Abstract
Principal mechanisms of resistance to azole antifungals include the upregulation of multidrug transporters and the modification of the target enzyme, a cytochrome P450 (Erg11) involved in the 14α-demethylation of ergosterol. These mechanisms are often combined in azole-resistant Candida albicans isolates recovered from patients. However, the precise contributions of individual mechanisms to C. albicans resistance to specific azoles have been difficult to establish because of the technical difficulties in the genetic manipulation of this diploid species. Recent advances have made genetic manipulations easier, and we therefore undertook the genetic dissection of resistance mechanisms in an azole-resistant clinical isolate. This isolate (DSY296) upregulates the multidrug transporter genes CDR1 and CDR2 and has acquired a G464S substitution in both ERG11 alleles. In DSY296, inactivation of TAC1, a transcription factor containing a gain-of-function mutation, followed by sequential replacement of ERG11 mutant alleles with wild-type alleles, restored azole susceptibility to the levels measured for a parent azole-susceptible isolate (DSY294). These sequential genetic manipulations not only demonstrated that these two resistance mechanisms were those responsible for the development of resistance in DSY296 but also indicated that the quantitative level of resistance as measured in vitro by MIC determinations was a function of the number of genetic resistance mechanisms operating in any strain. The engineered strains were also tested for their responses to fluconazole treatment in a novel 3-day model of invasive C. albicans infection of mice. Fifty percent effective doses (ED50s) of fluconazole were highest for DSY296 and decreased proportionally with the sequential removal of each resistance mechanism. However, while the fold differences in ED50 were proportional to the fold differences in MICs, their magnitude was lower than that measured in vitro and depended on the specific resistance mechanism operating.
Azole antifungal agents are widely used to treat fungal infections. Over the past 2 decades, several agents of this class, including fluconazole (FLC), itraconazole (ITR), voriconazole (VRC), and posaconazole (POS), have been made available for clinical use in life-threatening, invasive fungal disease. Each of these compounds has a specific clinical utility, which depends on its spectrum of activity, route of administration, and pharmacodynamic properties. Several fungal species have counteracted the action of azoles by developing a number of resistance mechanisms. The fungal target of azole antifungals is a cytochrome P450 (encoded by ERG11) involved in the 14α-demethylation of lanosterol, an essential step in the biosynthesis of ergosterol. One of the azole resistance mechanisms, therefore, involves alteration of the target enzyme by amino acid substitutions caused by nonsynonymous mutations in ERG11. Such modifications have been documented in ERG11 from Candida albicans, Candida tropicalis, and Cryptococcus neoformans and in related ERG11-like genes, such as Cyp51A from Aspergillus fumigatus (reviewed by Sanglard et al. [24]).
In addition to target alterations, azole resistance mechanisms include transport phenomena. Azole drugs have been shown to be actively effluxed by transport systems. These include ABC transporters and major facilitator superfamily (MFS) members, which are overexpressed in azole-resistant isolates. In C. albicans, both the ABC transporter genes CDR1 and CDR2 and the MFS gene MDR1 can be upregulated, although not simultaneously, indicating the existence of separate regulatory circuits for the two transporter classes. Regulators of these transporters, including TAC1 and MRR1, which control the expression of C. albicans CDR1/CDR2 and MDR1, respectively, have been identified (7, 17). Mutations in these transcription factors are responsible for the constitutively high expression of these transporters in clinical isolates (4-6, 9). Other azole resistance mechanisms have been reported; however, their occurrence is relatively rare. For example, loss-of-function mutations in the ERG3 gene, encoding sterol Δ5,6 desaturase, have been reported in C. albicans and Candida dubliniensis as a cause of azole resistance (3, 22). Molecular epidemiology of azole resistance mechanisms has often demonstrated the existence of several resistance mechanisms in the same yeast isolate. In these studies, known mechanisms were probed either by testing the expression levels of multidrug transporter genes or by comparing ERG11 nucleotide sequences between pairs of related azole-susceptible and azole-resistant isolates. In many cases, azole resistance correlated with the occurrence of transporter gene upregulation and nonsynonymous ERG11 mutations (20, 21). Few attempts have been made to evaluate the role of individual mechanisms in strains with multiple azole resistance mechanisms. Wirsching et al. (30) inactivated MDR1 in a C. albicans strain upregulating this transporter and also containing a G464S mutation in both ERG11 alleles. The mutant strain exhibited lower azole MICs than the azole-resistant isolate, but these values were not as low as those measured for the related azole-susceptible strain. The remaining difference in the MIC was attributed to the G464S substitution still present in the mdr1Δ strain. Selmecki et al. (28) have evaluated the relative roles of ERG11 and TAC1, which were present on an isochromosome originating from chromosome 5 (Chr. 5). ERG11 contributed to azole resistance by the effect of gene copy number variation rather than by a mutation.
We previously investigated an azole-resistant clinical isolate, DSY296, and its azole-susceptible parent (DSY294) from the same patient. Coste et al. (7) showed that the transcription factor TAC1 was involved in the regulation of the ABC transporters CDR1 and CDR2 when C. albicans was exposed to drugs, including fluphenazine and estradiol. TAC1 in the azole-resistant isolate DSY296 is homozygous and contains a gain-of-function (GOF) mutation (N977D), resulting in constitutively high expression of CDR1 and CDR2 (6). We showed earlier that the mutation was homozygous at its Chr. 5 location, along with the mating type locus (MATa/a) (7). Deletion of TAC1 in DSY296 decreased the expression of CDR1 and CDR2 to basal levels and consistently decreased azole resistance in the tac1Δ/Δ strain (DSY3083), thus demonstrating the relevance of TAC1 in the development of azole resistance in C. albicans (6). No MDR1 overexpression was noticed in DSY296 (27).
