Abstract
H5N1 highly pathogenic avian influenza virus (HPAIV) has posed a great threat not only for the poultry industry but also for human health. However, an effective vaccine to provide a full spectrum of protection is lacking in the poultry field. In the current study, a novel prime-boost vaccination strategy against H5N1 HPAIV was developed: chickens were first orally immunized with a hemagglutinin (HA) DNA vaccine delivered by attenuated Salmonella enterica serovar Typhimurium, and boosting with a killed vaccine followed. Chickens in the combined vaccination group but not in single vaccination and control groups were completely protected against disease following H5N1 HPAIV intranasal challenge, with no clinical signs and virus shedding. Chickens in the prime-boost group also generated significantly higher serum hemagglutination inhibition (HI) titers and intestinal mucosal IgA titers against avian influenza virus (AIV) and higher host immune cellular responses than those from other groups before challenge. These results demonstrated that the prime-boost vaccination strategy provides an effective way to prevent and control H5N1 highly pathogenic avian influenza virus.
The highly pathogenic avian influenza virus (HPAIV) H5N1 strain can cause severe clinical signs in poultry, which may result in a mortality rate up to 100% (1, 2). The first outbreak of the H5N1 HPAIV was in Hong Kong in 1997. Subsequently, the virus spread to several other countries in Asia, Europe, and Africa (2). Not only did infections with H5N1 strains result in production losses and high mortality in poultry, but this virus also infected humans, causing severe public health problems. Since 2003, there have been 467 confirmed human cases of H5N1 virus infection, and its fatality rate reached 60.4% (as of 30 December 2009) (http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_12_30/en/index.html).
Vaccination is an important measure to prevent and control H5N1 HPAIV infections in the poultry industry. Killed vaccines have been used to control the spread of highly pathogenic H5 avian influenza viruses (AIV) in some countries (9). In China, a killed vaccine derived from A/turkey/England/N-28/73 (H5N2) was first used for buffer zone vaccination during H5N1 outbreaks in 2004. Killed vaccines against H5N1 AIV can eliminate clinical signs of illness, but they do not completely prevent infection and virus shedding (5, 29). DNA vaccination has been explored as an alternative approach to protect chickens against H5N1 HPAIV (15, 19, 28). However, to our knowledge, there is no known poultry vaccine that can provide a full spectrum of protection against H5N1 HPAIV.
The lack of an effective vaccine against H5N1 HPAIV is most likely due to H5N1 HPAIV transmission through mucosal sites (12); the conventional killed vaccines and DNA vaccines are administered usually via parenteral pathways, leading to a weak mucosal immunity. Recently, a variety of Gram-positive and Gram-negative bacteria (such as Salmonella, Shigella, and Listeria) have been used as carriers for efficient delivery of either DNA vaccine constructs or vaccine antigens (10). In particular, attenuated Salmonella enterica serovar Typhimurium has been used to deliver DNA vaccines encoding immunogens of pathogenic microorganisms, including AIV, infectious bursal disease virus (IBDV), porcine reproductive and respiratory syndrome virus (PRRSV), and transmissible gastroenteritis virus (TGEV) (14, 22, 31, 38). This strategy allows administration of DNA vaccines via mucosal surfaces as well as delivery of the plasmid DNA directly to professional antigen-presenting cells (APC), which can elicit humoral and cellular responses against the protective antigens at both mucosal and systemic levels (4, 27, 37).
In a previous study, we have reported that Salmonella carrying hemagglutinin (HA) DNA vaccine could provide partial protection from H5N1 HPAIV challenge in chickens (31). To seek a more effective method of vaccination against H5N1 HPAIV, in the current study we tested the ability of different vaccination schedules to suppress viral shedding and resist homologous avian influenza virus challenge. As a number of studies have reported the effects of a DNA prime-protein or killed vaccine boost immunization against protozoal, bacterial, and viral infections (8, 20, 23, 24, 36), we determined whether priming with a DNA vaccine delivered by attenuated Salmonella Typhimurium and boosting with a killed vaccine could enhance the immune response and the protective efficacy against the challenge by H5N1 HPAIV.
MATERIALS AND METHODS
Virus strain.
Avian influenza virus subtype A/Goose/Jiangsu/1/2000 H5N1 was obtained from the Animal Infectious Diseases Laboratory of the Ministry of Agriculture, Yangzhou, China. This isolate proved to be HPAIV with an intravenous pathogenicity index (IVPI) in 6-week-old chickens of 2.92. The titer of the viral stock is 8.7 log10 50% egg infective doses (EID50)/ml, determined by titration on 9-day-old specific-pathogen-free (SPF) embryonated eggs.
Killed vaccine.
For preparation of killed vaccine, virus was inoculated into the allantoic cavities of 10-day-old embryonated eggs (Shandong Institute of Poultry Science, Jinan, China) and was harvested after 72 h of incubation at 35°C. The virus was inactivated by the addition of 0.2% (vol/vol) formalin and kept at 37°C for 24 h. Inactivation was confirmed by the absence of detectable infectivity after two blind passages of formalin-treated allantoic fluid in embryonated eggs. One part of the inactivated allantoic fluid was emulsified in two parts (vol/vol) of paraffin oil (Hangzhou Oil Refining Company, Hangzhou, China), which is currently used commercially as an adjuvant for veterinary vaccine production. The virus concentration in the final vaccine was 640 HA units per dose.
Chickens.
