Abstract
Intracellular pathogens such as Mycobacterium tuberculosis have adapted to a life inside host cells, in which they utilize host nutrients to replicate and spread. Ineffective methods for the evaluation of growth of intracellular pathogens in their true environment pose an obstacle for basic research and drug screening. Here we present the validation of a luminometry-based method for the analysis of intramacrophage growth of M. tuberculosis. The method, which is performed in a medium-throughput format, can easily be adapted for studies of other intracellular pathogens and cell types. The use of host cells in drug-screening assays dedicated to find antimicrobials effective against intracellular pathogens permits the discovery of not only novel antibiotics but also compounds with immunomodulatory and virulence-impairing activities, which may be future alternatives or complements to antibiotics.
One of the major health issues today, in a global perspective, is the high prevalence of tuberculosis. The causative agent, Mycobacterium tuberculosis, infects one-third of the global population, mainly in developing countries, and causes 1 to 2 million deaths annually. The spread of drug-resistant strains and the increasing synergy with human immunodeficiency virus further emphasize the urgent need for efficient tools to manage this global emergency (13).
M. tuberculosis is an intracellular pathogen which primarily targets alveolar macrophages of the host. Several recent reports have shown that there is a transcriptional shift of M. tuberculosis genes upon uptake by host macrophages, an event that may alter the susceptibility to drugs (10, 15, 19). Therefore, potential drugs identified in screens using broth cultures may prove to be ineffective for bacteria inside cells (7). Importantly, novel drug-screening methods, where the host cells of intracellular pathogens are included, will allow for the discovery of substances that act differently from standard antibiotics, e.g., virulence blockers that interfere with the capacity of the bacterium to cause infection and immunomodulators that enhance the capacity of the host cell to kill the invader.
The determination of the number of viable bacteria, which is very central to studies of the interaction of intracellular pathogens with the host cell, is generally performed using the labor-intensive method known as CFU plating. The method is based on the culturing of defined volumes of serial dilutions of lysates from infected cells on agar plates for subsequent analysis of the number of CFU. The slow growth of M. tuberculosis (2 to 3 weeks until visible colonies appear) and the laborious handling and evaluation of the plates pose a major bottleneck for effective analysis of intracellular growth of the bacterium. We have previously reported on the use of a virulent strain of M. tuberculosis, H37Rv, harboring the pSMT1 plasmid carrying the genes for Vibrio harveyi luciferase, for analysis of the growth of M. tuberculosis and Mycobacterium avium subsp. paratuberculosis in mouse spleen, liver, and lungs (9, 16, 20). The flash luminescence emitted upon the addition of the substrate for V. harveyi luciferase, n-decanal, is dependent on the presence of a cofactor, reduced flavin mononucleotide (FMNH2), which is present only in living bacteria. In addition, the substrate for bacterial luciferase is very inexpensive compared to the firefly luciferase substrate luciferin, which has been used by others (6). In this work, we present the validation of the use of luciferase-expressing M. tuberculosis for the analysis of intramacrophage growth of the bacterium in a medium-throughput format. We also show how the viability of the host cells can be easily assessed in a large number of samples by a simple procedure. Effective methods for the evaluation of the success of intracellular pathogens in their true environment will be important for future research on host-pathogen interactions and will enable the screening of compound libraries for the discovery of drugs that impair intracellular growth of pathogens in a cost- and labor-effective manner.
MATERIALS AND METHODS
Cells and bacteria.
