Abstract
We identified two overlapping neutralizing epitopes within residues 151 to 172 of the central unglycosylated region of the respiratory syncytial virus (RSV) attachment protein. In ∼40% of hospitalized and outpatient adults infected with RSV subtype A, these contiguous residues are the target of ≥4-fold increases in IgG response between acute- and convalescent-phase sera.
A hallmark of the respiratory syncytial virus (RSV) attachment (G) protein is the central unglycosylated region that typically comprises amino acids (aa) 151 to 190 (9). Residues 164 to 176 are invariantly conserved among G sequences from all RSV isolates, and there is a loop structure comprising two cystine disulfide bonds (Fig. 1). Genetic selection for RSV that can replicate in the presence of subtype-independent, partially neutralizing anti-G monoclonal antibodies (MAbs) such as L9 yields strains bearing amino acid changes localized mostly within the central unglycosylated region of G (8, 15).
FIG. 1.
Central unglycosylated region of the RSV G protein. Residues 151 to 190 of the RGH subtype A5 RSV strain and those from the prototypical subtype A (A2) and B (B1) strains are identified. The positions of relevant residues are shown by numbers above the amino acid alignment. Note that invariant residues 164 to 176 are shown in bold, and the predicted disulfide bonds are shown as solid lines connecting the relevant cysteine residues. The dotted line underlies residues 151 to 172 (the PCC; see the text) bearing the epitopes for L9 and K6 MAbs.
To better define the cognate epitopes for L9, as well as for the K6 MAb with similar neutralizing activities, we constructed a series of plasmids, each encoding a glutathione S-transferase (GST)-RSV G fusion protein bearing a portion of the central unglycosylated region of G (16). The fusion proteins were expressed in bacteria, purified to >95% homogeneity (data not shown), resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels under reducing and denaturing conditions, and tested for recognition by L9 and K6 MAbs in immunoblots.
(Portions of this work were presented previously at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, September 2009 [10].)
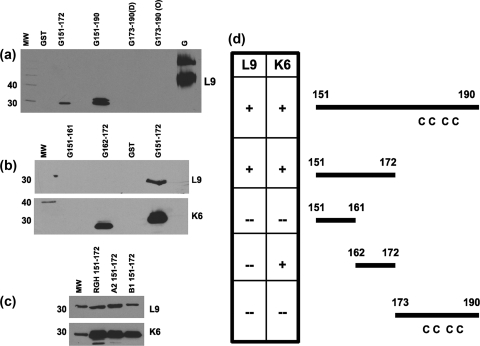
L9 and K6 did not recognize GST alone or the fusion protein consisting of GST and G residues 173 to 190 (GST-G173-190) but bound to purified, full-length G protein, GST-G151-190, and three forms of GST-G151-172 bearing corresponding residues from A2, B1, and RGH strains (Fig. 2). Neither MAb recognized GST-G151-161, but GST-G155-172, GST-G157-172, and GST-G162-172 were detected by K6 but not by L9 (Fig. 2 and data not shown). Consistent with these immunoblot results, all three forms of GST-G151-172 were recognized by both MAbs and GST-G162-172 was bound by K6 but not L9 in enzyme-linked immunosorbent assays (ELISAs) under nonreducing/nondenaturing conditions (data not shown). These results suggest that (i) RSV G residues 151 to 172 are required for recognition by L9; (ii) consistent with the subtype-independent neutralizing activities of both MAbs, K6 and L9 both recognized subtype A- and B-derived residues 151 to 172; and (iii) the K6 epitope involves aa 162 to 172. Based on the proximal location of residues 151 to 172 with respect to the G loop, we have termed these residues the proximal central core (PCC) region of the RSV G protein.
FIG. 2.
The cognate epitopes of L9 and K6 MAbs are localized within the PCC domain of the RSV G protein. (a to c) Representative immunoblots. Purified RSV G protein (subtype A), GST alone, or GST-RSV G fusion proteins (∼0.5 μg of protein/sample; G-derived residues are listed above the corresponding lanes) were resolved on denaturing SDS-12 or 6% PAGE gels, transferred onto nitrocellulose, probed with L9 or K6 MAbs at a 1:5,000 dilution followed by goat anti-mouse horseradish peroxidase-conjugated antibodies (diluted 1:20,000), and analyzed by using chemiluminescence. For each gel, molecular weight (MW) markers are resolved in the leftmost lane and the corresponding molecular masses (in kilodaltons) are indicated to the left. Note that in the analysis depicted in panel a, two independent preparations of GST-G173-190, one from bacterial strain DH5α (D) and another from strain Origami 2 (O), were used to test reactogenicity and that we occasionally observed a doublet in GST-G151-190 lanes. Note also that in the analysis depicted in panel c, fragments of aa 151 to 172 derived from the RGH, A2, and B1 RSV strains (as indicated by lane labels) were tested for recognition by L9 and K6 MAbs. (d) The L9 and K6 MAb epitopes within RSV G residues 151 to 172 are overlapping. On the right are various schematic diagrams, each representing a portion of the RSV G unglycosylated region that was expressed as a fusion protein in bacteria. Where relevant, the relative locations of the four cysteines (C) are shown. On the left is a table summarizing the immunoblot data obtained using both MAbs. + indicates reproducible MAb recognition of the RSV G-derived residues, while − indicates the absence of detectable MAb binding.