One-day-old white Leghorn SPF chickens were purchased from the Shandong Institute of Poultry Science. Animals were housed, handled, and immunized following approval by the institutional animal experimental committee.
Plasmids and attenuated Salmonella strain.
The construction process for plasmid DNA encoding HA protein pmcDNA3.1-HA is detailed below. The HA gene was amplified from the genome of the AIV A/Goose/Jiangsu/1/2000 strain by reverse transcription-PCR (RT-PCR). The 5′ primer was 5′-ACAGCTAGCAAAATGGAGAAAATAGTG-3′, and the 3′ primer was 5′-CACAAGCTTTACAATCTGAACTCACA-3′. There is an NheI site in the upstream 5′ primer and a HindIII site in the downstream 3′ primer (in boldface). The 1,748-bp PCR product was cloned into pGEM-T by TA cloning, and DNA sequencing was performed for confirmation. Then, the HA gene was extracted by using NheI and HindIII and subcloned into pmcDNA3.1+ at the same sites to result in the plasmid pmcDNA3.1-HA (31). The pmcDNA3.1+ is a modified vector originated from pcDNA3.1+ by removing the promoter region of the ampicillin resistance bla gene (promoterless bla gene was amplified from pcDNA3.1+ by using upstream primer 5′-TCATGAGAAAAAGGAAGAGTATGAGTAT-3′ and downstream primer 5′-AACGCGTGGTCTGACAGTTACCAATGC-3′; underlined are the MluI and PagI sites, respectively; the pmcDNA3.1+ was constructed by replacing the whole bla gene with the 900-bp promoterless bla gene through the MluI and PagI sites). The pmcDNA3.1+ has a much-enhanced stability within Salmonella Typhimurium compared to pcDNA3.1+ due to the downregulation of the bla gene (39).
Both the empty and recombinant pmcDNA3.1+ plasmids were purified from transformed Escherichia coli DH5α using the Qiafilter plasmid purification kit from Qiagen (Qiagen, Valencia, CA) according to the manufacturer's instructions. The attenuated S. Typhimurium aroA mutant strain SL7207 (S. Typhimurium 2337-65 derivative hisG46 del407 [aroA::Tn10 (Tcs)]) (13) was kindly provided by B. A. D. Stocker (Stanford University, CA).
Transformation of the HA gene into S. Typhimurium.
Attenuated S. Typhimurium SL7207 cells were grown at 37°C in LB broth to an optimal density at 600 nm (OD600) of 0.6 to 0.8 and resuspended in ice-cold ultrapure H2O. The plasmid pmcDNA3.1-HA or control vector pmcDNA3.1+ was transformed into S. Typhimurium cells by electroporation (2.5 KV, 25 μF, and 200 to 400 Ω). Positive transformants were selected on LB agar containing 50 μg/ml ampicillin and verified by PCR and restriction enzyme digestion. The recombinant Salmonella strains containing plasmid pmcDNA3.1-HA or pmcDNA3.1+ were designated SL(pHA5) and SL(p), respectively.
Transfer of plasmids from S. Typhimurium to chicken macrophage cells in vitro.
The chicken macrophage (HD11) cell line was inoculated in 6-well plates at 3 × 107 cells per well at 41°C. The next day, the cells were washed twice with antibiotic-free medium and infected with SL(pHA5) and SL(p) at a multiplicity of infection (MOI) of 50. The cultures were further incubated at 41°C for 30 min. After washing twice, the remaining extracellular bacteria were killed by addition of gentamicin in RPMI medium (50 mg/ml) supplemented with 10% fetal calf serum (FCS). After incubation for 4 h at 41°C, the intracellular bacterial multiplication was inhibited by addition of tetracycline (10 mg/ml). The cultures were incubated at 41°C. Cells were harvested at 48 and 72 h and lysed by freezing and thawing three times. After centrifugation at 16,000 rpm for 10 min, the supernatants were pooled and used for the measurement of HA antigen.
Measurement of HA antigen.
The HA antigen was measured using an H5 subtype AIV HA antigen enzyme-linked immunosorbent assay (ELISA) kit (Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Beijing, China) according to the manufacturer's instructions. A total of 100 μl of each cell lysate was added to an ELISA plate precoated with anti-HA antibodies. The plate was incubated at 37°C for 1 h. After the wells were washed with washing solution five times, 100 μl peroxidase-conjugated anti-HA antibody was added to the wells and incubated at 37°C for 30 min. After the wells were washed with washing solution five times, 100 μl diluted 3,3%,5,5%-tetramethylbenzidine (TMB) was added to each well and incubated at 37°C for 30 min. A total of 50 μl of 2 M H2SO4 was added, and the absorbance of each well was measured at 450 nm, using TMB buffer as a blank. Each sample was tested in duplicate, and the mean absorbance was calculated.
Preparation of S. Typhimurium cultures carrying recombinant eukaryotic plasmids for vaccination.
The recombinant S. Typhimurium strains SL(pHA5) and SL(p) were grown to an optical density of 0.6 to 0.8. The bacterial cells were collected by centrifugation at 5,000 × g for 10 min and resuspended in phosphate-buffered saline (PBS) to the expected cell concentrations, as determined by plating serial dilutions on LB agar plates.
Vaccination of chickens and challenge experiment.