We used primary human monocyte-derived macrophages (hMDMs) prepared as previously described (21) and seeded them in 96-well plates (Sarstedt) in medium free of antibiotics (105 cells/well) 1 day before infection. To achieve nonopsonic conditions, the medium was replaced with serum-free medium immediately before infection. The previously developed virulent M. tuberculosis strain H37Rv (American Type Culture Collection) harboring a pSMT1 plasmid (17) carrying the gene for Vibrio harveyi luciferase, the avirulent counterpart H37Ra, and the vaccine strain Mycobacterium bovis bacillus Calmette-Guérin (BCG) (ATCC) were grown in Middlebrook 7H9 broth supplemented with Tween 80 and oleic acid-albumin-dextrose-catalase (OADC) (Becton Dickinson) for 2 to 3 weeks at 37°C with 100 μg/ml hygromycin for selection before being reinoculated in fresh broth and incubated for 7 days to reach early log phase. Cultures of H37Rv-lux do not differ from cultures of the original H37Rv strain with regard to colony morphology, generation time, and the pattern of intracellular growth, assessed by CFU plating as for the parental strain. To test the susceptibility of H37Rv-lux to standard antimycobacterial drugs, we used Bactec MGIT 960 as previously described (2). In brief, dilution series of rifampin, streptomycin, and isoniazid were prepared in MGIT tubes, as was a control with no antibiotics present. H37Rv-lux from culture was then added to every antibiotic-containing tube and a 1:100 dilution of H37Rv-lux culture to the control tube. Tubes were incubated in a Bactec MGIT 960, and the growth of H37Rv-lux with antibiotics was compared to that of the 1:100 dilution control.
Experimental infection.
The bacterial suspension was centrifuged twice at 3,000 × g for 10 min in phosphate-buffered saline (PBS) with 0.05% Tween 80, the pellet was resuspended in plain Dulbecco's modified Eagle medium (DMEM), and a single-bacillus suspension was obtained by two sets of 10 passages through a sterile syringe equipped with a 27-gauge needle. The concentration was determined by using optical density at 600 nm (OD600) as a function of CFU/ml obtained from a standard curve developed by plating three 100-μl portions of bacterial suspension on three individual agar plates from four different OD values. The bacteria were added to hMDMs in 96-well plates at a multiplicity of infection (MOI) of 10. The preparations were incubated at 37°C for 1 h, and the medium was changed to serum-containing but antibiotic-free DMEM (pulse-chase approach) before incubation for the indicated times. For evaluating the effect of antimycobacterial drugs, streptomycin (100 μg/ml), isoniazid (10 μg/ml), or rifampin (10 μg/ml) was added after the 1-h pulse-chase and was present for the remainder of the experiment. For the amikacin experiment, amikacin was added after the pulse-chase at a concentration of 200 μg/ml for 2 h before the medium was changed, and the amikacin concentration was lowered to 20 μg/ml for the remainder of the experiment.
Adaptation of CFU plating assay to the 96-well format.
hMDMs were subjected to hypotonic lysis in sterile water without additives in order to avoid the possible interference of chemical substances with the subsequent growth of bacteria. Complete lysis as verified by microscopy was achieved after 10 min of incubation, and the cell-associated fraction was subjected to serial dilutions and plating of defined volumes (5 μl) of the different dilutions on an individual square of a 100-mm Middlebrook 7H10 agar plate on which an six-by-six grid was printed. The grid made it possible to plate 36 individual 5-μl samples on the same plate, allowing the handling of large numbers of samples. The plates were sealed with Parafilm and incubated for 2 weeks before evaluation of the number of colonies. All samples were analyzed in triplicate.
Analysis of bacterial growth by luminometry.
To allow the measurement of flash luminescence in the biosafety level 3 (BSL3) facility, a Modulus Microplate Multimode Reader (Turner BioSystems, Sunnyvale, CA) was used. The instrument was equipped with luminescence, fluorescence, and absorbance functions and an injector module allowing the measurement of flash luminescence in 96-well plates. During measurements, the instrument was placed inside a biosafety cabinet in the BSL3 facility to avoid spread of aerosols arising during injection. A 25-μl portion of the medium supernatant was transferred to white 96-well plates (Sarstedt) containing 200 μl of pure water before cells in each well were subjected to hypotonic lysis by addition of 70 μl of pure water and incubation at room temperature for 10 min. The bottom of every well was scraped before 25 μl of the cell-associated fraction of bacteria was transferred in the same manner to white 96-well plates. To both the supernatant and cell-associated fractions, substrate (1% decanal [Sigma]) was added by the instrument injector before luminescence was measured for 1 s (3 s postinjection). The procedure was performed in triplicates for every time point to compensate for experimental variation. When comparing the different strains of mycobacteria and the effects of antibiotics, the data for the supernatant and cell-associated fractions were standardized to compensate for dilution factors before being added together. The fold increase was calculated by dividing the luminescence value on each day by the uptake of bacteria (day 0 value = 1).
