol. 2003;23:7719–31. doi: 10.1128/MCB.23.21.7719-7731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Link KA, Burd CJ, Williams E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–15. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millar EK, Dean JL, McNeil CM, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009 doi: 10.1038/onc.2009.13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang G, Wang J, Sadar MD. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res. 2008;68:9918–27. doi: 10.1158/0008-5472.CAN-08-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shand RL, Gelmann EP. Molecular biology of prostate-cancer pathogenesis. Curr Opin Urol. 2006;16:123–31. doi: 10.1097/01.mou.0000193384.39351.64. [DOI] [PubMed] [Google Scholar]
- 48.Cochrane DR, Wang Z, Muramaki M, Gleave ME, Nelson CC. Differential regulation of clusterin and its isoforms by androgens in prostate cells. J Biol Chem. 2007;282:2278–87. doi: 10.1074/jbc.M608162200. [DOI] [PubMed] [Google Scholar]
- 49.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–26. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabbah M, Courilleau D, Mester J, Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc Natl Acad Sci U S A. 1999;96:11217–22. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veldscholte J, Ris-Stalpers C, Kuiper GG, et al. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem Biophys Res Commun. 1990;173:534–40. doi: 10.1016/s0006-291x(05)80067-1. [DOI] [PubMed] [Google Scholar]
- 52.Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) Polymorphism Is Associated with Increased Cancer Risk in Human Populations: A Meta-Analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:2773–81. doi: 10.1158/1055-9965.EPI-08-0169. [DOI] [PubMed] [Google Scholar]
- 53.Xing D, Tan W, Lin D. Genetic polymorphisms and susceptibility to esophageal cancer among Chinese population (review). Oncol Rep. 2003;10:1615–23. [PubMed] [Google Scholar]
- 54.Holley SL, Parkes G, Matthias C, et al. Cyclin D1 polymorphism and expression in patients with squamous cell carcinoma of the head and neck. Am J Pathol. 2001;159:1917–24. doi: 10.1016/S0002-9440(10)63038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sathyan KM, Nalinakumari KR, Abraham T, Kannan S. CCND1 polymorphisms (A870G and C1722G) modulate its protein expression and survival in oral carcinoma. Oral Oncol. 2008;44:689–97. doi: 10.1016/j.oraloncology.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Wang L, Habuchi T, Takahashi T, et al. Cyclin D1 gene polymorphism is associated with an increased risk of urinary bladder cancer. Carcinogenesis. 2002;23:257–64. doi: 10.1093/carcin/23.2.257. [DOI] [PubMed] [Google Scholar]
- 57.Yu CP, Yu JC, Sun CA, Tzao C, Ho JY, Yen AM. Tumor susceptibility and prognosis of breast cancer associated with the G870A polymorphism of CCND1. Breast Cancer Res Treat. 2008;107:95–102. doi: 10.1007/s10549-007-9522-y. [DOI] [PubMed] [Google Scholar]
- 58.Benzeno S, Lu F, Guo M, et al. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006;25:6291–303. doi: 10.1038/sj.onc.1209644. [DOI] [PubMed] [Google Scholar]
- 59.Betticher DC, Heighway J, Thatcher N, Hasleton PS. Abnormal expression of CCND1 and RB1 in resection margin epithelia of lung cancer patients. Br J Cancer. 1997;75:1761–8. doi: 10.1038/bjc.1997.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dworakowska D, Jassem E, Jassem J, et al. Prognostic value of cyclin D1 overexpression in correlation with pRb and p53 status in non-small cell lung cancer (NSCLC). J Cancer Res Clin Oncol. 2005;131:479–85. doi: 10.1007/s00432-004-0661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umekita Y, Ohi Y, Sagara Y, Yoshida H. Overexpression of cyclinD1 predicts for poor prognosis in estrogen receptor-negative breast cancer patients. Int J Cancer. 2002;98:415–8. doi: 10.1002/ijc.10151. [DOI] [PubMed] [Google Scholar]
- 62.Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–8. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Habuchi T, Mitsumori K, et al. Increased risk of prostate cancer associated with AA genotype of cyclin D1 gene A870G polymorphism. Int J Cancer. 2003;103:116–20. doi: 10.1002/ijc.10793. [DOI] [PubMed] [Google Scholar]
- 64.Koike H, Suzuki K, Satoh T, et al. Cyclin D1 gene polymorphism and familial prostate cancer: the AA genotype of A870G polymorphism is associated with prostate cancer risk in men aged 70 years or older and metastatic stage. Anticancer Res. 2003;23:4947–51. [PubMed] [Google Scholar]
- 65.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narod SA, Seth A, Nam R. Fusion in the ETS gene family and prostate cancer. Br J Cancer. 2008;99:847–51. doi: 10.1038/sj.bjc.6604558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–9. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 68.Shen H, Powers N, Saini N, et al. The SWI/SNF ATPase Brm is a gatekeeper of proliferative control in prostate cancer. Cancer Res. 2008;68:10154–62. doi: 10.1158/0008-5472.CAN-08-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo Z, Yang X, Sun F, et al. A Novel Androgen Receptor Splice Variant Is Up-regulated during Prostate Cancer Progression and Promotes Androgen Depletion-Resistant Growth. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.