Six groups (10 chickens per group) of chickens were used for the present study. The doses and times of immunization in each group are shown in Table 1. Chickens were primed orally with recombinant S. Typhimurium at 1 day or 2 weeks of age, and this was followed by boosting with recombinant S. Typhimurium or killed vaccine at 4 weeks of age. All chickens except those in the negative control (NC) group were intranasally challenged with 105 EID50 of HPAIV H5N1 A/Goose/Jiangsu/1/2000 in 0.1 ml at 6 weeks of age. Oropharyngeal and cloacal swabs were collected from chickens at 3, 5 and 7 days postchallenge (p.c.). Each swab was washed in 1 ml of cold PBS, and virus titration was conducted as described previously (30). The limit of detection in this study was 101.0 50% egg lethal dose (ELD50)/ml. Chickens were observed daily for disease signs for 2 weeks. Serum samples were collected from chickens at 28, 42, and 52 days after priming with recombinant S. Typhimurium for the detection of hemagglutination inhibition (HI) antibody. HI assays were performed according to international standards (26).
TABLE 1.
Experimental design
| Groupa | Day 1 | Wk 2 | Wk 4 | Wk 6 (challenge) |
|---|---|---|---|---|
| NC | PBS | PBS | PBS | No |
| CC | PBS | PBS | PBS | Yes |
| SL(p) | 109 CFU | 109 CFU | 109 CFU | Yes |
| SL(p)+K | 109 CFU | 109 CFU | K | Yes |
| SL(pHA5) | 109 CFU | 109 CFU | 109 CFU | Yes |
| SL(pHA5)+K | 109 CFU | 109 CFU | K | Yes |
NC, negative control; CC, challenge control; SL(p), SL7207(pmcDNA3.1); K, killed vaccine of H5N1 AIV; SL(pHA5), SL7207(pmcDNA3.1-HA).
Mucosal antibody titers to AIV measured by ELISA.
To evaluate the mucosal antibody response, 5 chickens per group were humanely euthanized at 28, 42, and 52 days after priming with recombinant S. Typhimurium. The abdominal cavity was opened aseptically, and the entire small intestine, including the duodenum, jejunum, and ileum, was collected. Pancreas, connective tissue, and fat were removed, and the intestine was cut longitudinally and then cut into 1-cm-long sections. The intestinal antibodies were extracted using a PBS solution containing Tween 20 (0.05%), soybean trypsin inhibitor (0.1 mg/ml) (Sigma-Aldrich, St. Louis, MO), EDTA (0.05 mg/ml), and phenylmethylsulfonylfluoride (PMSF) (0.35 mg/ml) (Sigma-Aldrich, St. Louis, MO). Intestinal lavage solutions were mixed with extraction solution and shaken for 2 h at 4°C. After centrifugation at 20,000 × g for 30 min at 4°C, the supernatant was collected. Bovine serum albumin was added to a final concentration of 0.1%, and samples were preserved at −20°C. The level of antibodies against AIV in intestinal secretions was measured using an ELISA as described by Desmidt et al. (7). Briefly, 96-well plates were coated with AIV (10 μg/ml) in 0.05 M carbonate-bicarbonate buffer (pH 9.6) and incubated overnight at 4°C. After tapping out the liquid contents from the wells, plates were blocked with 300 μl skim milk for 1 h at 37°C. Subsequently, the plates were washed and incubated with 100-μl volumes of dilutions of intestinal extract. Intestinal secretion samples were diluted 1:5. Goat anti-chicken IgA Fc HRP conjugate (Sigma-Aldrich, St. Louis, MO) was employed to detect chicken IgA that had bound to AIV antigens. The plates were developed using the chromogenic substrate OPD according to the manufacturer's directions. The reaction was stopped with 0.5 M H2SO4, and the optical density was determined at 490 nm. Results of mucosal antibody were expressed as endpoint titers, calculated as the reciprocal values of the last dilution with an optical density of 0.2.
Lymphocyte proliferation assay.
The T-cell proliferation assay was performed as described previously (3) from spleen samples taken at 28, 42, and 52 days following priming with recombinant S. Typhimurium. Briefly, assays were established at 106 splenocytes/well in U-bottomed 96-well microtiter plates and cocultured with RPMI 1640 5% fetal bovine serum (FBS) supplemented with either 10 μg/ml AIV antigen, 10 μg/ml concanavalin A ConA (Sigma-Aldrich, St. Louis, MO), or RPMI-FBS alone in a final volume of 200 μl/well. Microtiter plates were incubated at 40°C in an atmosphere of 5% CO2 in air for 24 h prior to addition of 0.5 μCi/well of [3H]thymidine (Amersham, Little Chalfont, United Kingdom) and incubation for a further 18 h. Cells were harvested with a Tomtec Mach IIIM cell harvester (Perkin-Elmer, Bridgeport, CT), and incorporation of [3H]thymidine was determined using a 1450 Microbeta Trilux scintillation counter (Perkin-Elmer, Bridgeport, CT). Lymphocyte proliferation is expressed as the stimulation index (SI), which is defined as the mean of the experimental data divided by the mean of the data from the unstimulated control (6).
Statistical analysis.
Data were analyzed using the Statistical Package for the Social Sciences (SPSS version 13.0; SPSS, Chicago, IL). Antibody titers, the SI value, and virus titers were analyzed for significance (P < 0.05) by using the one-way analysis of variance (ANOVA), and means of treatments were compared using Duncan's multiple range test. Frequency of virus shedding and survival were analyzed for significance (P < 0.05) using Fisher's exact test.
RESULTS
HA expression in HD11 cells infected with recombinant S. Typhimurium.
