Abstract
Etravirine (ETV) is a second-generation nonnucleoside reverse transcriptase (RT) inhibitor (NNRTI) introduced recently for salvage antiretroviral treatment after the emergence of NNRTI-resistant human immunodeficiency virus type 1 (HIV-1). Following its introduction, two naturally occurring mutations in HIV-1 RT, V106I and V179D, were listed as ETV resistance-associated mutations. However, the effect of these mutations on the development of NNRTI resistance has not been analyzed yet. To select highly NNRTI-resistant HIV-1 in vitro, monoclonal HIV-1 strains harboring V106I and V179D (HIV-1V106I and HIV-1V179D) were propagated in the presence of increasing concentrations of efavirenz (EFV). Interestingly, V179D emerged in one of three selection experiments from HIV-1V106I and V106I emerged in two of three experiments from HIV-1V179D. Analysis of recombinant HIV-1 clones showed that the combination of V106I and V179D conferred significant resistance to EFV and nevirapine (NVP) but not to ETV. Structural analysis indicated that ETV can overcome the repulsive interactions caused by the combination of V106I and V179D through fine-tuning of its binding module to RT facilitated by its plastic structure, whereas EFV and NVP cannot because of their rigid structures. Analysis of clinical isolates showed comparable drug susceptibilities, and the same combination of mutations was found in some database patients who experienced virologic NNRTI-based treatment failure. The combination of V106I and V179D is a newly identified NNRTI resistance pattern of mutations. The combination of polymorphic and minor resistance-associated mutations should be interpreted carefully.
Human immunodeficiency virus type 1 (HIV-1) sequences differ among infected individuals, and there are a number of naturally occurring amino acid changes commonly found in treatment-naïve patients (3, 23, 28). These polymorphic changes can occur even in genes that encode drug target proteins, and in fact, some drug resistance-associated mutations in protease genes are often present in treatment-naïve individuals, especially in non-subtype B clade-infected individuals (13, 15, 22). Minor resistance mutations, which are considered to compensate for the impaired replication fitness of viruses containing major resistance mutations, do not have a substantial effect on the viral phenotype by themselves (14, 27). In the reverse transcriptase (RT) coding region, drug resistance-associated mutations were detected at a low frequency in treatment-naïve individuals regardless of the HIV-1 clade. However, etravirine (ETV), a second-generation nonnucleoside RT inhibitor (NNRTI), has been available in the clinical setting and the following have been listed as ETV resistance-associated mutations in RT: V90I, A98G, L100I, K101E/H/P, V106I, E138A, V179D/F/T, Y181C/I/V, G190S/A, and M230L (14). V106I and V179D are often found in treatment-naïve individuals but are considered to have no substantial impact on NNRTI-containing treatment by themselves. ETV exhibits activity against many viruses that are resistant to first-line NNRTIs, including efavirenz (EFV) and nevirapine (NVP), and shows clinical efficacy in salvage treatment after NNRTI treatment failure (18, 21). However, it is possible that EFV- and NVP-resistant viruses derived from HIV-1 harboring V106I or V179D could compromise the efficacy of ETV. To determine the impact of these polymorphic mutations on the mutation patterns of NNRTI resistance, EFV-resistant HIV-1 strains were selected in vitro from monoclonal viruses harboring V106I and V179D, respectively. The virologic effects of selected specific mutation patterns were analyzed by constructing recombinant HIV-1 clones, and their clinical relevance was confirmed by analysis of isolates from infected individuals.
MATERIALS AND METHODS
HIV-1 sequences and clinical isolates from treatment-naïve individuals.
HIV-1 RT sequences were analyzed in 364 antiretroviral treatment-naïve infected individuals who visited the outpatient clinic of the AIDS Clinical Center, International Medical Center of Japan, in 2007 and 2008 and gave written informed consent to this study. Viral RNA was extracted from stocked plasma samples, and the HIV-1 RT coding region was amplified by RT-PCR and nested PCR using previously published primer pairs (7, 10, 11). Direct sequencing was performed using dye terminators, and the HIV-1 subtypes of the sequences obtained were determined by the neighbor-joining method. Clinical HIV-1 isolates were obtained using MAGIC-5 cells (CCR5- and CD4-expressing HeLa-LTR-β-D-gal cells) from fresh plasma samples collected from seven of the above-mentioned treatment-naïve patients and stored at −80°C until use (9).