Genetic tools for C. albicans have improved greatly, and it is now possible to use almost any clinical strain for genetic manipulation. We have therefore further investigated the clinical isolate DSY296, with its dual azole resistance mechanisms (7, 27). The genetic mediators of these mechanisms (TAC1 and ERG11) were sequentially removed by genetic manipulation or were replaced by wild-type genes, and the resulting azole susceptibilities were measured. Moreover, the azole susceptibilities of the different engineered strains were also determined in animal models with mice given fluconazole treatment.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study are listed in Table 1. Strains DSY294 and DSY296 were shown to be indistinguishable by several typing methods using a Ca3 repetitive probe (2) and by multilocus sequence typing (MLST) using recent standards (19). Both strains belong to clade 11 (D. Sanglard and F. Odds, unpublished data). Yeasts were grown in a complete medium, yeast extract-peptone-dextrose (YEPD) (1% Bacto peptone [Difco Laboratories, Basel, Switzerland], 0.5% yeast extract [Difco], and 2% glucose [Fluka, Buchs, Switzerland]), or in a defined medium, 0.67% yeast nitrogen base (YNB; Difco) with 2% glucose (Fluka). To prepare inocula for experimental infections, yeasts were grown in NGY medium (0.1% Neopeptone [Difco], 0.4% glucose, 0.1% yeast extract [Difco]). When strains were grown on a solid medium, 2% agar (Difco) was added. Escherichia coli DH5α was used as a host for plasmid constructions and propagation. DH5α was grown in Luria-Bertani (LB) broth or on LB plates, supplemented with ampicillin (0.1 mg/ml) when required.
TABLE 1.
Strains used in this study
| Strain | Parent | Genotype | Reference |
|---|---|---|---|
| DSY294 | TAC1-3/TAC1-4 | 7 | |
| ERG11-3/ERG11-4 | |||
| DSY296 | DSY294 | TAC1-5/TAC1-5 | 7 |
| ERG11-5/ERG11-5 | |||
| DSY3040 | DSY294 | ura3Δ::FRT/ura3Δ::FRT | 7 |
| DSY3041 | DSY296 | ura3Δ::FRT/ura3Δ::FRT | 7 |
| DSY3083 | DSY296 | tac1-5Δ::hisG/tac1-5Δ::hisG- URA3-hisG | 7 |
| ERG11-5/ERG11-5 | |||
| DSY3090 | DSY3083 | tac1-5Δ::hisG/tac1-5Δ::hisG | 7 |
| ERG11-5/ERG11-5 | |||
| DSY3586 | DSY3090 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::CtURA3/ERG11-5 | |||
| DSY3593 | DSY3090 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-5 | |||
| DSY3595 | DSY3593 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-1::CtURA3 | |||
| DSY3603 | DSY3595 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| DSY3604 | DSY3593 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-5 | |||
| RPS1::CIp10 | |||
| DSY3706 | DSY3603 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| RPS1::CIp10 | |||
| DSY3606 | DSY3603 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| RPS1::TAC1-5 | |||
| DSY3608 | DSY3603 | tac1-5Δ::hisG/tac1-5Δ::hisG | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| RPS1::TAC1-1 | |||
| DSY3671 | DSY3041 | TAC1-5/TAC1-5 | This study |
| ERG11-1::SAT1/ERG11-5 | |||
| DSY3745 | DSY3671 | TAC1-5/TAC1-5 | This study |
| ERG11-1::SAT1/ERG11-1::CtURA3 | |||
| DSY3750 | DSY3745 | TAC1-5/TAC1-5 | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| DSY3752 | DSY3750 | TAC1-5/TAC1-5 | This study |
| ERG11-1::SAT1/ERG11-1::SAT1 | |||
| RPS1::CIp10 | |||
| DSY294-CIp10 | DSY3040 | RPS1::CIp10 | This study |
| DSY296-CIp10 | DSY3041 | RPS1::CIp10 | This study |
| DSY3083-CIp10 | DSY3090 | RPS1::CIp10 | This study |
Yeast transformation.
C. albicans cells from 0.2 ml of stationary-phase cultures were resuspended in 0.1 ml of a solution containing 200 mM lithium acetate (LiAc) (pH 7.5), 40% (wt/vol) polyethylene glycol (PEG) 8000, 15 mg/ml dithiothreitol (DTT), and 250 μg/ml denatured salmon sperm DNA. Transforming DNA (1 to 5 μg) was added to the yeast suspension, which was incubated for 60 min at 44°C. Transformation mixtures were plated directly onto selective plates. The SAT1 marker, which is responsible for nourseothricin resistance, was selected with YEPD at a concentration of 200 μg/ml nourseothricin (Werner BioAgents, Germany).
Plasmid constructions.
To replace ERG11 from DSY296 with wild-type copies, two plasmids were constructed, one containing a SAT1 marker (pDS1453) and the other containing a URA3 marker from C. tropicalis (pDS1454). pDS1453 was obtained by inserting the SAT1 marker, amplified from pSFS2 with primers SAT1-StuI (5′-ATAAGAATAGGCCTGTCAAAACTAGAGAATAATAAAG-3′) and SAT1-NruI (5′-GCGCAAATCGCGAGGACCACCTTTGATTGTAAATAGT-3′), into the PmeI site of pDS501. Plasmid pDS501 contains the entire ERG11 open reading frame (ORF) and flanking regions from C. albicans strain SC5314, produced by cloning ERG11 as a 3.5-kb EcoRV-ClaI blunt-ended fragment from pDS271 (26) into pMLT21, which had first been digested by SmaI and AccI followed by blunt ending. For C. albicans transformations, the ERG11-SAT1 cassette was liberated from pDS1453 by XhoI and KpnI digestion. Plasmid pDS1454 was constructed by inserting the C. tropicalis URA3 (CtURA3) gene as a 1.3-kb ApaI-BamHI fragment from pQDS62 in a PCR-amplified ERG11 fragment from pDS501. Plasmid pQDS62 contains the C. tropicalis URA3 gene as a 1.3-kb HindIII fragment cloned into the HindIII site of pBluescript KS(+) (D. Sanglard, unpublished data). The PCR fragment was obtained by using primers pSP2-Apa (5′-AATCCTGGGCCCATATTAGCAGATGATGC-3′) and pSP2-BamBI (5′-TTCATCGGATCCTCACCATGCCTTATT-3′) with pDS501 as a template. pDS1454 is similar to pDS1453 but contains CtURA3 at the site of SAT1 insertion. For C. albicans transformations, the ERG11-CtURA3 cassette was liberated from pDS1454 by digestion with KpnI and StuI. pDS1663 and pDS1665 were obtained by subcloning TAC1-1 and TAC1-5 as BamHI-XhoI fragments from pDS1097 and pDS1099 (7) into the XhoI-BglII sites of CIp10 (18).
PCR and sequencing.