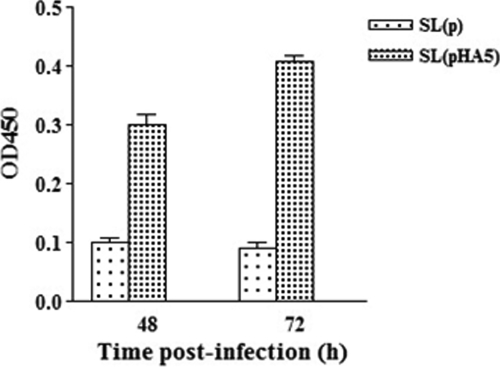
To justify that S. Typhimurium can transfer HA DNA vaccine into chicken cells, the chicken HD11 macrophage cells were infected with SL(pHA5) or control strain SL(p). The HA antigen can be detected from SL(pHA5)-infected cells 48 and 72 h postinfection (Fig. 1). Cells infected with the control strain gave negative results. These results demonstrated that it is feasible to use S. Typhimurium to deliver DNA vaccines.
FIG. 1.
HA expression in chicken macrophage HD11 cells infected with recombinant S. Typhimurium. HD11 cells were infected with recombinant S. Typhimurium SL(pHA5) or SL(p), and cell lysates were harvested at 48 and 72 h postinfection. HA antigen expression (expressed as OD450) was determined from the cell lysates.
Protection efficacy by prime-boost vaccination against challenge with H5N1 avian influenza virus.
To test that DNA vaccine delivered by S. Typhimurium alone or in combination with a killed vaccine can provide a better protection against HPAIV infection, four groups of chickens were immunized with SL(pHA5), SL(pHA5)+killed vaccine, SL(p), and SL(p)+killed vaccine. In addition, two nonvaccinated (PBS) groups were included: one was a negative control and another was a challenge control (CC) (Table 1). Following three immunizations as detailed in Table 1, chickens were challenged intranasally with 105 EID50 H5N1 HPAIV in a volume of 0.1 ml. To measure virus shedding, oropharyngeal and cloacal swabs were obtained from chickens on days 3, 5, and 7 after challenge. As shown in Table 2, five of 10 chickens in the SL(pHA5) group shed virus on day 3, and four of them died during the observation period. Five chickens in the SL(p)+K group shed virus, and one died on day 5 p.c. Chickens in the CC and SL(p) groups started to show disease signs 3 days p.c. and died within a week after challenge. In contrast, chickens in the SL(pHA5)+K group were completely protected following viral challenge, showing no signs of disease, detectable virus shedding, or death. For oropharyngeal swabs, the reduction in the number of the SL(pHA5)+K group chickens shedding was significantly less compared with those of the SL(pHA5) and SL(p)+K groups on days 3 and 5, respectively. For cloacal swabs, the reduction was not significant for the SL(pHA5)+K group versus the SL(pHA5) and SL(p)+K groups on days 3 and 5, respectively, but a significant difference was found between the SL(pHA5)+K group and the SL(p) or CC group (Table 2). Furthermore, the SL(pHA5)+K group significantly reduced the quantity of challenge virus shedding compared with that of another group (Table 2). These results indicate that chickens immunized with a DNA vaccine transported by Salmonella and a killed vaccine can be effectively protected from the development of H5N1 highly pathogenic avian influenza.
TABLE 2.
Protective efficacy of prime-boost strategies in chickens against H5N1 virus challengea
| Group | No. of shedding birds/total no. (log10 EID50/ml) p.c.b |
No. of surviving birds/total no. | |||||
|---|---|---|---|---|---|---|---|
| Day 3 |
Day 5 |
Day 7 |
|||||
| Oropharyngeal | Cloacal | Oropharyngeal | Cloacal | Oropharyngeal | Cloacal | ||
| NC | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 10/10 |
| CC | 10/10 A (4.0) c | 10/10 A (3.0) c | 5/5 A (3.6) c | 5/5 A (3.1) c | / | / | 0/10 A |
| SL(p) | 10/10 A (3.7) c | 10/10 A (3.4) c | 4/4 A (4.2) d | 4/4 A (3.5) c | / | / | 0/10 A |
| SL(p)+K | 5/10 B (2.4) d | 3/10 B (2.0) d | 4/9 A (1.6) e | 3/9 AB (1.7) d | 2/9 A (1.4) c | 1/9 A (1.4) c | 9/10 B |
| SL(pHA5) | 5/10 B (2.9) d | 2/10 B (1.5) cd | 3/6 A (1.8) e | 2/6 AB (1.4) d | 1/6 A (1.1) c | 1/6 A (1.0) c | 6/10 B |
| SL(pHA5)+K | 0/10 C (NI) e | 0/10 B (NI) e | 0/10 B (NI) f | 0/10 B (NI) e | 0/10 A (NI) c | 0/10 A (NI) c | 10/10 B |
All chickens except chickens in the NC group were challenged intranasally with 105 EID50 of A/Goose/Jiangsu/1/2000 virus in a 100-μl volume at 42 days of age. The swabs were suspended in 1 ml PBS and were titrated for virus shedding in eggs at an initial dilution of 1:10. The minimum virus titer detected by virus isolation procedures in this study was 101.0 ELD50/ml.
/, all chickens died; NI, none isolated. Different uppercase letters (A, B, C) denote significance between treatment groups; Fisher's exact test, P < 0.05. Different lowercase letters (c, d, e, f) denote significance (P < 0.05) between treatment groups using one-way ANOVA. For statistical purposes, all oropharyngeal and cloacal swabs from which virus was not isolated were given a numeric value of 100.9 EID50/ml, which represents the lowest detectable level of virus.