Measurement of cell viability.
In order to assess the proportions of viable cells at the different times of infection, the infection procedure was carried out with hMDMs seeded on black 96-well plates (Sarstedt). The cells were washed three times with PBS and incubated with 4 μM calcein (Invitrogen) diluted in PBS for 30 min. Fluorescence was measured in the Modulus Microplate Reader using a 490-nm filter. Methanol-treated cells were used as negative controls.
Statistical analysis.
To ensure that our data were normally distributed, the Kolmogorov-Smirnov test was used. A one-way repeated-measures analysis of variance (ANOVA) was performed to evaluate the growth of H37Rv-lux as reported by the two different methods. A P value of <0.05 was considered significant.
RESULTS AND DISCUSSION
Luminometry-based versus viable-count-based assessment of intracellular mycobacterial growth.
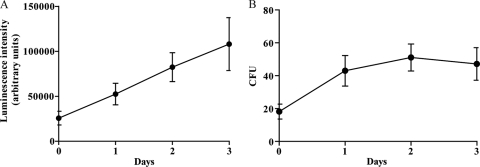
hMDMs from different donors were infected with H37Rv-lux in several independent experiments (n = 6) at an MOI of 10 using a pulse-chase approach. The phagocytosis (uptake) resulting from the infection was 0.5 bacterium/cell as determined by luminometry (not shown). After 0 to 3 days postinfection, the cells were lysed and the cell-associated fraction of bacteria was subjected to luminometry by automated addition of the luciferase substrate (Fig. 1A) (the relative luminescence intensity is given in arbitrary light units [ALU]) or serially diluted and plated on solid medium for determination of CFU numbers (Fig. 1B). When determining the difference in relative bacterial growth, there was a 4.2-fold increase in bacterial growth between days 0 and 3 as determined by the luminometry assay and a 2.6-fold increase with CFU plating. This growth was statistically significant for both luminometry (P = 0.0156) and CFU plating (P = 0.0103). The ALU data correlate well with microscopy data, where the number of intracellular green fluorescent protein (GFP)-expressing H37Rv cells was evaluated at every time point (not shown), indicating that the signal from the cell-associated fraction is mainly from intracellular bacteria.
FIG. 1.
Growth of H37Rv-lux in cells from day 0 to 3. Cells from six different donors were infected at an MOI of 10 and incubated for up to 3 days. At days 0 to 3 the cells were lysed, and the number of intracellular H37Rv-lux cells was determined by both luminometry (A) and CFU plating (B). Both the luminometry and CFU plating show a significant increase in bacterial numbers (P < 0.05). Error bars show standard errors of the means (SEM).
From the mean value for every time point, the coefficient of variation between the experiments was calculated by dividing the standard deviation by the mean (interassay variability). Furthermore, the coefficient of variation was calculated within the triplicates of every experiment and time point (intra-assay variability). The variabilities of the luminometry-based and CFU plating-based methods are summarized in Table 1, and the coefficient of variation is presented as a percentage. The intra-assay variability obtained with the luminometry was lower (1.06 to 13.99% at day 2) than the variability obtained with CFU plating (18.8 to 68.27% at day 2). The high interassay variability obtained using both luminometry and CFU plating (>30%) may be due to different capacities of the cells from individual donors to control infection as well as to differences in bacterial fitness between the different bacterial inoculates.
TABLE 1.
Inter- and intra-assay variability of the luminometry and CFU plating methods at days 0 to 3
| Day | Variability (%) |
|||
|---|---|---|---|---|
| Interassay |
Intra-assay |
|||
| Luminometry | CFU | Luminometry | CFU | |
| 0 | 72.76 | 60.84 | 4.91-17.52 | 7.87-54.30 |
| 1 | 55.89 | 52.87 | 7.31-35.44 | 8.32-38.68 |
| 2 | 47.76 | 39.50 | 1.06-13.99 | 18.80-68.27 |
| 3 | 66.71 | 51.66 | 7.69-38.20 | 36.96-83.22 |
Sensitivity and range of the luminometry-based assay.