Generation of recombinant HIV-1 strains.
The desired mutations were introduced into the XmaI-NheI region of pTZNX, which encodes Gly-15 to Ala-267 of HIV-1 RT (strain BH 10), by the oligonucleotide-based mutagenesis method (10, 16). The XmaI-NheI fragment was inserted into pNLH219Q, which was modified from pNL101 and encoded the full genome of HIV-1 strain BH 10. pNLH219Q harbors the H219Q mutation in the HIV-1 Gag region, which facilitated HIV-1 replication in MT-2 and H9 cells (6, 8). HIV-1 derived from pNLH219Q was used as the wild type. Determination of the nucleotide sequences of the plasmids confirmed that each clone had the desired mutations but was devoid of unintended mutations. Each molecular clone was transfected into COS-7 cells with the GenePORTER Transfection Reagent (Gene Therapy Systems, San Diego, CA), and the virions obtained were harvested 48 h after transfection and stored at −80°C until use.
Selection of EFV-resistant HIV-1.
The infectious HIV-1 clones harboring the V106I (HIV-1V106I) and V179D (HIV-1V179D) mutations in their RTs were propagated in MT-2 cells in the presence of increasing concentrations of EFV (7, 8, 31). Briefly, MT-2 cells (1 × 105) were exposed to 500 blue-cell-forming units (BFU) in MAGIC-5 cells containing HIV-1V106I and HIV-1V179D and cultured in the presence of EFV at an initial concentration of 3 nM. Viral replication was monitored by observation of the cytopathic effect in MT-2 cells. The culture supernatant was harvested on day 7 of culture and used to infect fresh MT-2 cells for the next round of culture. When the virus began to propagate in the presence of the drug, the drug concentration was increased by 0.5-log-fold. This selection was carried out for a total of 14 passages. The proviral HIV-1 RT coding region in infected MT-2 cells was amplified and sequenced at several passages.
Drug susceptibility assay.
EFV, NVP, and ETV were generously provided by Merck Co., Inc. (Rahway, NJ), Boehringer Ingelheim Pharmaceutics Inc. (Ridgefield, CT), and Tibotec Pharmaceuticals (Little Island, Co., Cork, Ireland), respectively. Recombinant and isolated HIV-1 susceptibility to EFV, NVP, and ETV was determined in triplicate using MAGIC-5 cells (10, 12, 16). Briefly, MAGIC-5 cells were infected with an adjusted virus stock (300 BFU) in various concentrations of NNRTIs, cultured for 48 h, fixed, and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Takara Shuzo, Ohtsu, Japan). The blue-stained cells were counted under a light microscope. The drug concentrations that inhibited 50% of the stained cells of a drug-free control (EC50s) were determined by referring to the dose-response curve. The drug susceptibility assay was performed in triplicate and repeated three times. Fold resistance was calculated by comparing the viral EC50 with that of monoclonal wild-type HIV-1 (HIV-1WT). Drug resistance was judged significant when it was higher than threefold.
Viral replication kinetics assay.
MT-2 cells (1 × 105) were exposed to each infectious virus preparation (500 BFU) for 2 h, washed twice with phosphate-buffered saline (PBS), and cultured in the presence or absence of 10 nM EFV, 100 nM NVP, or 10 nM ETV. The culture supernatants were harvested every other day, and p24 Gag amounts were determined with a chemiluminescence enzyme immunoassay kit (Fuji-Rebio, Tokyo, Japan). Replication assays were performed in triplicate and repeated three times using independently generated virus preparations (7).
Competitive HIV-1 replication assay.
Freshly prepared H9 cells (3 × 105) were exposed to mixtures of paired virus preparations (300 BFU each) to be examined for their replication ability for 2 h, washed twice with phosphate-buffered saline (PBS), and cultured in the absence or presence of 10 nM EFV or 100 nM NVP as described previously (7, 17). On day 1, one-third of the infected H9 cells were harvested and washed twice with PBS, and proviral DNAs were sequenced (0 week). Every 7 days, the supernatant of the virus culture was transferred to new uninfected H9 cells; the cells harvested at the end of each passage were subjected to direct DNA sequencing of the HIV-1 RT coding region, and the change in the viral population was determined by the relative peak height on the sequencing electrogram. The persistence of the original amino acid substitution was confirmed at the end of the assay.