Virus-specific serum and intestinal antibody responses.
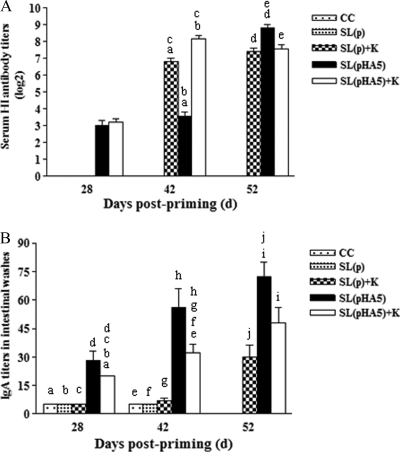
Serum and intestinal samples were collected at 28, 42, and 52 days and tested for serum antibody titers by HI tests and for intestinal IgA titers by ELISA. On day 28, 2 weeks after the second immunization, the difference in HI antibody titers between the SL(pHA5)+K and SL(pHA5) groups was not significant. On day 42, 2 weeks after the third immunization, chickens in the SL(pHA5)+K group had the highest serum antibody titers, and these were significantly higher than those of chickens in the SL(pHA5) group. On day 52, 10 days after challenge, the mean HI antibody titers of the SL(pHA5)+K group decreased, whereas those of the SL(pHA5) and SL(p)+K groups increased. In addition, chickens in the SL(pHA5) group had serum antibody titers significantly higher than those of the chickens in the SL(pHA5)+K and SL(p)+K groups (Fig. 2A).
FIG. 2.
Specific antibody titers in chickens before challenge at 28 and 42 days of age and 10 days postchallenge (day 52) with strain A/Goose/Jiangsu/1/2000 of AIV. (A) Data represent the sample mean (n = 5) ± standard deviation (SD) for titers of anti-AIV antibodies as determined in triplicate by a hemagglutination inhibition assay. Data points sharing a common letter were significantly different from one another at the following P values: a and b, P < 0.001; c and d, P = 0.001; e, P = 0.003. (B) Data represent the sample mean (n = 5) ± SD for titers of anti-AIV IgA in intestinal washes as determined in triplicate by an end-point dilution ELISA. All chickens of the CC and SL(p) groups were dead at 52 days. Data points sharing a common symbol were significantly different from one another at the following P values: a, b, and c, P < 0.001; e and f, P = 0.001; g, P = 0.002; h and i, P = 0.003; d, P = 0.018; j, P = 0.043.
Intestinal AIV-specific IgA antibodies were also measured (Fig. 2B). On days 28 and 42, chickens in the SL(pHA5) group had the higher IgA antibody titers, and these were significantly higher than those of the SL(pHA5)+K group, but significant difference could also be found between the SL(pHA5)+K group and the SL(p)+K, SL(p) or CC group. On day 52, all chickens in the challenged groups showed increases in AIV-specific IgA antibody titers, and again, chickens in the SL(pHA5) group had significantly higher virus-specific IgA responses than those of chickens in the SL(p)+K group (Fig. 2B).
These results suggest that a killed H5N1 HPAIV vaccine can induce a high level of systemic HI antibody but a minimal amount of mucosal IgA antibody, while a DNA vaccine given orally can induce a high level of mucosal IgA antibody but a low level of systemic HI antibody. The high mucosal IgA response induced by oral DNA vaccine alone reflects that this immunization pathway can efficiently activate mucosal immunity by directly targeting and activating mucosal antigen-presenting cells through the bacterial carrier. IgA at mucosal sites, whose function could be further enforced by systemic HI antibodies, is critical in defending against viral invasions. This is the likely explanation for the better protection achieved by the combination of oral DNA vaccine and killed vaccine.
Lymphocyte proliferation assay.
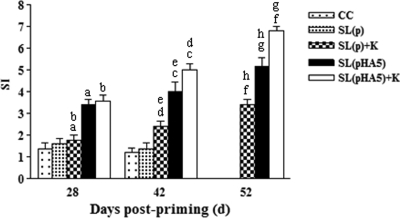
Representative results of a proliferation study using spleen cells from various groups are shown in Fig. 3. On day 28, chickens in the SL(pHA5)+K and SL(pHA5) groups had higher SI in spleen lymphocytes stimulated with AIV than chickens in the SL(p)+K groups. Furthermore, the SI of the SL(pHA5)+K and SL(pHA5) groups increased at 42 days, and there was significant difference between these two groups. On day 52, the proliferative responses in the SL(pHA5)+K and SL(pHA5) groups increased further, while chickens in the SL(pHA5)+K group showed an SI markedly higher than those of the other groups. These results indicate that chickens immunized with a combination of a mucosal DNA vaccine and a killed vaccine also induce the highest HPAIV-specific cellular proliferation, in addition to the humoral responses discussed above.
FIG. 3.
Lymphocyte proliferation in chickens before challenge at 28 and 42 days of age and 10 days postchallenge (day 52). Lymphocytes were isolated from the spleens of animals of all groups and stimulated in vitro with or without AIV antigens. ConA served as a positive control. Each bar represents the sample mean (n = 5) ± SD of the SIs determined in triplicate. All chickens of the CC and SL(p) groups were dead at 52 days. Data points sharing a common symbol were significantly different from one another at the following P values: a, b, d, and f, P < 0.001; e and h, P = 0.001; g, P = 0.002; c, P = 0.03.