The RSV G PCC contains most of the conserved amino acid sequence HFEVFNFVPCSIC (residues 164 to 176). Our data suggest that the K6 epitope likely exists within these residues and that the L9 epitope requires nonconserved residues 151 to 163 for correct epitope conformation. Also within the PCC is the motif (Y/F)XFXXFXF (residues 163 to 170), in which F163, F165, F168, and F170 are present in G proteins from subtype A strains (e.g., A2 and RGH) while Y163 is present in the G protein from the B1 strain (5, 17) (Fig. 1). This amino acid motif is also found in RSV-related viruses with different host specificities (e.g., ovine and bovine RSVs), suggesting an evolutionarily conserved structural and/or immunological function (6, 7). Alanine substitution for F163 but not for F165 abolishes K6 binding to GST-G162-172 in immunoblots, suggesting that the F163 residue is likely to be involved in K6-epitope interactions (data not shown).
To determine the clinical and immunological relevance of the PCC-embedded epitopes in human RSV infections, we assayed serum reactogenicity to GST-G151-172 in ELISAs using paired acute- and convalescent-phase sera from RSV subtype A-infected hospitalized and outpatient adults (3). Among paired sera from 32 RSV-infected hospitalized adults, samples from 14 patients (44%) showed a ≥4-fold increase in the anti-RSV G PCC titers from acute- to convalescent-phase sera. Similarly, among serum samples from 19 outpatient adults, samples from 8 (42%) showed a ≥4-fold increase in the anti-G PCC antibody response. For both populations, the increase in the mean anti-G PCC titer (expressed as the log2 reciprocal serum dilution ± the standard deviation) after RSV infection was statistically significant (for hospitalized patients, the mean titer increased from 8.3 ± 1.8 to 10.1 ± 2.8; for outpatients, it increased from 8.1 ± 1.5 to 9.9 ± 1.6; for both populations, the P value was <0.005 by the Wilcoxon signed-rank test). These data suggest that (i) an acute rise in anti-G PCC titers is found in ∼40% of RSV subtype A-infected adults and (ii) such titer increases are observed with no obvious correlation to the severity of illness as defined by the initial clinical evaluation in hospital versus outpatient settings.
Our results have implications for the structure, function, and immunogenicity of the RSV G PCC region. Due to hydrophobic interactions involving F165, F168, F170, V171, P155, and P156, residues 149 to 177 of RSV G likely form a disk-like structure with two hydrophobic faces (4). Within the G central unglycosylated domain, residues 166 to 170 (EVFNF) may be involved in the multimerization of RSV G and/or interactions with a cellular RSV G protein receptor (4). Our results suggest that ≥1 neutralizing epitope is found within and flanking RSV G residues 166 to 170 and raise the possibility that L9 and other MAbs recognizing the G PCC-embedded epitopes may directly or indirectly affect RSV G structure (e.g., by destabilizing multimerization) and/or function (i.e., by blocking interactions with the host target cell). Previously, we reported the isolation of an L9-resistant virus that bore mutations outside the G PCC region; perhaps these mutations represent second-site/compensatory changes that counteract the action(s) of L9 MAb on RSV G structure/function (15).
Within the RSV G protein, short “protective” B-cell epitopes have been identified, of which two (aa 152 to 163 and 165 to 172) are located within the RSV G PCC region (13). It should be noted that L9 and K6 are both neutralizing and subtype independent but that none of the protective epitope-recognizing MAbs were neutralizing and that it is unclear whether the protective effect against viral challenge was subtype independent. These differences in the functional profiles of the various MAbs may be due to the immunogen (purified, native RSV G protein from RSV-infected mammalian cells versus a bacterially derived, refolded RSV G fragment) used to generate the respective MAbs.
The human serological characterization of RSV G epitopes, especially those within the G PCC domain, remains incomplete. A very limited number of adult human sera (n = 2) were used to study reactogenicity to RSV G-derived protective epitopes (13). Other RSV G-based human serological screening studies utilized bacterially synthesized, genetically hypervariable regions flanking the central unglycosylated region or G-derived peptides (overlapping or nonoverlapping) to screen adult or pediatric sera (1, 2, 11, 12, 14). In this study, we demonstrate a ≥4-fold increase in serum reactogenicity to the PCC domain of RSV G in a significant proportion of RSV subtype A-infected adults. These data suggest that the G PCC domain is immunologically significant in human RSV infections and may be a target of prophylactic and/or therapeutic agents against RSV.