Structure modeling.
We constructed 18 structural models of the HIV-1 RT and NNRTI complex by computational analysis. First, we constructed the initial models of wild-type RT with one of three NNRTIs by homology modeling using Molecular Operating Environment 2007.09.02 (Chemical Computing Group, Montreal, Quebec, Canada). The crystal structures of RT with NNRTIs (Protein Data Bank codes 1IKW [20], 1VRT [26], and 1SV5 [4]) were used as template structures. The homology modeling enabled the building of missing atoms in template structures. The ff94 force field and distance-dependent electrostatic energy function were applied in the modeling. Next, we refined the initial models by energy minimization using the sander module of the AMBER9 software package in two steps. In the first step, energies for the NNRTIs in the complex models were minimized in the gas phase by the conjugated gradient method. When the energy was not converged until 10,000 steps, this step was ignored. In the second step, energies of whole structures were converged up to 0.5 kcal/mol/Å by 50 steps of the steepest-descent method and the subsequent conjugated gradient method under implicit water solvent conditions. In each minimization, the AMBER ff03 (5, 19), the general AMBER force field (30), and the generalized Born implicit solvent surface area method (IGB = 2) (24) were applied for potential energy calculations. The cutoff for long-distance interaction energy was set at 15.0 Å. The charge and type of every atom in NNRTIs were automatically assigned using the AMBER9 Antechamber module. We also constructed five mutant RTs with the NNRTIs by considering every possible conformer of the respective mutant models. The possible conformers were generated from the wild-type homology models using PyMOL ver. 0.99rc6 (http://www.pymol.org). V106A, V106I, and V179D mutants had one, three to five, and five possible conformers, respectively. The structural model of each conformer was refined by a method similar to that used in the wild-type models. Among the refined conformers, we selected the conformer with the lowest energy as each mutant model.
RESULTS
Frequencies of NNRTI resistance mutations in treatment-naïve individuals.
To determine the frequency of NNRTI resistance-associated mutations, the HIV-1 RT coding region was analyzed and the viral subtype was determined in 364 treatment-naïve infected individuals. The most frequent subtype was clade B (n = 334; 91.8%), followed by clade AE (n = 20; 5.5%). Clades C and G were also found and at similar low frequencies (n = 5; 1.4%). Variable amino acid substitutions were identified at the positions of NNRTI resistance-associated mutations, including the 90th, 98th, 101st, 103rd, 106th, 108th, 138th, and 179th, though only the wild-type amino acids were observed at the 100th, 181st, 188th, 190th, 225th, and 230th positions of HIV-1 RT (Table 1). K103N and V108I are listed as EFV and NVP resistance-associated mutations and V90I, K101E, V106I, E138A, V179D, and V179T are listed as ETV resistance-associated mutations in the Drug Resistance Mutation List of the International AIDS Society (IAS)-USA (14). Of these NNRTI resistance-associated mutations, V106I (2.5%) and V179D (5.8%) were frequently observed in treatment-naïve patients, while the other six mutations were less common (0.3 to 0.8%). These data indicate that V106I and V179D occur naturally in treatment-naïve individuals at significant frequencies and are clinically important as polymorphic mutations. Accordingly, we focused on these two polymorphic mutations and analyzed their effects on the development of NNRTI resistance. There was no overt linkage among the amino acids at the positions of NNRTI resistance-associated mutations, though one patient harbored both V106I and V179D without any other resistance-associated mutations. There was no correlation between clades and the 106th and 179th amino acids.
TABLE 1.
Frequencies of amino acids at positions associated with NNRTI resistance mutations in HIV-1 RT in treatment-naïve patients
| Position | Amino acid, frequency [n (%)/364]a | ||||||
|---|---|---|---|---|---|---|---|
| 90 | V, 361 (99.2) | I, 3 (0.8) | |||||
| 98 | A, 344 (94.5) | S, 20 (5.5) | |||||
| 101 | K, 352 (96.7) | Q, 8 (2.2) | R, 3 (0.8) | E, 1 (0.3) | |||
| 103 | K, 353 (97.0) | R, 7 (1.9) | N, 2 (0.5) | Q, 2 (0.5) | |||
| 106 | V, 355 (97.5) | I, 9b (2.5) | |||||
| 108 | V, 362 (99.5) | I, 2 (0.5) | |||||
| 138 | E, 361 (99.2) | K, 1 (0.3) | A, 1 (0.3) | G, 1 (0.3) | |||
| 179 | V, 312 (85.7) | D, 21c (5.8) | I, 21 (5.8) | E, 4 (1.1) | A, 4 (1.1) | T, 1 (0.3) | N, 1 (0.3) |
Only wild-type amino acids (L, Y, Y, G, P, and M, respectively) were identified at the 100th, 181st, 188th, 190th, 225th, and 230th positions of HIV-1 RT.