DISCUSSION
H5N1 HPAIV is a highly contagious virus which has caused great economic loss in the poultry industry. What is more severe is that this virus has spread to human beings and resulted in a high mortality rate. Vaccination is an effective way to prevent and control AIV diseases, particularly from chickens and relevant birds. Given the limited efficacy of currently available H5N1 HPAIV vaccines, the development of novel vaccines and/or optimized vaccination protocols against H5N1 HPAIV is urgent. In the current study, we have developed a novel prime-boost vaccination regimen which completely protected chickens against H5N1 HPAI. This strategy utilized a mucosal DNA vaccine delivered by Salmonella and a boost with conventional killed vaccines. This regimen combines the strengths of DNA vaccine and traditional protein vaccine and fully activates the whole immune system, including cellular and humoral as well as mucosal and systemic immune responses.
Although DNA-based vaccination has been investigated extensively for influenza virus immunization in a variety of species, the ability of DNA vaccines to generate humoral immunity remains controversial. Following intramuscular immunization with HA-encoding DNA, strong or weak serum HI antibodies have been observed (16, 17, 34). Similarly, the authors (31) and others (32) have shown that Salmonella or Shigella HA DNA vaccination induced low levels of HI antibodies, an observation in agreement with this study. But when a killed vaccine boost was used, we found that a significant rise in HI antibody titers occurred among the Salmonella DNA vaccine-primed chickens, as this was also seen in previous studies (20, 21, 27). These results indicated that a strong B-cell memory response was generated after Salmonella HA DNA vaccination. The level of increase in HI titers after challenge can be a parameter to indicate the level of viral replication and accordingly, reflect the level of protection afforded by vaccination (35). Consistently, none of the chickens showed clinical signs and virus shedding in the SL(pHA5)+K group during the challenge period, but there was death and virus shedding in the SL(pHA5) and SL(p)+K groups (Table 2). These results clearly demonstrated that only the chickens receiving the combination of DNA vaccine and killed vaccine, but not either alone, were fully protected from virulent H5N1 HPAIV challenge.
As H5N1 HPAIV is transmitted mainly at mucosal sites, solid mucosal immunity is a critical parameter in evaluating a good vaccine candidate. Previous studies showed that DNA vaccine transported by attenuated bacteria can induce mucosal immune responses following mucosal administration and protect from virus challenge (32, 37). In the current study, we demonstrated that oral DNA vaccine delivered by Salmonella can induce a high level of mucosal IgA response. Mucosal IgA can form a critical first line of host defense against influenza infection by preventing viral attachment to epithelial cells and exerting protective functions in mucosal districts far from the site of immunization. It is interesting to find that even though mucosal IgA is important, it is not sufficient to provide solid protection against viral challenge. Only in the combination with killed vaccine was superior protection observed. This suggested that a systemic immune response, such as HI antibody, also contributes to antiviral immunity, possibly acting at a later stage of antiviral responses.
In addition to humoral responses, cellular immunity also plays an important role against pathogenic influenza virus infections (18). In the current study, the Salmonella DNA vaccine not only generated the mucosal IgA response but also stimulated a high level of the AIV-specific cellular response. In contrast, the killed vaccine was not efficient in inducing the cellular response, but when Salmonella DNA vaccine and killed vaccine were combined, a high cellular proliferation was observed. This supports the hypothesis that killed vaccines can enhance cellular responses (11).
In conclusion, we have demonstrated in this study that a novel prime-boost immunization strategy not only induced a strong immune responses but also effectively protected immunized chickens from a high dose of homologous virus challenge. The Salmonella-based DNA vaccine is easy to prepare and to administrate on a large scale, and killed vaccine is readily available. The superior vaccination efficacy of this combination has provided a valuable means of protecting against H5N1 HPAIV, particularly in the absence of a fully effective vaccine. DNA vaccines have been shown to provide better cross-protection against challenge with heterologous strains of influenza viruses (25, 33). It will be valuable to evaluate the protective efficacy of the prime-boost strategy against heterologous virus challenges and in particular to extend it to other animal models against the emerging H1N1 influenza virus infection.
Acknowledgments
This work was supported by grants from the 973 program (no. 2006CB504404), National Nature Science Foundation of China (no. 30871860), the 863 program (NO. 2006AA10A206), and the Government of Jiangsu Province (no. BK2007511 and BK2008011).