Acknowledgments
This work was supported by Public Health Service grants from the National Institute of Allergy and Infectious Diseases (grants R21 AI076781 to Y.M. and R01 AI045969 to E.E.W.).
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Cane, P. A. 1997. Analysis of linear epitopes recognised by the primary human antibody response to a variable region of the attachment (G) protein of respiratory syncytial virus. J. Med. Virol. 51:297-304. [DOI] [PubMed] [Google Scholar]
- 2.Cane, P. A., H. M. Thomas, A. F. Simpson, J. E. Evans, C. A. Hart, and C. R. Pringle. 1996. Analysis of the human serological immune response to a variable region of the attachment (G) protein of respiratory syncytial virus during primary infection. J. Med. Virol. 48:253-261. [DOI] [PubMed] [Google Scholar]
- 3.Falsey, A. R., P. A. Hennessey, M. A. Formica, C. Cox, and E. E. Walsh. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 352:1749-1759. [DOI] [PubMed] [Google Scholar]
- 4.Gorman, J. J., J. L. McKimm-Breschkin, R. S. Norton, and K. J. Barnham. 2001. Antiviral activity and structural characteristics of the nonglycosylated central subdomain of human respiratory syncytial virus attachment (G) glycoprotein. J. Biol. Chem. 276:38988-38994. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. U. S. A. 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langedijk, J. P., R. H. Meloen, G. Taylor, J. M. Furze, and J. T. van Oirschot. 1997. Antigenic structure of the central conserved region of protein G of bovine respiratory syncytial virus. J. Virol. 71:4055-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langedijk, J. P., W. G. Middel, W. M. Schaaper, R. H. Meloen, J. A. Kramps, A. H. Brandenburg, and J. T. van Oirschot. 1996. Type-specific serologic diagnosis of respiratory syncytial virus infection, based on a synthetic peptide of the attachment protein G. J. Immunol. Methods 193:157-166. [DOI] [PubMed] [Google Scholar]
- 8.Martinez, I., J. Dopazo, and J. A. Melero. 1997. Antigenic structure of the human respiratory syncytial virus G glycoprotein and relevance of hypermutation events for the generation of antigenic variants. J. Gen. Virol. 78(Pt. 10):2419-2429. [DOI] [PubMed] [Google Scholar]
- 9.Melero, J. A., B. Garcia-Barreno, I. Martinez, C. R. Pringle, and P. A. Cane. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J. Gen. Virol. 78(Pt. 10):2411-2418. [DOI] [PubMed] [Google Scholar]
- 10.Murata, Y., P. M. Lightfoote, A. R. Falsey, and E. E. Walsh. 2009. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1565.
- 11.Norrby, E., M. A. Mufson, H. Alexander, R. A. Houghten, and R. A. Lerner. 1987. Site-directed serology with synthetic peptides representing the large glycoprotein G of respiratory syncytial virus. Proc. Natl. Acad. Sci. U. S. A. 84:6572-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palomo, C., B. Garcia-Barreno, C. Penas, and J. A. Melero. 1991. The G protein of human respiratory syncytial virus: significance of carbohydrate side-chains and the C-terminal end to its antigenicity. J. Gen. Virol. 72(Pt. 3):669-675. [DOI] [PubMed] [Google Scholar]
- 13.Plotnicky-Gilquin, H., L. Goetsch, T. Huss, T. Champion, A. Beck, J. F. Haeuw, T. N. Nguyen, J. Y. Bonnefoy, N. Corvaia, and U. F. Power. 1999. Identification of multiple protective epitopes (protectopes) in the central conserved domain of a prototype human respiratory syncytial virus G protein. J. Virol. 73:5637-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shinoff, J. J., K. L. O'Brien, B. Thumar, J. B. Shaw, R. Reid, W. Hua, M. Santosham, and R. A. Karron. 2008. Young infants can develop protective levels of neutralizing antibody after infection with respiratory syncytial virus. J. Infect. Dis. 198:1007-1015. [DOI] [PubMed] [Google Scholar]
- 15.Walsh, E. E., A. R. Falsey, and W. M. Sullender. 1998. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J. Gen. Virol. 79(Pt. 3):479-487. [DOI] [PubMed] [Google Scholar]
- 16.Walsh, E. E., C. B. Hall, J. J. Schlesinger, M. W. Brandriss, S. Hildreth, and P. Paradiso. 1989. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J. Gen. Virol. 70(Pt. 11):2953-2961. [DOI] [PubMed] [Google Scholar]
- 17.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. U. S. A. 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]