Including two cases with V90/A98/K101Q/K103/V106I/V108/E138/V179, two cases with V90/A98/K101/K103/V106I/V108/E138/V179I, one case with V90/A98/K101/K103/V106I/V108/E138/V179D, and four cases with V90/A98/K101/K103/V106I/V108/E138/V179.
Including 2 cases with V90/A98S/K101/K103/V106/V108/E138/V179D, 1 case with V90/A98/K101E/K103R/V106/V108/E138/V179D, 1 case with V90/A98/K101/K103R/V106/V108/E138/V179D, 1 case with V90/A98/K101/K103/V106I/V108/E138/V179D, and 16 cases with V90/A98/K101/K103/V106/V108/E138/V179D.
Selection of EFV-resistant HIV-1 from HIV-1V106I and HIV-1V179D.
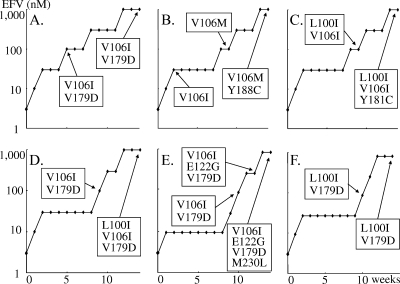
To analyze the effects of V106I and V179D on the resistance pattern of mutations, EFV-resistant HIV-1 strains were selected from monoclonal HIV-1 strains harboring V106I (HIV-1V106I) or V179D (HIV-1V179D). These selection experiments were performed independently in triplicate. Interestingly, in one of three selection experiments with HIV-1V106I, V179D emerged when the EFV concentration reached 100 nM, and it was persistently identified until the end of the passages without additional mutations (Fig. 1A). In two other experiments with HIV-1V106I, I106M emerged, followed by Y188C, and L100I emerged, followed by Y181C. These four mutations are already known NNRTI resistance-associated mutations (Fig. 1B and C) (1, 14, 25). In one of three experiments with HIV-1V179D, V106I emerged when the EFV concentration reached 100 nM, and L100I further emerged at the end of the experiments (Fig. 1D). In another experiment, V106I emerged when the EFV concentration was 100 nM, and E122G and M230L followed subsequently (Fig. 1E). In the last experiment, L100I emerged when the EFV concentration reached 100 nM, and it remained until the end of the passages without additional mutations (Fig. 1F). In summary, selected by EFV, V179D emerged in one of three experiments from HIV-1V106I and V106I emerged in two of three experiments from HIV-1V179D, suggesting that the combination of two polymorphic mutations, V106I and V179D, alters viral susceptibility to EFV.
FIG. 1.
Selected amino acid substitutions under selective pressure from EFV. HIV-1V106I (A to C) and HIV-1V179D (D to F) were propagated in MT-2 cells in the presence of increasing concentrations of EFV. The selected amino acid substitutions were analyzed at several passages by sequencing the proviral HIV-1 RT coding region in MT-2 cells. Amino acid substitutions compared with wild-type strain BH 10 are shown.
NNRTI susceptibility of recombinant HIV-1 strains.
To analyze the effects of V106I, V179D, and their combination on NNRTI susceptibility, a panel of recombinant HIV-1 clones was constructed and their EFV, NVP, and ETV EC50s were determined. As expected, the single mutation V106I or V179D did not confer significant resistance to EFV and NVP (Table 2). HIV-1V106A was generated as a reference, and it showed high-fold resistance to NVP but not to EFV, in agreement with previous studies (10, 14). The addition of V179D to HIV-1V106A (HIV-1V106A/V179D) increased its resistance to NVP and conferred significant resistance to EFV. The combination of the two polymorphic mutations, V106I and V179D, which emerged in resistance selection experiments with EFV, conferred significant resistance not only to EFV but also to NVP. In the susceptibility assay with ETV, it exhibited potent anti-HIV-1 activity in all of the HIV-1 strains examined, including the NNRTI-resistant clones described above, indicating that ETV has a different binding formulation with RT molecules than EFV and NVP do.