ERG11 alleles from C. albicans isolates were PCR amplified from genomic DNA. The primers used (CYPCB [5′-GCGGATCCTTAAAACATACAAGTTTCTCTTTT-3′] and CYPNS2 [5′-ACGCGTCGACAATATGGCTATTGTTGAAACTGTC-3′]) amplified the entire ERG11 ORF. Primer ERG11-3B (5′-CCCATTAAGAATCCCTGAA-3′) was used for sequencing in order to reveal the A1390G mutation, which leads to a G464S substitution. Sequencing was performed on a 3130XL genetic analyzer (Applied Biosystems). Geneious software (Biomatters Ltd., New Zealand) was used for sequence analysis.
SNP sequencing.
To determine the single nucleotide polymorphisms (SNPs) along Chr. 5, several chromosomal positions were amplified by PCR and were subjected to sequencing. PCR fragments were obtained from primer pairs (listed in Table S1 in the supplemental material), each containing a forward or reverse sequence tag that was used in subsequent sequencing reactions. Geneious software (Biomatters Ltd., New Zealand) was used for SNP analysis.
Construction of DSY296-derivative strains.
Plasmid pDS1453 was first used to transform the ura3 derivative of DSY3083 (DSY3090), a tac1Δ/Δ strain from DSY296, to obtain nourseothricin-resistant colonies heterozygous for ERG11-1 (DSY3593 [ERG11-1::SAT1/ERG11-5]). Sequencing of ERG11 PCR products from DSY3593 showed an ambiguity at nucleotide 1390 in ERG11, consistent with the presence of the two different alleles (data not shown). Next, DSY3593 was transformed with pDS1454 to yield Ura+ colonies. One of these colonies, DSY3595 (ERG11-1::SAT1/ERG11-1::CtURA3) was chosen for further work. To restore the C. albicans URA3 gene, this strain was first plated onto 5-fluoroorotic acid (5-FOA), resulting in the loss of the ERG11-1::CtURA3 allele and its replacement by the ERG11-1::SAT1 allele, thus producing strain DSY3603 (ERG11-1::SAT1/ERG11-1::SAT1). DSY3603 was next transformed with CIp10 to introduce URA3 at the neutral RPS1 locus (18), resulting in DSY3706. The loss of ERG11-1::CtURA3 in DSY3603 most probably occurred by mitotic recombination between the two homozygous left arms of Chr. 5 of this DSY296-derived strain. This conclusion was supported by the presence of SNPs (see above) in DSY3603 identical to those detected in DSY296 (data not shown) (see also Results). CIp10-derived plasmids containing the hyperactive TAC1-5 allele (pDS1665) and the wild-type TAC1-1 allele (pDS1663) were also used to transform DSY3603, yielding DSY3606 and DSY3608, respectively. This restored TAC1 in these strain backgrounds. To restore URA3 in the ERG11-1/ERG11-5 background, DSY3593 was transformed with CIp10, yielding DSY3604.
Both ERG11-5 alleles from strain DSY296 were also replaced by wild-type ERG11-1 alleles in a similar fashion. The final isolate, in which URA3 was introduced by CIp10, was named DSY3752 (TAC1-5/TAC1-5 ERG11-1::SAT1/ERG11-1::SAT1) (Table 1). This isolate, therefore, exhibits only one azole resistance mechanism mediated by both TAC1-5 alleles.
Drug susceptibility testing.
Azole MICs were measured by broth microdilution according to EUCAST standards with slight modifications (10, 11). Briefly, C. albicans strains were cultivated overnight at 30°C under constant agitation in YEPD. Cultures were diluted to a density of 5 × 104 cells per ml in RPMI 1640 medium (Sigma) with l-glutamine, without bicarbonate, and with phenol red as the pH indicator. RPMI 1640 medium was buffered to pH 7 with 0.165 M morpholinepropanesulfonic acid (MOPS) and was supplemented with glucose to a final concentration (wt/vol) of 2% and with 1% dimethyl sulfoxide (DMSO). Serial drug dilutions were made from stocks dissolved in DMSO for VRC, ITR, and POS and from stocks dissolved in H2O for FLC. Drug dilutions were made for each azole in the corresponding solvent at 100-fold strength and were diluted 100-fold in RPMI 1640 medium. The inoculum was distributed in 100-μl volumes in 96-well flat-bottom microdilution plates; then 100-μl volumes of serially diluted drugs were added. Drug-free cultures and sterility controls were included in each 96-well plate. Plates were incubated at 35°C for 24 h, and then MICs were read with a spectrophotometer plate reader set at 450 nm. The MIC was defined as the drug concentration at which the optical density decreased more than 50% from that of the drug-free culture. Drug-free cultures contained 1% DMSO. Fluconazole was obtained from Sigma. Itraconazole, voriconazole, and posaconazole were gifts from Janssen, Pfizer, and Schering Plough, respectively.
RNA and DNA analysis.
Expression of genes was analyzed by Northern blotting as described by Coste et al. (5). Labeled probes were generated by random priming with [α-32P]dATP by using the Mega Labeling kit (GE Healthcare) according to the manufacturer's instructions. CDR1, CDR2, ERG11, and ACT1 probes were obtained as described previously (7). Southern blotting was carried out as described previously (7). Signals resulting from Northern and Southern blotting were obtained by phosphorimaging on a Typhoon Trio system (GE Healthcare).
Animal experiments.
The effect of fluconazole treatment was assessed in mice experimentally infected with five of the C. albicans isolates used in this study: DSY294-CIp10, DSY296-CIp10, DSY3083-CIp10, DSY3606, and DSY3706. All animal experimentation conformed to the requirements of United Kingdom Home Office legislation and of the Ethical Review Committee of the University of Aberdeen. We have previously shown a strong correlation between loss of mouse body weight over 3 days after intravenous (i.v.) challenge with C. albicans and survival times up to 28 days (14). Kidney fungal burdens at 3 days also showed an association with strain virulence (13). The present experiments were therefore designed with a 3-day end point to minimize their duration.
Female BALB/c mice (Harlan, United Kingdom) weighing 18 to 22 g, housed in groups of as many as 10 animals and individually marked for identification, were supplied with food and water ad libitum. Mice were infected i.v. with a saline suspension of C. albicans cells from cultures grown overnight in NGY medium (15). Fluconazole treatment was started by intraperitoneal injection 1 h after challenge and was repeated 24 h and 48 h after challenge. At 72 h postchallenge, animals were humanely terminated; the left kidneys were removed with aseptic precautions and were homogenized in saline; and C. albicans viable counts in the homogenates were determined as log10 CFU per g of kidney.