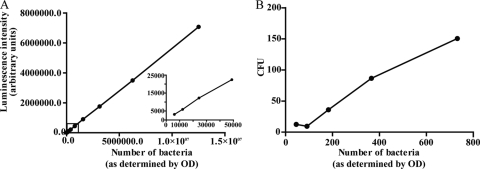
To determine the sensitivity and the range of detection of the luminometry-based assay, a bacterial culture was prepared as described above and the concentration was determined by optical density. The suspension was diluted 1:1 in 16 steps ranging from >1 × 107 to <1 × 104, and the luminescence of the samples was determined (Fig. 2A). From Fig. 2A, 1 ALU was determined to correspond to approximately two bacteria (y = 0.5647x). The linear relationship between the number of bacteria and ALU was stable down to 5,000 bacteria. Further dilutions rendered detectable but nonlinear signals (not shown). CFU plating was more sensitive, allowing the observer to detect a very small number of bacteria, but had a narrow range (at above 200 CFU, separate colonies were indistinguishable) (Fig. 2B), requiring dilution of the initial sample down to this range. Furthermore, single colonies were difficult to discriminate due to aggregates (not shown).
FIG. 2.
Correlation between bacterial number and luminescence intensity/CFU. (A) The graph shows the relationship between bacterial number (as determined by OD) and the luminescence signal measured by luminometry. A dilution series (1:1) shows the linearity from <1 × 107 bacteria down to >1 × 104 bacteria (inset graph). One arbitrary unit corresponds to approximately two bacteria. (B) The different bacterial dilutions from panel A were also plated on Middlebrook 7H10 agar and incubated for 2 weeks before the number of CFU was counted. The graph shows that the bacterial number (as determined by OD) and CFU have a linear relationship down to ∼100 bacteria.
Cell-associated versus extracellular growth of H37Rv-lux.
To determine the ratio of bacterial growth inside the cells to that in the medium, we examined the possibility of measuring bacteria in the medium using the luminometric approach. A defined volume of supernatant from the samples to be tested was transferred to a luminescence plate before the rest of the medium was discarded, and the cells were subjected to lysis. Supernatant and cell-associated fractions were collected at days 0 to 3 and analyzed with respect to luminescence. The ALU values obtained were recalculated to correspond to the total amount of luminescence in the individual wells, and Fig. 3 shows the ratios of intracellular to free bacteria. Amikacin used at low doses has been shown to impair extracellular, but not intracellular, growth of M. tuberculosis (18). However, as shown in Fig. 3, we observed an effect of amikacin on both the cell-associated fraction and the bacteria in the medium supernatant. Amikacin has been reported to accumulate in lysosomes (12), and aminoglycoside antibiotics may have effects on enzymes and membrane fusion in cells (3, 5). Since our setup involved long-term experiments with M. tuberculosis-infected macrophages, we decided not to use amikacin but instead consistently measured both the cell-associated and supernatant fractions.
FIG. 3.
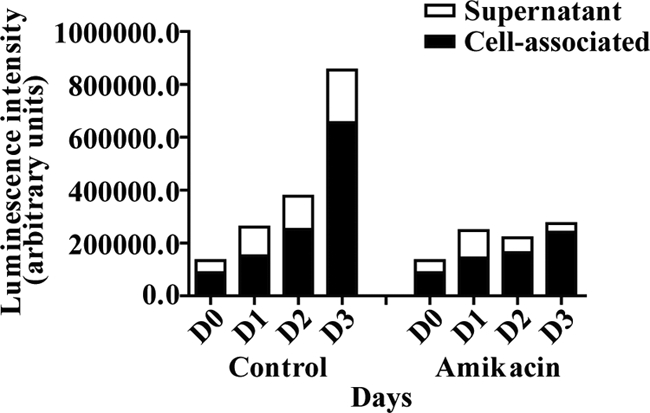
Measurement of cell-associated and extracellular bacteria. The diagram shows results from a representative measurement of H37Rv-lux over 3 days in the presence or absence of amikacin. Defined volumes of medium supernatant and cell-associated fractions were transferred to a luminescence plate and analyzed using luminometry. The measured values were then recalculated to correspond to the total volume of the sample.