TABLE 2.
NNRTI susceptibility of recombinant HIV-1 strains
| HIV-1 strain | Mean EC50 (μM) ±SD (fold resistance)a |
||
|---|---|---|---|
| EFV | NVP | ETV | |
| Wild type | 0.002 ± 0.0007 | 0.05 ± 0.01 | 0.0012 ± 0 |
| V106A clone | 0.003 ± 0.0009 (1.5) | 3.43 ± 0.98 (69) | 0.0005 ± 0.0001 (0.40) |
| V106I clone | 0.003 ± 0.0003 (1.5) | 0.02 ± 0.0012 (0.40) | 0.0015 ± 0.0004 (1.3) |
| V179D clone | 0.004 ± 0.0002 (2.0) | 0.13 ± 0.02 (2.6) | 0.0019 ± 0.0004 (1.6) |
| V106A V179D clone | 0.013 ± 0.004 (6.5) | 4.53 ± 0.72 (91) | 0.0014 ± 0.0004 (1.2) |
| V106I V179D clone | 0.029 ± 0.007 (15) | 0.37 ± 0.12 (7.0) | 0.0024 ± 0.0004 (2.0) |
The drug susceptibility assay was performed in triplicate and repeated three times (nine experiments). Data are means of nine experiments.
Replication kinetics of recombinant HIV-1 strains.
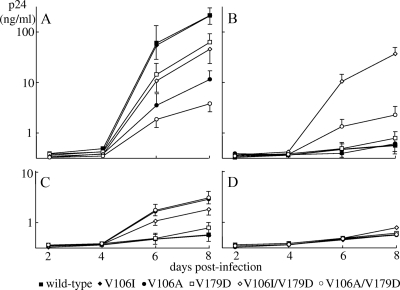
To analyze the effects of single mutations and their combinations on HIV-1 replication efficiency, we assayed the replication kinetics of recombinant HIV-1 strains in MT-2 cells in the absence or presence of an NNRTI. Each replication assay was performed in triplicate and repeated three times. In the absence of an NNRTI, V106I did not alter HIV-1WT replication while V179D significantly reduced HIV-1WT replication (p24 of HIV-1V179D versus HIV-1WT on day 8; P < 0.05) (Fig. 2A). The addition of V106I to HIV-1V179D (HIV-1V106I/179D) did not significantly alter its replication kinetics. V106A significantly reduced viral replication (p24 of HIV-1V106A versus HIV-1WT on day 8; P < 0.01), and the addition of V179D to it (HIV-1V106A/179D) further reduced the virus's replication ability (p24 of HIV-1V106A/V179D versus HIV-1V106A on day 8; P < 0.05).
FIG. 2.
Replication kinetics of recombinant HIV-1 clones in the absence and presence of NNRTIs. Recombinant HIV-1 clones were propagated in MT-2 cells in the absence (A) and presence of 10 nM EFV (B), 100 nM NVP (C), or 10 nM ETV (D). The concentration of p24 in the culture medium was measured every other day. The assay was performed in triplicate and repeated three times (nine experiments). The data are logarithmic mean p24 concentrations ± standard deviations (days 6 and 8 in panels A to C).
In the presence of 10 nM EFV, HIV-1WT, HIV-1V106I, and HIV-1V106A failed to propagate and HIV-1V179D exhibited reduced replication compared with that observed in the absence of an NNRTI (P < 0.01) (Fig. 2B). HIV-1V106I/V179D and HIV-1V106A/V179D showed efficient replication, though the replication of HIV-1V106A/V179D was slightly reduced compared with that observed in the absence of an NNRTI. In the presence of 100 nM NVP, HIV-1WT and HIV-1V106I failed to propagate and HIV-1V179D exhibited reduced replication compared with that observed in the absence of an NNRTI (P < 0.01) (Fig. 2C). HIV-1V106A and HIV-1V106I/V179D showed efficient replication, though the replication of HIV-1V106I/V179D was reduced significantly compared with that observed in the absence of an NNRTI (P < 0.05). HIV-1V106A/V179D exhibited replication comparable to that observed in the absence of an NNRTI. In the presence of 10 nM ETV, all of the HIV-1 strains examined exhibited severely compromised replication (Fig. 2D). The results of the replication kinetics experiments were in agreement with the drug susceptibility data.