The 50% effective dose (ED50) was defined as the fluconazole dose effective at reducing the pathological effects of i.v. C. albicans challenge significantly relative to those for saline-treated controls. The two parameters measured were the percentage of weight change over 3 days postchallenge, for which ED50 was the dose that reduced the mean weight change below 50% of that for controls, and log CFU/g, for which ED50 was the dose that reduced mean kidney burdens more than 1.5 log units below those of the controls. A third definition of ED50 was based on an “outcome score,” devised to allow combination of the 3-day weight loss and kidney burden data. The outcome score is calculated as log CFU/g of kidney − (0.5 × percent weight change). This formula provided for approximately equal contributions of the two parameters to the score.
A pilot experiment with DSY294-CIp10, DSY296-CIp10, and DSY3083-CIp10 showed that all three strains had similar virulence for mice, and lower virulence than strains SC5314 and CAF2-1, which we have tested extensively previously (13, 15). We characterized the virulence of the five strains in terms of 3-day outcome parameters, with groups of 3 mice each receiving a test challenge dose of 5 × 104, 1 × 105, or 2 × 105 CFU/g of body weight. For these and several other C. albicans isolates, we have established a correlation between the logarithm of the challenge dose and the 3-day outcome score (D. M. MacCallum and F. C. Odds, unpublished data). The actual challenge doses used to investigate the efficacy of fluconazole treatment were based on the detailed results of these experiments. To minimize the numbers of mice involved in the experiments, fluconazole treatments with doses of 1.0, 3.5, 10, 35, 120, and 200 mg/kg of body weight were tailored by initial selection of two of these doses for groups of 6 mice according to the known in vitro susceptibility of the infecting strain. Further experiments were then carried out with other fluconazole doses when the pair of doses initially tested was insufficient to establish ED50s.
RESULTS
Engineering DSY296-derivative strains.
This study focused on two clinical isolates from the same patient, DSY294 and DSY296; the latter has been described as fluconazole resistant. In addition to the TAC1 GOF mutation in DSY296, a mutation was also found in the ERG11 gene of this isolate (G464S), which has been reported previously to be involved in azole resistance (25). The effect of this mutation was observed in strain DSY3083, which was tac1Δ/Δ and was directly derived from DSY296: the fluconazole (FLC) MIC for this mutant was 4 μg/ml, which was greater than the FLC MIC for the azole-susceptible parental isolate, DSY294 (0.25 μg/ml). Interestingly, ERG11 is located at the left extremity of Chr. 5, and the G464S mutation was shown to be homozygous in DSY296. An SNP analysis of Chr. 5 performed in a previous study (6) indicated that the entire region spanning the telomere of Chr. 5 up to a region 100 kb downstream of TAC1 was homozygous, thus suggesting that loss of heterozygosity occurred by mitotic recombination in this region. Further analysis of this region delimited the loss of heterozygosity within a minimal 348-bp sequence of orf19.3178, a gene situated 13 kb from TAC1 (see Table S2 in the supplemental material).
The difference in azole susceptibility between DSY3083, lacking TAC1, and DSY294 led us to replace the mutant ERG11 alleles with wild-type alleles in DSY3083. To achieve this purpose, starting from a strain in which TAC1 was inactivated by the use of the “Ura-blaster” method (DSY3083), two different genetic markers (SAT1 and CtURA3) were used to selectively replace ERG11-5 alleles with ERG11 wild-type alleles. In order to achieve these replacements, we constructed two different plasmids (pDS1453 and pDS1454), each containing the wild-type ERG11 allele from the C. albicans isolate SC5314 (orf19.922) but possessing SAT1 or CtURA3 as a genetic marker, respectively. For practical purposes, we will name this allele ERG11-1. The strategy adopted in this study (see also Fig. 1) was first to obtain Ura+ and nourseothricin-resistant colonies (ERG11-1/ERG11-1 genotype) and next to regenerate ura3 by gene conversion of the URA3-containing allele with the SAT1-containing allele. This allowed the regeneration of the ura3 auxotrophic marker and the subsequent reintroduction of the C. albicans URA3 gene at the neutral RPS1 locus using CIp10.
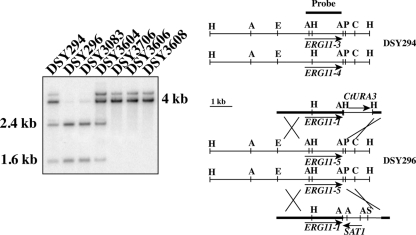
FIG. 1.
Southern blot analysis of ERG11 allele replacement. (Right) Restriction maps of genomic ERG11 loci. Abbreviations: A, AciI; H, HindIII; P, PstI; C, ClaI; S, SalI; E, EcoRI. ERG11 genomic elements from pDS1453 and pDS1454 are indicated by thick lines. (Left) Fragments expected from the hybridization with the labeled probe are 1.6 and 2.4 kb in ERG11-3 and ERG11-5 and 4.0 kb in ERG11-4. DSY294 shows the pattern of two different ERG11 alleles, while DSY296 exhibits the pattern of only one allele. The weak signal above 4.0 kb is probably due to partial digestion of genomic DNA.
DSY294 possesses two distinct ERG11 alleles (ERG11-3 and ERG11-4), while DSY296 possesses a single ERG11 allele (ERG11-5, derived from ERG11-3) (25). ERG11-1 and ERG11-3 differ by two nucleotides, resulting in the A129G and E266D substitutions. Since these two substitutions are not associated with azole resistance (25), we used ERG11-1 for allele replacements. ERG11-5 contains a single A1390G change compared to ERG11-3, resulting in a G464S substitution. The engineering of DSY296 derivatives in which ERG11 alleles were replaced is detailed in Materials and Methods. Southern blot analysis of the ERG11 loci demonstrated ERG11 allele replacement starting from strain DSY3090 (ERG11-5/ERG11-5): while DSY3604 (ERG11-5/ERG11-1) exhibited AciI restriction profiles identical to those of DSY294, DSY3706 showed restriction profiles consistent with the presence of ERG11-1 only (see Fig. 1 for details). Analysis of ERG11 from this strain showed that nucleotide position 1390 was homozygous for “A,” thus indicating that ERG11 is, as expected, wild type at this site (data not shown).