Detecting changes in mycobacterial growth.
To establish that the method was able to detect changes in the growth of H37Rv-lux, hMDMs were infected by the pulse-chase approach, followed by addition of antimycobacterial drugs (100 μg/ml streptomycin, 10 μg/ml isoniazid, or 10 μg/ml rifampin) before cell-associated and supernatant fractions were analyzed by luminometry at days 0, 1, and 3. Because the initial bacterial load varied between donors due to differences in phagocytosis, the values from days 0, 1, and 3 were normalized to the initial phagocytosis (day 0 values). Figure 4 shows that the presence of antibiotics impairs the growth of H37Rv-lux, and for drug-screening purposes it is possible, from similar experiments including antibiotics, to determine a cutoff value, where the growth can be considered impaired. The pSMT1 plasmid harboring the luxAB gene also carries a hygromycin resistance gene for growth selection. To determine that H37Rv-lux is suitable for drug-screening purposes and that hygromycin resistance did not induce overlapping resistance, we analyzed the MIC values for the three different antibiotics used in Fig. 4. The MIC values for H37Rv-lux were determined using Bactec MGIT 960 and serial dilution and were 0.5 μg/ml for streptomycin, 0.032 μg/ml for isoniazid, and 0.064 μg/ml for rifampin. These levels were very similar to those previously reported for H37Rv, which is highly susceptible to the first-line drugs against tuberculosis (8). To further test the ability of the method to detect different kinetics of mycobacterial growth, the avirulent H37Ra strain and the BCG strain carrying the pSMT1 plasmid (H37Ra-lux and BCG-lux) were prepared as described above and allowed to infect macrophages. Growth was monitored by luminometry of both cell-associated and supernatant fractions at days 0 to 3. With luminometry, both H37Ra and BCG showed impaired growth compared to H37Rv (Fig. 5), indicating that the assay is capable of detecting differences in growth kinetics.
FIG. 4.
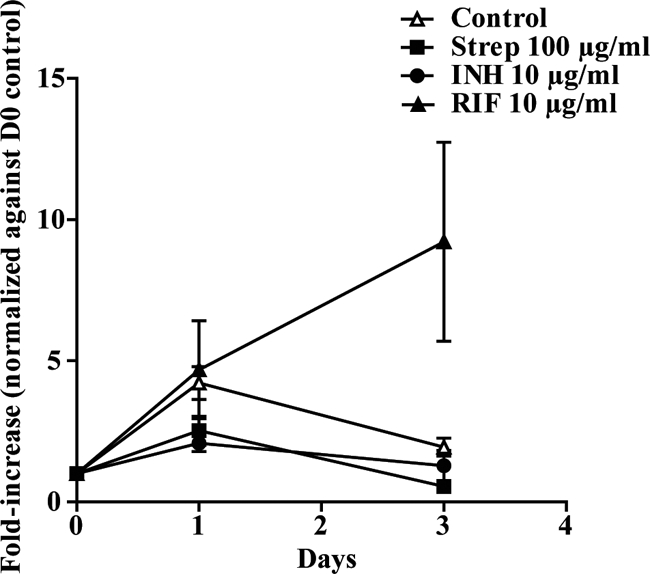
Antibiotics impair growth of M. tuberculosis in the luminometry-based assay. hMDMs were incubated with streptomycin (Strep), isoniazid (INH), or rifampin (RIF) at the indicated concentrations during the infection or were left untreated. The infection was followed by luminometry at days 0, 1, and 3 for both the cell-associated and supernatant fractions. The figure shows the fold increase of the number of bacteria compared to that at day 0, i.e., uptake of bacteria (n = 4). Error bars show SEM.
FIG. 5.