To analyze the precise roles of V106I and V179D in HIV-1V106I/V179D, a competitive HIV-1 replication assay was performed using H9 cells (7, 17). The assay of HIV-1V106I and HIV-1V106I/V179D indicated that the addition of V179D compromised the replication fitness of HIV-1V106I but conferred resistance to EFV and NVP (see Fig. S1A to C in the supplemental material). The assay of HIV-1V179D and HIV-1V106I/V179D indicated that the addition of V106I slightly reduced the replication ability of HIV-1V179D but conferred resistance to EFV and NVP (see Fig. S1D to F in the supplemental material). In the presence of 10 nM ETV, the HIV-1 clones examined could not be passaged efficiently because ETV efficiently suppressed viral replication.
NNRTI susceptibility of HIV-1 clinical isolates.
Analysis of the recombinant HIV-1 clones indicated that the combination of V106I and V179D conferred significant resistance to EFV and NVP but not to ETV, although each single mutation did not alter viral susceptibility to NNRTIs (Table 2). To determine the clinical relevance of the results of recombinant HIV-1 analysis, HIV-1 clinical isolates were obtained using MAGIC-5 cells from seven treatment-naïve individuals (cases 1 to 7). In cases 1 and 2, only wild-type amino acids (valine) were detected at the 106th and 179th codons of the HIV-1 RT coding region. In cases 3 and 4, V106I was detected and wild-type valine was found at the 179th codon. In cases 5 and 6, wild-type valine was found at the 106th codon and V179D was identified. In case 7, both V106I and V179D were identified, and none of the other patients harbored both mutations in HIV-1 RT. Subclonal analysis determined that V106I and V179D were on the same virus and that they were highly dominant in case 7 (Table 1). All seven of the patients were infected with HIV-1 subtype B, and no other known resistance-associated mutations were detected at any RT codon other than the 106th and 179th. The six isolates derived from cases 1 to 6 did not show significant resistance to NNRTIs (Table 3). The isolate from case 7, however, exhibited significant resistance to EFV and NVP but not to ETV. These data confirmed the results obtained by recombinant HIV-1 analysis.
TABLE 3.
NNRTI susceptibility of clinical HIV-1 isolates
| HIV-1 strain (case no.) | Mean EC50 (μM) ±SD (fold resistance)a |
||
|---|---|---|---|
| EFV | NVP | ETV | |
| Wild type (V106 V179) | 0.002 ± 0.0001 | 0.05 ± 0.004 | 0.0013 ± 0.0001 |
| V106 V179 isolate (1) | 0.002 ± 0.0004 (1.0) | 0.04 ± 0.004 (0.8) | 0.002 ± 0.0004 (1.5) |
| V106 V179 isolate (2) | 0.002 ± 0.0003 (1.0) | 0.04 ± 0.004 (0.8) | 0.003 ± 0.0002 (2.3) |
| V106I V179 isolate (3) | 0.002 ± 0.0002 (1.0) | 0.03 ± 0.01 (0.6) | 0.0012 ± 0.0002 (0.9) |
| V106I V179 isolate (4) | 0.004 ± 0.001 (2.0) | 0.09 ± 0.01 (1.8) | 0.0024 ± 0.0002 (1.8) |
| V106 V179D isolate (5) | 0.006 ± 0.001 (3.0) | 0.07 ± 0.02 (1.4) | 0.0015 ± 0.0002 (1.2) |
| V106 V179D isolate (6) | 0.004 ± 0.002 (2.0) | 0.07 ± 0.004 (1.4) | 0.0011 ± 0.0001 (0.8) |
| V106I V179D isolate (7) | 0.01 ± 0.001 (7.0) | 0.19 ± 0.02 (3.8) | 0.002 ± 0.0003 (1.5) |
The drug susceptibility assay was performed in triplicate and repeated three times (nine experiments). Data are means of nine experiments.