We next verified the gene expression of major determinants of azole resistance, including CDR1, CDR2, and ERG11, in the engineered strains. As observed in Fig. 2, and as expected, CDR1 and CDR2 were not upregulated in the tac1Δ/Δ strain (DSY3083) or in other strains derived from this strain background (DSY3604 and DSY3706). CDR1 and CDR2 were, however, upregulated in strain DSY3606, into which the hyperactive TAC1-5 allele had been reintroduced. In contrast, CDR1 and CDR2 levels in DSY3608, into which TAC1-1 had been reintroduced, were equal to those in DSY294. ERG11 expression in DSY296-derived isolates did not change significantly from that in the parent. This indicates that in the engineered strains, the presence of a genetic marker flanking ERG11 did not influence the expression of ERG11. We also noticed that ERG11 expression was approximately 2-fold higher in DSY296 than in DSY294, suggesting that some factor in DSY296 may contribute to this property. No isochromosome formation has been detected in this strain background (5).
FIG. 2.
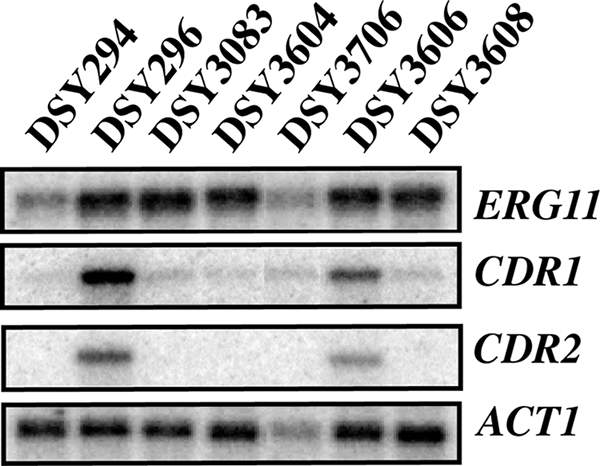
Expression of azole resistance genes in DSY294, DSY296, and DSY296-derived strains. Signals were obtained by phosphorimaging and were quantified by ImageQuant software (GE Healthcare). ERG11 expression was normalized to ACT1 expression and to ERG11 expression levels in DSY294. Differences in ERG11 expression levels between DSY296 and derivative strains fluctuate from 1.7- to 2.4-fold.
Drug susceptibility assays.
Clinical strains and their derivatives were tested for susceptibilities to the four azole antifungals used in therapy of invasive fungal diseases (FLC, ITR, VRC, and POS), according to EUCAST protocols. FLC and VRC are close structural relatives, bearing structures different from those of ITR and POS, both of which have a series of heterocyclic rings attached to the azole pharmacophore via a dioxolane or furan ring. Table 2 summarizes the results obtained. Strikingly, the replacement of ERG11-5 alleles by wild-type alleles in a tac1Δ/Δ mutant background increased azole susceptibility. When ERG11 was in the heterozygous state, as in DSY3604 (ERG11-5/ERG11-1), the FLC MIC decreased 2-fold (to 2 μg/ml) from that for the tac1Δ/Δ parent strain (4 μg/ml). A further 8-fold MIC decrease (from 2 μg/ml to 0.25 μg/ml FLC) was obtained by restoring two wild-type ERG11-1 alleles in the background of DSY3706. Interestingly, this MIC corresponded to that measured for DSY294, thus confirming by genetic evidence that only two azole resistance mechanisms contribute to the high azole MIC for DSY296, i.e., the G464S ERG11 mutation and ABC transporter upregulation. FLC MICs did not change in this strain background when a wild-type TAC1 allele was restored, consistent with previous observations (7). TAC1 functionality was verified by CDR1/CDR2 inducibility with fluphenazine exposure (data not shown). However, when TAC1-5 was reintroduced in the DSY3603 background, FLC MICs were increased 16-fold (MIC, 4 μg/ml) over that for DSY294 or DSY3706. These values are consistent with the restoration of CDR1/CDR2 upregulation, which was verified by Northern blot analysis. Besides confirming by genetic evidence the presence of two resistance mechanisms in DSY296, our results also highlight the fact that C. albicans requires the accumulation of several resistance mechanisms to obtain elevated levels of azole resistance. Both TAC1-5- and ERG11-5-mediated resistance mechanisms elevate the FLC MIC 16-fold, as deduced from the comparison between FLC MICs of 4 μg/ml and 0.25 μg/ml (DSY294), whereas the presence of both resistance mechanisms in DSY296 yields a 256-fold increase in the FLC MIC over that for DSY294 or DSY3706.
TABLE 2.
MICs of azole antifungals against clinical and engineered C. albicans strains
| Strain | MIC (μg/ml) (fold increase in MIC relative to that for DSY294) |
|||
|---|---|---|---|---|
| FLC | ITR | VRC | POS | |
| DSY294 | 0.25 (1) | 0.0156 (1) | 0.0078 (1) | 0.0312 (1) |
| DSY296 | 64 (256) | 0.25 (16) | 2 (256) | 0.25 (8) |
| DSY3083 tac1Δ/Δ | 4 (16) | 0.031 (2) | 0.125 (16) | 0.031 (1) |
| DSY3604 tac1Δ/Δ ERG11-1/ERG11-5 | 2 (8) | 0.031 (2) | 0.0625 (8) | 0.031 (1) |
| DSY3706 tac1Δ/Δ ERG11-1/ERG11-1 | 0.25 (1) | 0.0156 (1) | 0.0078 (1) | 0.031 (1) |
| DSY3606 tac1Δ/Δ ERG11-1/ERG11-1 (TAC1-5) | 4 (16) | 0.125 (8) | 0.125 (16) | 0.25 (8) |
| DSY3608 tac1Δ/Δ ERG11-1/ERG11-1 (TAC1-1) | 0.25 (1) | 0.031 (2) | 0.0078 (1) | 0.031 (1) |
| DSY3752 (TAC1-5/TAC1-5ERG11-1/ERG11-1) | 4 (16) | 0.25 (16) | 0.125 (16) | 0.25 (8) |
A similarly graded MIC hierarchy, related to the engineered presence and absence of one or two genes encoding azole resistance, was seen in the results with VRC (Table 2). The same was less true with ITR and POS, with which DSY296 and DSY3606 were inhibited at azole levels between 0.125 and 0.25 μg/ml, while all the other strains were inhibited at levels between 0.0156 and 0.0312 μg/ml. The MIC differences among strains thus ranged from 4- to 16-fold for ITR and POS, compared with 4- to 256-fold for FLC and VRC. In particular, replacement of ERG11-5 by ERG11-1 alleles in tac1Δ/Δ strains did not change ITR and POS MICs. Resistance to ITR and POS in DSY296 is mediated primarily by CDR1 and CDR2, since the replacement of ERG11-5 alleles with wild-type ERG11 alleles did not increase the relative POS MIC. These data are confirmed by POS (and ITR) MIC testing of isolate DSY3752: the replacement of ERG11-5 alleles by wild-type alleles in this strain did not decrease POS and ITR MIC values from those for DSY296, as opposed to FLC and VRC MIC values, thus confirming the differentiated effect of ERG11-mediated azole resistance on azoles.