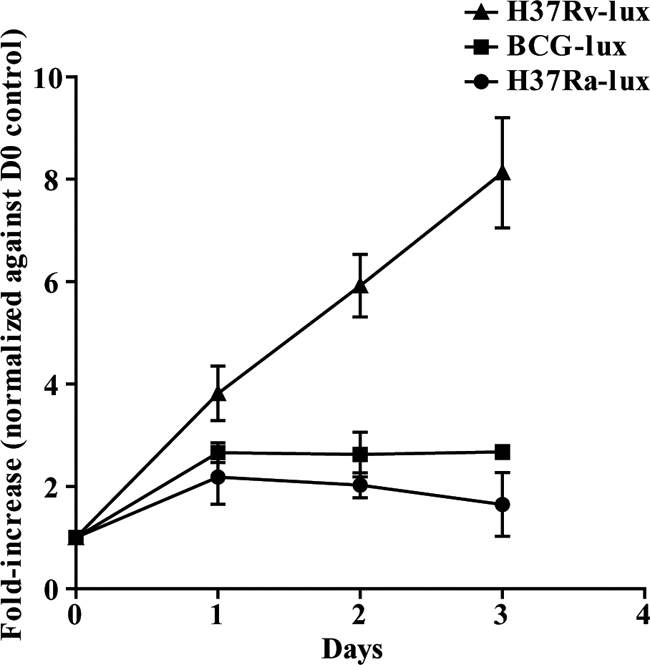
H37Ra-lux and BCG-lux display impaired growth kinetics compared to H37Rv-lux. hMDMs were infected at an MOI of 10 with either BCG-lux or H37Ra-lux, and the number of bacteria in each well was analyzed by luminometry over 3 days of incubation. The growth of both H37Ra-lux and BCG-lux was impaired compared to that of H37Rv-lux. The figure shows the fold increase of the number of bacteria compared to that at day 0, i.e., uptake of bacteria (n = 3). Error bars show SEM.
Analysis of cell viability.
The replication of M. tuberculosis within the human macrophage eventually causes cell death (14). This makes the testing of cell viability in the individual wells a very important issue for any method used for measuring intracellular growth of M. tuberculosis. In addition, the measurement of cell viability is central for drug-screening purposes, since a cytotoxic substance is not a good drug candidate. To evaluate the number of living hMDMs in our experimental setup, we used calcein as a fluorescent probe of cell viability. The method is based on the conversion of nonfluorescent, cell-permeative calcein acetoxymethyl to highly fluorescent calcein by living cells. Cells from five different donors were either infected, treated with methanol for 10 min, or left untreated in black 96-well plates. As shown in Fig. 6, both infected and uninfected cells lost viability during the course of the experiment; however, the infected hMDMs died more rapidly (50% of infected cells were dead at day 1, while uninfected cells remained unaffected). At day 3, fewer than 20% of the infected cells were viable, while 41% of the uninfected cells still displayed viability. It is known that mock-infected monocytes/macrophages in culture display apoptotic signs to some degree after 24 to 48 h in culture (1, 4), and this is consistent with our data.
FIG. 6.
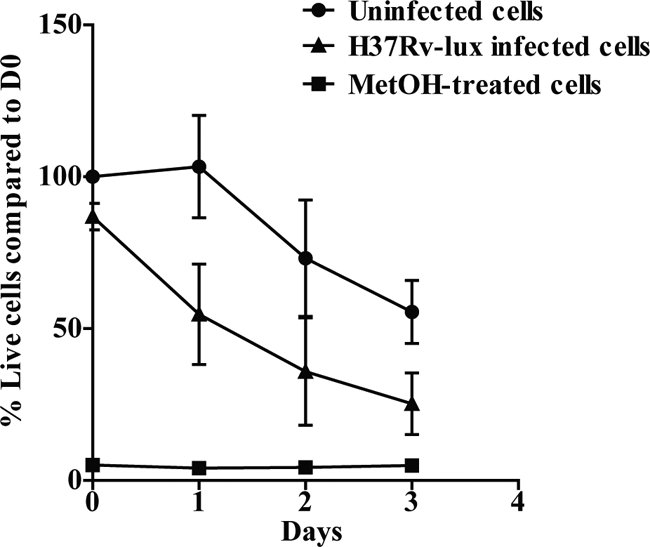
Comparison of cell death levels in noninfected and M. tuberculosis H37Rv-lux infected hMDMs. The viability of the cells was evaluated at every given time point (n = 5) by adding 4 μM calcein and measuring fluorescence after 30 min. The graph shows the percentage of viable cells in H37Rv-lux infected, untreated, and methanol (MetOH)-treated hMDMs compared to the day 0 value for uninfected hMDMs. Error bars show SEM.