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Alexander, D. J. 2003. Report on avian influenza in the Eastern Hemisphere during 1997-2002. Avian Dis. 47:792-797. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 2007. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002-2006. Avian Dis. 51:161-166. [DOI] [PubMed] [Google Scholar]
- 3.Beal, R. K., C. Powers, P. Wigley, P. A. Barrow, and A. L. Smith. 2004. Temporal dynamics of the cellular, humoral and cytokine responses in chickens during primary and secondary infection with Salmonella enterica serovar Typhimurium. Avian Pathol. 33:25-33. [DOI] [PubMed] [Google Scholar]
- 4.Benitez, A. J., N. McNair, and J. R. Mead. 2009. Oral immunization with attenuated Salmonella typhimurium encoding Cryptosporidium parvum Cp23 and Cp40 antigens induces a specific immune response in mice. Clin. Vaccine Immunol. 16:1272-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bublot, M., F. X. Le Gros, D. Nieddu, N. Pritchard, T. R. Mickle, and D. E. Swayne. 2007. Efficacy of two H5N9-inactivated vaccines against challenge with a recent H5N1 highly pathogenic avian influenza isolate from a chicken in Thailand. Avian Dis. 51:332-337. [DOI] [PubMed] [Google Scholar]
- 6.Denizot, F., and R. Lang. 1986. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J. Immunol. Methods 89:271-277. [DOI] [PubMed] [Google Scholar]
- 7.Desmidt, M., R. Ducatelle, J. Mast, B. M. Goddeeris, B. Kaspers, and F. Haesebrouck. 1998. Role of the humoral immune system in Salmonella enteritidis phage type four infection in chickens. Vet. Immunol. Immunopathol. 63:355-367. [DOI] [PubMed] [Google Scholar]
- 8.Dunachie, S. J., M. Walther, J. E. Epstein, S. Keating, T. Berthoud, L. Andrews, R. F. Andersen, P. Bejon, N. Goonetilleke, I. Poulton, D. P. Webster, G. Butcher, K. Watkins, R. E. Sinden, G. L. Levine, T. L. Richie, J. Schneider, D. Kaslow, S. C. Gilbert, D. J. Carucci, and A. V. Hill. 2006. A DNA prime-modified vaccinia virus Ankara boost vaccine encoding thrombospondin-related adhesion protein but not circumsporozoite protein partially protects healthy malaria-naive adults against Plasmodium falciparum sporozoite challenge. Infect. Immun. 74:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, T. M., C. Y. Leung, M. K. Chow, L. A. Bissett, W. Wong, Y. Guan, and J. S. Malik Peiris. 2004. Vaccination of chickens against H5N1 avian influenza in the face of an outbreak interrupts virus transmission. Avian Pathol. 33:405-412. [DOI] [PubMed] [Google Scholar]
- 10.Gentschev, I., G. Dietrich, S. Spreng, A. Kolb-Maurer, V. Brinkmann, L. Grode, J. Hess, S. H. Kaufmann, and W. Goebel. 2001. Recombinant attenuated bacteria for the delivery of subunit vaccines. Vaccine 19:2621-2628. [DOI] [PubMed] [Google Scholar]
- 11.Gorse, G. J., M. J. Campbell, E. E. Otto, D. C. Powers, G. W. Chambers, and F. K. Newman. 1995. Increased anti-influenza A virus cytotoxic T cell activity following vaccination of the chronically ill elderly with live attenuated or inactivated influenza virus vaccine. J. Infect. Dis. 172:1-10. [DOI] [PubMed] [Google Scholar]
- 12.Hasegawa, H., T. Ichinohe, A. Ainai, S. Tamura, and T. Kurata. 2009. Development of mucosal adjuvants for intranasal vaccine for H5N1 influenza viruses. Ther. Clin. Risk Manag. 5:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, P., W. Jiang, Y. Li, S. Wu, and J. Xu. 2004. Humoral immune response induced by oral administration of S. typhimurium containing a DNA vaccine against porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 102:321-328. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, Y., K. Yu, H. Zhang, P. Zhang, C. Li, G. Tian, Y. Li, X. Wang, J. Ge, Z. Bu, and H. Chen. 2007. Enhanced protective efficacy of H5 subtype avian influenza DNA vaccine with codon optimized HA gene in a pCAGGS plasmid vector. Antiviral Res. 75:234-241. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, P. A., M. A. Conway, J. Daly, C. Nicolson, J. Robertson, and K. H. Mills. 2000. Plasmid DNA encoding influenza virus haemagglutinin induces Th1 cells and protection against respiratory infection despite its limited ability to generate antibody responses. J. Gen. Virol. 81:1737-1745. [DOI] [PubMed] [Google Scholar]
- 17.Kadowaki, S., Z. Chen, H. Asanuma, C. Aizawa, T. Kurata, and S. Tamura. 2000. Protection against influenza virus infection in mice immunized by administration of hemagglutinin-expressing DNAs with electroporation. Vaccine 18:2779-2788. [DOI] [PubMed] [Google Scholar]
- 18.Kreijtz, J. H., R. Bodewes, G. van Amerongen, T. Kuiken, R. A. Fouchier, A. D. Osterhaus, and G. F. Rimmelzwaan. 2007. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 25:612-620. [DOI] [PubMed] [Google Scholar]
- 19.Laddy, D. J., J. Yan, N. Corbitt, D. Kobasa, G. P. Kobinger, and D. B. Weiner. 2007. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine 25:2984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen, D. L., A. Karasin, and C. W. Olsen. 2001. Immunization of pigs against influenza virus infection by DNA vaccine priming followed by killed-virus vaccine boosting. Vaccine 19:2842-2853. [DOI] [PubMed] [Google Scholar]
- 21.Le Gall-Reculé, G., M. Cherbonnel, N. Pelotte, P. Blanchard, Y. Morin, and V. Jestin. 2007. Importance of a prime-boost DNA/protein vaccination to protect chickens against low-pathogenic H7 avian influenza infection. Avian Dis. 51:490-494. [DOI] [PubMed] [Google Scholar]
- 22.Li, L., W. Fang, J. Li, Y. Huang, and L. Yu. 2006. Oral DNA vaccination with the polyprotein gene of infectious bursal disease virus (IBDV) delivered by the attenuated Salmonella elicits protective immune responses in chickens. Vaccine 24:5919-5927. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzo, G., R. Martin-Folgar, F. Rodriguez, and A. Brun. 2008. Priming with DNA plasmids encoding the nucleocapsid protein and glycoprotein precursors from Rift Valley fever virus accelerates the immune responses induced by an attenuated vaccine in sheep. Vaccine 26:5255-5262. [DOI] [PubMed] [Google Scholar]
- 24.Lu, S. 2009. Heterologous prime-boost vaccination. Curr. Opin. Immunol. 21:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery, D. L., J. W. Shiver, K. R. Leander, H. C. Perry, A. Friedman, D. Martinez, J. B. Ulmer, J. J. Donnelly, and M. A. Liu. 1993. Heterologous and homologous protection against influenza A by DNA vaccination: optimization of DNA vectors. DNA Cell Biol. 12:777-783. [DOI] [PubMed] [Google Scholar]
- 26.OIE. 2005. Manual of diagnostic tests for avian influenza. OIE, Paris, France. http://www.oie.int/eng/normes/mmanual/A_00037.htm.