Experimental infections and fluconazole efficacy.
In order to evaluate the effects of azole resistance mechanisms on the efficacy of azole treatment in vivo, selected strains were used in a mouse model of systemic candidiasis, and the infected animals were treated with at least two fluconazole doses to allow determination of ED50s. The selected strains contained CIp10 to restore URA3 at the neutral RPS1 locus: DSY294-CIp10 and DSY296-CIp10 (originating from the wild-type clinical strains DSY294 and DSY296), DSY3083-CIp10 (originating from the homozygous tac1Δ/Δ mutant DSY3083), DSY3706 (in which ERG11-5 alleles were replaced by wild-type ERG11-1 alleles), and finally DSY3606 (in which the TAC1-5 allele was reintroduced into a background similar to that of DSY3706). The azole susceptibilities of CIp10-transformed derivatives of DSY294, DSY296, and DSY3083 were identical to those of their parents (data not shown). The results of the animal experiments are summarized in Tables 3 and 4. The challenge inocula were based on pilot dose-finding experiments and were intended to give a 3-day outcome score between 8 and 16.
TABLE 3.
Effect of FLC treatment on experimental infection outcome
| Strain | Challenge inoculum (CFU/g body wt) | FLC Rx (mg/kg/day) | No. of animals | Infection outcome parameter on day 3 (mean ±SD) |
||
|---|---|---|---|---|---|---|
| Kidney fungal burden (log CFU/g kidney) | 3-day wt change (%) | Outcome scorea | ||||
| DSY294-CIp10 | 1.2 × 105 | Saline | 12 | 5.7 ± 0.6 | −7.0 ± 4.5 | 9.2 ± 2.6 |
| 1.0 | 6 | 4.8 ± 0.1 | −2.1 ± 3.0 | 5.9 ± 1.5 | ||
| 3.5 | 6 | 4.7 ± 0.2 | 3.2 ± 3.7 | 3.1 ± 1.8 | ||
| 10 | 6 | 4.3 ± 0.2 | 4.3 ± 2.7 | 2.2 ± 1.3 | ||
| 35 | 6 | 4.3 ± 0.1 | 5.5 ± 0.9 | 1.5 ± 0.4 | ||
| DSY296-CIp10 | 2.2 × 105 | Saline | 6 | 7.3 ± 0.2 | −10.9 ± 2.6 | 12.7 ± 1.5 |
| 120 | 6 | 6.3 ± 0.1 | −4.4 ± 18 | 8.4 ± 1.0 | ||
| 200 | 6 | 6.3 ± 0.1 | 0.2 ± 2.6 | 6.2 ± 1.2 | ||
| DSY3083-CIp10 | 3.3 × 105 | Saline | 12 | 7.3 ± 0.7 | −16.2 ± 2.6 | 15.4 ± 1.6 |
| 3.5 | 6 | 6.3 ± 0.5 | −14.2 ± 2.2 | 13.4 ± 1.4 | ||
| 10 | 6 | 5.5 ± 0.3 | −2.1 ± 2.8 | 6.6 ± 1.6 | ||
| 35 | 6 | 5.3 ± 0.4 | −0.7 ± 1.7 | 5.6 ± 1.1 | ||
| 120 | 6 | 5.2 ± 0.2 | 0.7 ± 2.3 | 4.8 ± 1.2 | ||
| DSY3606 | 5.6 × 104 | Saline | 6 | 6.1 ± 0.2 | −9.3 ± 3.6 | 10.7 ± 1.9 |
| 120 | 6 | 4.4 ± 0.2 | −0.8 ± 2.8 | 4.8 ± 1.4 | ||
| 200 | 6 | 4.5 ± 0.2 | 1.2 ± 2.2 | 3.9 ± 1.3 | ||
| DSY3706 | 2.7 × 105 | Saline | 12 | 6.8 ± 0.6 | −13.7 ± 3.0 | 13.6 ± 2.0 |
| 1.0 | 6 | 5.7 ± 0.2 | −5.9 ± 3.4 | 8.7 ± 1.9 | ||
| 3.5 | 6 | 5.0 ± 0.3 | −0.4 ± 2.9 | 5.2 ± 1.6 | ||
| 10 | 6 | 4.7 ± 0.2 | 1.4 ± 5.7 | 4.0 ± 3.0 | ||
| 35 | 6 | 4.5 ± 0.1 | 1.8 ± 2.6 | 3.6 ± 1.3 | ||
Calculated as kidney burden − (0.5 × weight change).
TABLE 4.
In vivo ED50 scores
| Strain | FLC MIC (μg/ml) | ED50 (mg/kg) determined on the basis of: |
Median ED50 (mg/kg) | ||
|---|---|---|---|---|---|
| Kidney burden | Wt change | Outcome score | |||
| DSY294-CIp10 | 0.25 | 10 | 1.0 | 3.5 | 3.5 |
| DSY296-CIp10 | 64 | >200 | 120 | 200 | 200 |
| DSY3083-CIp10 | 4 | 10 | 10 | 10 | 10 |
| DSY3606 | 4 | 120 | 120 | 120 | 120 |
| DSY3706 | 0.25 | 3.5 | 1 | 3.5 | 3.5 |
For DSY294-CIp10 (FLC MIC, 0.25 μg/ml), the ED50 was 1.0 mg/kg based on weight change, 3.5 mg/kg based on the outcome score, and 10 mg/kg based on kidney burdens (Table 4), according to the ED50 criteria defined in Materials and Methods. The result, based on the median of the three ED50 calculations, was the same as that based on the outcome score. For DSY296-CIp10 (FLC MIC, 64 μg/ml), the ED50 was 120 mg/kg based on weight change, 200 mg/kg based on the outcome score, and >200 mg/kg based on kidney burdens (Tables 3 and 4). The median ED50 in vivo was thus 57-fold greater for DSY296-CIp10 than for DSY294-CIp10—less than the 1,024-fold measured in vitro, but substantial nevertheless.