The major bottleneck of the luciferase-based measurement of intramacrophage bacterial growth is the cost and handling of the primary human macrophages. The problem can be partially solved by using monocytic cell lines, which may be cultivated in a large-scale format and differentiated to macrophage-like cells. We tested whether the human macrophage-like cell line THP-1 could replace the hMDMs in the luciferase assay and found that the inter- and intra-assay variabilities using THP-1 cells were similar to the values obtained with hMDMs (not shown).
By including the host cell in drug screens aiming at identifying antimicrobial agents against intracellular pathogens, drug candidates acting via enhancement of cellular functions or by preventing the microorganism from establishing intracellular infection can be found. This may be crucial for the identification of antimicrobial drugs that act differently from classical antibiotics, since the phenotype of an infecting microorganism can differ remarkably from that of one growing in artificial medium. Also, it is possible that microorganisms are less prone to develop resistance against virulence blockers and immunomodulators (11).
Scaling up an assay is often associated with problems, and the variability obtained with the CFU plating method used in the 96-well format would probably be lower using larger sample volumes for dilution and plating of the bacteria. Nevertheless, the adaptation of a protocol to assays allowing the analysis of a larger number of samples is a prerequisite for efficient analysis and is of interest not only for basic investigation but also for industrial approaches such as drug screens. The luciferase-based method described here meets these requirements, and in addition, it solves some major problems with CFU plating of mycobacteria, where colonies do not appear until 2 to 3 week after plating and the aggregated morphology of the colonies makes it difficult to evaluate the number of colonies.
In conclusion, the luminometry-based method described here is a faster, less laborious, and more exact method than classical CFU plating. The described method can be used for drug-screening purposes and adapted to any intracellular pathogen, which may be transformed with plasmids carrying the Vibrio harveyi luciferase genes under the control of a suitable promoter. The opportunity to identify antimicrobial agents that are effectively bactericidal in their true environment makes the inclusion of host cells in drug-screening assays an attractive approach for the development of effective therapies against intracellular pathogens.
Acknowledgments
We are grateful to Kristina Orselius for excellent technical assistance, to Michaela Nordvall for MIC analyses, and to Douglas Young (Imperial College, London, United Kingdom) for providing the pSMT1 plasmid.
This project was funded by the Bill & Melinda Gates Foundation through the Grand Challenges Exploration Initiative (M.L.) and by grants from the Swedish Research Council (2003-5994 and 2007-2673 [M.L.] and 2005-6020 and 2008-3101 [O.S.]), the Ekhaga Foundation (M.L.), the Carl Trygger Foundation (M.L.), the Swedish Heart Lung Foundation (O.S.), SIDA (Swedish International Development Cooperation Agency) (O.S.), the County Council of Östergötland (O.S. and M.L.), and the Research Council of Southeast Sweden (FORSS) (T.S.). This work was also partially funded by grants G.0376.05 and G.0063.09 from FWO-Vlaanderen (K.H.).
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Baran, J., K. Guzik, W. Hryniewicz, M. Ernst, H. D. Flad, and J. Pryjma. 1996. Apoptosis of monocytes and prolonged survival of granulocytes as a result of phagocytosis of bacteria. Infect. Immun. 64:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann, J. S., and G. L. Woods. 1997. Mycobacterial growth indicator tube for susceptibility testing of Mycobacterium tuberculosis to isoniazid and rifampin. Diagn. Microbiol. Infect. Dis. 28:153-156. [DOI] [PubMed] [Google Scholar]
- 3.Carlier, M. B., G. Laurent, P. J. Claes, H. J. Vanderhaeghe, and P. M. Tulkens. 1983. Inhibition of lysosomal phospholipases by aminoglycoside antibiotics: in vitro comparative studies. Antimicrob. Agents Chemother. 23:440-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, A. R. 2002. In vitro detection of apoptosis in monocytes/macrophages infected with human coronavirus. Clin. Diagn. Lab. Immunol. 9:1392-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean, R. T., W. Jessup, and C. R. Roberts. 1984. Effects of exogenous amines on mammalian cells, with particular reference to membrane flow. Biochem. J. 217:27-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deb, D. K., K. K. Srivastava, R. Srivastava, and B. S. Srivastava. 2000. Bioluminescent Mycobacterium aurum expressing firefly luciferase for rapid and high throughput screening of antimycobacterial drugs in vitro and in infected macrophages. Biochem. Biophys. Res. Commun. 279:457-461. [DOI] [PubMed] [Google Scholar]
- 7.Dick, T. 2009. In vitro-in vivo disconnect in TB drug discovery, p. 56. In Tuberculosis: biology, pathology and therapy. Keystone Symposia Global Health Series, 25 to 30 January, Keystone, CO.