Structural modeling analysis.
To obtain structural insight into the molecular mechanisms through which RT mutations alter susceptibility to NNRTIs, we conducted structural analyses by computational methods. A total of 18 structural models of RT-NNRTI complexes were constructed with six RTs (wild-type RT and V179D, V106A, V106I, V106A/V179D, and V106I/V179D mutant RTs) and three NNRTIs (EFV, NVP, and ETV), and the differences in binding energy between the mutant and wild-type complexes (ΔΔGb) were calculated. Notably, ΔΔGb was proportionally related to the logarithm of fold resistance in RT (Table 2); i.e., the ΔΔGb values for each RT-NNRTI set were well compatible with the in vitro resistance data described above, suggesting that our modeling appropriately reflects the actual mode of binding between the RT molecule and the NNRTI.
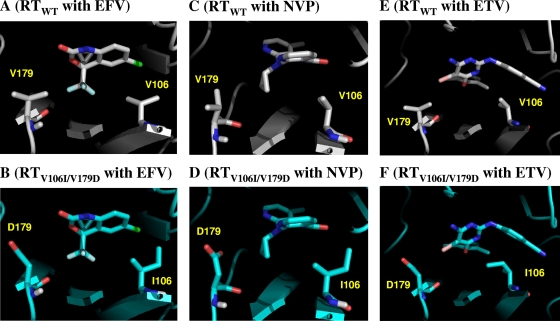
In these models, EFV and NVP were predicted to bind to the hydrophobic pocket of RT, as demonstrated in the crystal structures (20, 26). In the wild-type RT, V106 and V179 contributed to the stabilization of the binding of EFV and NVP through hydrophobic interactions (Fig. 3A and C). However, the V106I and V179D mutations attenuated this stabilization by the following mechanisms. In the case of EFV, the V106I mutation caused a steric clash with the chlorine atom on one side of EFV and the V179D mutation caused electrostatic repulsion on the other side of EFV between the carbonyl oxygen atom of EFV and the carboxyl oxygen atoms of D179 in RT (Fig. 3B). In the case of NVP, the V106I mutation caused a steric clash with the aromatic ring of NVP, whereas the V179D mutation reduced hydrophobic contacts with NVP and the charged carboxyl atoms of D179 showed unfavorable contacts with the hydrophobic three-member ring of NVP (Fig. 3D). The effect of a single mutation on binding affinity was relatively moderate because a slight positional shift in EFV and NVP reduced unfavorable contacts. However, V106I V179D double mutations coincidentally caused repulsive interactions at the distinct sites of EFV and NVP which significantly attenuated the affinity of EFV and NVP for RT (Fig. 3B and D).
FIG. 3.
Interactions of NNRTIs with the 106th and 179th residues of RTs. Interaction sites of NNRTIs and RTs in the models are shown. (A) Wild-type RT (RTWT) with EFV. (B) RTV106I/V179D with EFV. (C) RTWT with NVP. (D) RTV106I/V179D with NVP. (E) RTWT with ETV. (F) RTV106I/V179D with ETV. In the RTWT and RTV106I/V179D models, carbon atoms appear gray and cyan, respectively. NNRTIs and the 106th and 179th residues are highlighted by the stick configuration. Blue sticks, nitrogen; red sticks, oxygen; light blue, fluorine; light green, chlorine; pink sticks, bromine atoms.
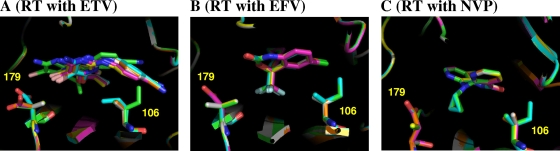
ETV has rotatable bonds that link aromatic rings. Therefore, it is conceivable that conformational plasticity allowed fine-tuning of ETV conformations for stable binding (Fig. 3E and F). In fact, superposition of the structural models of ETV-RT complexes showed that ETV changed its conformation and position depending on the mutations (Fig. 4A). In contrast, the conformations of EFV and NVP were more rigid due to a lack of rotatable bonds. Therefore, the conformations of EFV and NVP remained similar with various mutant RTs (Fig. 4B and C). These results suggest that the plasticity of the conformation of ETV plays a key role in maintaining its binding affinity for various mutant RTs, as reported in the crystal structural study (4).