- 27.Pan, Z., X. Zhang, S. Geng, N. Cheng, L. Sun, B. Liu, J. Huang, and X. Jiao. 2009. Priming with a DNA vaccine delivered by attenuated Salmonella typhimurium and boosting with a killed vaccine confers protection of chickens against infection with the H9 subtype of avian influenza virus. Vaccine 27:1018-1023. [DOI] [PubMed] [Google Scholar]
- 28.Rao, S., W. P. Kong, C. J. Wei, Z. Y. Yang, M. Nason, D. Styles, L. J. DeTolla, A. Panda, E. M. Sorrell, H. Song, H. Wan, G. C. Ramirez-Nieto, D. Perez, and G. J. Nabel. 2008. Multivalent HA DNA vaccination protects against highly pathogenic H5N1 avian influenza infection in chickens and mice. PLoS One 3:e2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swayne, D. E., C. W. Lee, and E. Spackman. 2006. Inactivated North American and European H5N2 avian influenza virus vaccines protect chickens from Asian H5N1 high pathogenicity avian influenza virus. Avian Pathol. 35:141-146. [DOI] [PubMed] [Google Scholar]
- 30.Swayne, D. E., D. A. Senne, and C. W. Beard. 1998. Isolation and identification of avian pathogens, p. 150-155. In D. E. Swayne, J. R. Glisson, M. W. Jackwood, J. E. Pearson and W. M. Reed (ed.), Influenza, 4th ed. American Association of Avian Pathologists, Kennett Square, PA.
- 31.Tang, L. H., Z. M. Pan, N. N. Cheng, X. A. Jiao, and X. M. Zhang. 2007. Construction and immunogenicity of attenuated Salmonella typhimurium harbouring stable DNA vaccine against H5 subtype of avian influenza virus. Wei Sheng Wu Xue Bao (Beijing) 47:662-666. [PubMed] [Google Scholar]
- 32.Vecino, W. H., N. M. Quanquin, L. Martinez-Sobrido, A. Fernandez-Sesma, A. Garcia-Sastre, W. R. Jacobs, Jr., and G. J. Fennelly. 2004. Mucosal immunization with attenuated Shigella flexneri harboring an influenza hemagglutinin DNA vaccine protects mice against a lethal influenza challenge. Virology 325:192-199. [DOI] [PubMed] [Google Scholar]
- 33.Wang, S., C. Parker, J. Taaffe, A. Solorzano, A. Garcia-Sastre, and S. Lu. 2008. Heterologous HA DNA vaccine prime-inactivated influenza vaccine boost is more effective than using DNA or inactivated vaccine alone in eliciting antibody responses against H1 or H3 serotype influenza viruses. Vaccine 26:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong, J. P., M. A. Zabielski, F. L. Schmaltz, G. G. Brownlee, L. A. Bussey, K. Marshall, T. Borralho, and L. P. Nagata. 2001. DNA vaccination against respiratory influenza virus infection. Vaccine 19:2461-2467. [DOI] [PubMed] [Google Scholar]
- 35.Wood, J. M., Y. Kawaoka, L. A. Newberry, E. Bordwell, and R. G. Webster. 1985. Standardization of inactivated H5N2 influenza vaccine and efficacy against lethal A/Chicken/Pennsylvania/1370/83 infection. Avian Dis. 29:867-872. [PubMed] [Google Scholar]
- 36.Woodland, D. L. 2004. Jump-starting the immune system: prime-boosting comes of age. Trends Immunol. 25:98-104. [DOI] [PubMed] [Google Scholar]
- 37.Xie, C., J. S. He, M. Zhang, S. L. Xue, Q. Wu, X. D. Ding, W. Song, Y. Yuan, D. L. Li, X. X. Zheng, Y. Y. Lu, and Z. Shang. 2007. Oral respiratory syncytial virus (RSV) DNA vaccine expressing RSV F protein delivered by attenuated Salmonella typhimurium. Hum. Gene Ther. 18:746-752. [DOI] [PubMed] [Google Scholar]
- 38.Yang, H., S. Cao, X. Huang, J. Liu, Y. Tang, and X. Wen. 2009. Intragastric administration of attenuated Salmonella typhimurium harbouring transmissible gastroenteritis virus (TGEV) DNA vaccine induced specific antibody production. Vaccine 27:5035-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X. M., X. A. Jiao, L. H. Tang, Z. M. Pan, Y. L. Yin, and X. F. Liu. 2005. Enhanced stability of plasmid pcDNA3.1+ within Salmonella typhimurium by downregulation of the ampicillin resistance gene expression. Microbiology (Beijing) 32:51-55. [Google Scholar]