- 8.Donald, P. R., and A. H. Diacon. 2008. The early bactericidal activity of anti-tuberculosis drugs: a literature review. Tuberculosis (Edinb.) 88(Suppl. 1):S75-S83. [DOI] [PubMed] [Google Scholar]
- 9.D'Souza, S., V. Rosseels, O. Denis, A. Tanghe, N. De Smet, F. Jurion, K. Palfliet, N. Castiglioni, A. Vanonckelen, C. Wheeler, and K. Huygen. 2002. Improved tuberculosis DNA vaccines by formulation in cationic lipids. Infect. Immun. 70:3681-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontan, P., V. Aris, S. Ghanny, P. Soteropoulos, and I. Smith. 2008. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect. Immun. 76:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keyser, P., M. Elofsson, S. Rosell, and H. Wolf-Watz. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17-29. [DOI] [PubMed] [Google Scholar]
- 12.Maurin, M., and D. Raoult. 1994. Phagolysosomal alkalinization and intracellular killing of Staphylococcus aureus by amikacin. J. Infect. Dis. 169:330-336. [DOI] [PubMed] [Google Scholar]
- 13.Nathan, C. 2009. Taming tuberculosis: a challenge for science and society. Cell Host Microbe 5:220-224. [DOI] [PubMed] [Google Scholar]
- 14.Porcelli, S. A., and W. R. Jacobs, Jr. 2008. Tuberculosis: unsealing the apoptotic envelope. Nat. Immunol. 9:1101-1102. [DOI] [PubMed] [Google Scholar]
- 15.Rohde, K. H., R. B. Abramovitch, and D. G. Russell. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352-364. [DOI] [PubMed] [Google Scholar]
- 16.Rosseels, V., V. Roupie, D. Zinniel, R. G. Barletta, and K. Huygen. 2006. Development of luminescent Mycobacterium avium subsp. paratuberculosis for rapid screening of vaccine candidates in mice. Infect. Immun. 74:3684-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snewin, V. A., M. P. Gares, P. O. Gaora, Z. Hasan, I. N. Brown, and D. B. Young. 1999. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 67:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamm, L. M., J. H. Morisaki, L. Y. Gao, R. L. Jeng, K. L. McDonald, R. Roth, S. Takeshita, J. Heuser, M. D. Welch, and E. J. Brown. 2003. Mycobacterium marinum escapes from phagosomes and is propelled by actin-based motility. J. Exp. Med. 198:1361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tailleux, L., S. J. Waddell, M. Pelizzola, A. Mortellaro, M. Withers, A. Tanne, P. R. Castagnoli, B. Gicquel, N. G. Stoker, P. D. Butcher, M. Foti, and O. Neyrolles. 2008. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS One 3:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanghe, A., S. D'Souza, V. Rosseels, O. Denis, T. H. Ottenhoff, W. Dalemans, C. Wheeler, and K. Huygen. 2001. Improved immunogenicity and protective efficacy of a tuberculosis DNA vaccine encoding Ag85 by protein boosting. Infect. Immun. 69:3041-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welin, A., M. E. Winberg, H. Abdalla, E. Sarndahl, B. Rasmusson, O. Stendahl, and M. Lerm. 2008. Incorporation of Mycobacterium tuberculosis lipoarabinomannan into macrophage membrane rafts is a prerequisite for the phagosomal maturation block. Infect. Immun. 76:2882-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]