With regard to the engineered strains, the FLC ED50 for DSY3083-CIp10 was 10 mg/kg by all three parameters, a 3-fold increase over that for DSY294-CIp10, compared with a 64-fold difference in the MIC (Tables 3 and 4). The ED50 for DSY3706 was 1.0 mg/kg by weight change and 3.5 mg/kg by kidney burden and outcome score; thus, it was the same as for DSY294, as was also found in vitro. Finally, the ED50 for DSY3606 was 120 mg/kg by all three parameters (Table 4), a 35-fold difference from the FLC ED50 for DSY294-CIp10, compared with a 64-fold MIC difference (Table 4).
DISCUSSION
This study undertook the sequential replacement of drug resistance genes by wild-type copies. Starting from the azole-resistant isolate DSY296, we produced isolate DSY3706 with ERG11 wild-type alleles and with TAC1 deleted. Using SNP analysis of markers along Chr. 5, we showed that this strain was indistinguishable from DSY296 in terms of the SNP profile (see Table S2 in the supplemental material). We showed that after the replacement of the ERG11-5 allele by wild-type alleles, azole MICs for engineered strains reverted to those of the DSY294 isolate, the azole-susceptible parent of DSY296. MIC values were not different when TAC1-1 was reintroduced into the DSY3706 background, a finding consistent with results obtained in previous studies (7). Until now, few other studies have dissected azole resistance mechanisms by sequential genetic manipulations. In one study, MDR1 was inactivated in an azole-resistant isolate, but the resulting strains were not as susceptible as the azole-susceptible parent isolate, probably due to ERG11 mutations (30). Another study inactivated MRR1, a regulator of MDR1, in the same isolates, which also resulted in strains with intermediate azole resistance (17).
In this study, we have utilized technologies now available to introduce specific alleles of the resistance-associated genes TAC1 and ERG11 in a stepwise fashion, and we have confirmed the relationship between the FLC MIC and the number of resistance genes being expressed in a C. albicans strain. It was previously self-evident that the level of fluconazole resistance of a C. albicans isolate was proportional to the number of molecular resistance mechanisms expressed. White et al. (29) investigated the resistance mechanisms expressed in 17 sequential isolates from a single HIV-positive patient with persistently relapsing oropharyngeal C. albicans infection managed clinically with increasing doses of fluconazole. As the fluconazole MIC for successive isolates rose progressively from 0.25 μg/ml against the pretreatment isolate to 64 μg/ml against isolate 17, the number of detectable resistance mechanisms increased from overexpression of MDR1 through point mutations in ERG11, loss of ERG11 allelic variation, and finally overexpression of CDR1 (29). This isolate set thus represented a quantitative, stepwise rise in fluconazole resistance in which the FLC MIC depended on the number of different resistance mechanisms expressed in each isolate. The mechanisms detected in the previous publication were different from those in our pair of clinical isolates (DSY294 and DSY296), but the MIC difference measured was the same.
Our study shows that the impacts of resistance mechanisms on the MIC are dependent on the type of azole molecule investigated. These impacts differ for the two pairs of related azole antifungal agents, FLC and VRC versus ITR and POS. While ABC transporters confer cross-resistance to all the azoles investigated here, the ERG11 mutations had a differential impact. Because of the absence of POS MIC variation when ERG11-5 alleles were replaced with ERG11-1 alleles (see Table 2), our data suggest that POS probably interacts with the Erg11 structure by mechanisms different from those of FLC and VRC. A mutation in Erg11, such as G464S, has no effect on POS, and probably this position interacts weakly with this azole. With regard to these properties, ITR has an intermediate position between POS and FLC/VRC. Our data are consistent with previous models of Erg11 interactions with azoles, where ITR and POS stabilize their binding to mutated Erg11 proteins more efficiently than FLC and VRC (31). The differential effects of mutations in other ERG11-like genes have been documented in other fungal species. For example, the ERG11 ortholog in Aspergillus fumigatus, cyp51A, exhibits mutations as principal resistance mechanisms to azoles. However, the specific cyp51A mutations determine resistance to specific azoles (8, 16, 23). With the strains engineered in this study, it will be possible to test other azoles for their MIC profiles, allowing the impact of each resistance mechanism on specific azoles to be determined.
We have also shown here that the different levels of resistance, as measured by the FLC MIC in vitro, correlate with levels of resistance measured in the mouse i.v. challenge model of experimental C. albicans infection. Our in vivo data validate our experimental approach to antifungal treatment for mice: 48 h of treatment followed by assessment of kidney burdens and percentage of body weight change on day 3 postinfection. The ED50s calculated from the kidney burden and weight change data were proportional to the MIC differences for the strains tested, although the magnitudes of the differences in vivo were smaller than those measured by FLC MICs. The mean ED50 values for strains DSY294-CIp10 and DSY3706, from which two resistance mechanisms were removed, were calculated by three different approaches and were identical (3.5 mg/kg). The absence of the hyperactive TAC1-5 allele (strain DSY3083-CIp10) resulted in a 10-fold decrease in the mean ED50 value from that for DSY296-CIp10. This underscores the effect of the ERG11 mutation (G464S), which is apparent by comparison of DSY3083-CIp10 with DSY3706, with a 3-fold difference between ED50 values. The effect of TAC1-5 alone can be deduced from the comparison of mean ED50 values for DSY3606 and DSY3706 and is about 34-fold. This suggests that the relative contribution of azole resistance to these different ED50 values is much stronger for TAC1 than for ERG11. The ED50 values obtained in the present work are in good agreement with those available from other animal experiment studies, with the major difference that immunocompetent mice were used and the length of therapy was longer (3 days versus 24 h) in this study. For example, Louie et al. (12) gave a value of 4.56 mg/kg based on decreased kidney burden for an azole-susceptible isolate. Andes and van Ogtrop (1) reported a value of 1.9 mg/kg for a C. albicans isolate with a FLC MIC of 0.5 μg/ml. The ED50 values were 61 and 114 mg/kg for two other isolates with MIC values of 16 and 32 μg/ml, respectively. We reported a mean ED50 value of 3.5 mg/kg for isolates with a FLC MIC of 0.25 μg/ml, which lies within the values reported by both studies. The ED50 values (10 and 120 mg/kg) for isolates with higher MICs (8 μg/ml) depend on the resistance mechanism in operation, as mentioned above. These values are, however, in the range of those given by Andes and van Ogtrop (1).
In conclusion, this study provides genetic evidence for the participation of different resistance mechanisms in the development of increasing azole resistance. The engineered strains not only were useful for distinguishing the effect of an individual resistance mechanism on specific azoles but also helped to estimate the impact of the mechanism on treatment efficacy. Since a variety of other azole resistance mechanisms exist, other clinical strains could be subjected to the same genetic dissections and in vivo validation; this work is under way in our laboratory.
Supplementary Material
Acknowledgments
This study was supported by a grant from the European Commission under the acronym EURESFUN (LSHM-CT-2005-518199). D.S. is supported by the Swiss Research National Foundation (3100A0-114131/1).
Footnotes
Published ahead of print on 19 January 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Andes, D., and M. van Ogtrop.1999. Characterization and quantitation of the pharmacodynamics of fluconazole in a neutropenic murine disseminated candidiasis infection model. Antimicrob. Agents Chemother. 43:2116-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boerlin, P., F. Boerlin-Petzold, J. M. Goudet, C. Durussel, J. L. Pagani, J. P. Chave, and J. Bille.1996. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J. Clin. Microbiol. 34:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chau, A. S., M. Gurnani, R. Hawkinson, M. Laverdiere, A. Cacciapuoti, and P. M. McNicholas.2005. Inactivation of sterol Δ5,6-desaturase attenuates virulence in Candida albicans. Antimicrob. Agents Chemother. 49:3646-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coste, A., J. Crittin, C. Bauser, B. Rohde, and D. Sanglard.2009. Functional analysis of cis- and trans-acting elements of the Candida albicans CDR2 promoter with a novel promoter reporter system. Eukaryot. Cell 8:1250-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coste, A., A. Selmecki, A. Forche, D. Diogo, M. E. Bougnoux, C. d'Enfert, J. Berman, and D. Sanglard.2007. Genotypic evolution of azole resistance mechanisms in sequential Candida albicans isolates. Eukaryot. Cell 6:1889-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coste, A., V. Turner, F. Ischer, J. Morschhäuser, A. Forche, A. Semelcki, J. Berman, J. Bille, and D. Sanglard.2006. A mutation in Tac1p, a transcription factor regulating CDR1 and CDR2, is coupled with loss of heterozygosity at chromosome 5 to mediate antifungal resistance in Candida albicans. Genetics 172:2139-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard.2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela.2003. A point mutation in the 14α-sterol demethylase gene Cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunkel, N., J. Blass, P. D. Rogers, and J. Morschhäuser.2008. Mutations in the multidrug resistance regulator MRR1, followed by loss of heterozygosity, are the main cause of MDR1 overexpression in fluconazole-resistant Candida albicans strains. Mol. Microbiol. 69:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EUCAST-AFST (European Committee on Antimicrobial Susceptibility Testing-Subcommittee on Antifungal Susceptibility Testing).2008. EUCAST technical note on fluconazole. Clin. Microbiol. Infect. 14:193-195.18070130 [Google Scholar]
- 11.EUCAST-AFST (European Committee on Antimicrobial Susceptibility Testing-Subcommittee on Antifungal Susceptibility Testing).2008. EUCAST technical note on voriconazole. Clin. Microbiol. Infect. 14:985-987. [DOI] [PubMed] [Google Scholar]
- 12.Louie, A., G. L. Drusano, P. Banerjee, Q. F. Liu, W. Liu, P. Kaw, M. Shayegani, H. Taber, and M. H. Miller.1998. Pharmacodynamics of fluconazole in a murine model of systemic candidiasis. Antimicrob. Agents Chemother. 42:1105-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacCallum, D. M., L. Castillo, A. J. P. Brown, N. A. R. Gow, and F. C. Odds.2009. Early-expressed chemokines predict kidney immunopathology in experimental disseminated Candida albicans infections. PLoS One 4:e6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacCallum, D. M., L. Castillo, K. Nather, C. A. Munro, A. J. P. Brown, N. A. R. Gow, and F. C. Odds.2009. Property differences among the four major Candida albicans strain clades. Eukaryot. Cell 8:373-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacCallum, D. M., and F. C. Odds.2005. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses 48:151-161. [DOI] [PubMed] [Google Scholar]
- 16.Mellado, E., G. Garcia-Effron, L. Alcazar-Fuoli, W. J. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela.2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morschhäuser, J., K. S. Barker, T. T. Liu, B. W. J. Bla, R. Homayouni, and P. D. Rogers.2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. Brown.2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 19.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert.2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 6:1041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson.2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perea, S., J. L. Lopez-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson.2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinjon, E., G. P. Moran, C. J. Jackson, S. L. Kelly, D. Sanglard, D. C. Coleman, and D. J. Sullivan.2003. Molecular mechanisms of itraconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 47:2424-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodero, L., E. Mellado, A. C. Rodriguez, A. Salve, L. Guelfand, P. Cahn, M. Cuenca-Estrella, G. Davel, and J. L. Rodriguez-Tudela.2003. G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob. Agents Chemother. 47:3653-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanglard, D., A. Coste, and S. Ferrari.2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9:1029-1050. [DOI] [PubMed] [Google Scholar]
- 25.Sanglard, D., F. Ischer, L. Koymans, and J. Bille.1998. Amino acid substitutions in the cytochrome P450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contributing to the resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanglard, D., F. Ischer, M. Monod, and J. Bille.1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC-transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 27.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille.1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selmecki, A., M. Gerami-Nejad, C. Paulson, A. Forche, and J. Berman.2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68:624-641. [DOI] [PubMed] [Google Scholar]
- 29.White, T. C., K. A. Marr, and R. A. Bowden.1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirsching, S., S. Michel, and J. Morschhäuser.2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856-865. [DOI] [PubMed] [Google Scholar]
- 31.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas.2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.