FIG. 4.
Superposition of RTWT and five mutant models. (A) RTs with ETV. (B) RTs with EFV. (C) RTs with NVP. NNRTIs and the 106th and 179th residues are highlighted by the stick configuration. White sticks, RTWT; yellow, RTV106A; green, RTV106I; orange, RTV179D; purple, RTV106A/V179D; cyan, RTV106I/V179D.
DISCUSSION
The results of the present study indicated that the combination of two polymorphic mutations, V106I and V179D, alters the susceptibility of HIV-1 to EFV, as demonstrated in the resistance selection experiments (Fig. 1). Analysis of the recombinant monoclonal HIV-1 strains revealed that the combination confers significant resistance to EFV and NVP but not to ETV, although each mutation alone could not alter NNRTI susceptibility (Table 2). Furthermore, one clinical HIV-1 isolate from a treatment-naïve patient who harbored V106I and V179D without any other resistance-associated mutation showed significant resistance to EFV and NVP but not to ETV (Table 3). In a previous study, Tee et al. (29) analyzed HIV-1 RT and protease sequences in 36 antiretroviral-treated patients with detectable viral loads but they could not find any known resistance-associated mutation in 8 patients. In one of their patients on EFV treatment (04MYKL1665), V106I and V179D coexisted in the HIV-1 RT according to GenBank (accession no. AY960901; accessed in October 2009). In a clinical trial of tipranavir, the HIV-1 isolate from one patient (case 48-1084), who experienced NNRTI-treatment failure, harbored V106I and V179D without any other NNRTI resistance-associated mutations (DQ880530) (2). These data strongly indicate that the combination of V106I and V179D also confers significant resistance to NNRTIs in vivo.
Structural modeling indicated that V106I and V179D cooperatively reduce NNRTI binding to EFV and NVP. ETV, however, exhibits structural plasticity and can avoid any disturbance caused by the combination of V106I and V179D. This specific structure probably contributes to the efficacy of ETV against many NNRTI-resistant HIV-1 strains, resulting in an excellent rate of response to ETV-containing salvage treatment (18, 21).
Both V106I and V179D are listed as minor ETV resistance-associated mutations in the current version of the IAS-USA Drug Resistance Mutation List (14), but both are not recognized as EFV and NVP resistance-associated mutations. They are often identified individually but rarely coexist in treatment-naïve individuals (Table 1). The combination of V106I and V179D, however, can be found in patients whose baseline HIV-1 held either V106I or V179D after failure of EFV- or NVP-containing treatment (2, 29). Considering that either V106I or V179D was identified in a significant portion of treatment-naïve patients (29/364; 8%) (Table 1), the above information on NNRTI resistance caused by the mutation combination should be recognized by all clinical specialists involved in the interpretation of genotype drug resistance tests and those physicians responsible for changing antiretroviral treatment regimens. In a previous study, we selected EFV-resistant HIV-1 by culture of monoclonal HIV-1 harboring another common polymorphic mutation, K103R (HIV-1K103R), and found the additional emergence of V179D; we then confirmed that the combination of K103R and V179D conferred significant resistance to EFV and NVP (7). Considering these findings together, one assumes that the combinations of polymorphic mutations can reduce NNRTI susceptibility and that other combinations of polymorphic mutations can confer NNRTI resistance. Furthermore, mutations found to be important for one drug may actually have a greater effect on other drugs of the same class. Even polymorphic and minor resistance mutations should be considered carefully when interpreting the results of genotype testing.
Supplementary Material
Acknowledgments
This work was supported in part by a Grant-in-Aid for AIDS Research from the Ministry of Health, Labor, and Welfare (H20-AIDS-002) and the Global Center of Excellence Program (Global Education and Research Center Aiming at the Control of AIDS) from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Published ahead of print on 1 February 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski.2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers.2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, I. F., I. Pandrea, P. A. Marx, and C. Apetrei.2007. HIV genetic diversity: biological and public health consequences. Curr. HIV Res. 5:23-45. [DOI] [PubMed] [Google Scholar]
- 4.Das, K., A. D. Clark, Jr., P. J. Lewi, J. Heeres, M. R. De Jonge, L. M. Koymans, H. M. Vinkers, F. Daeyaert, D. W. Ludovici, M. J. Kukla, B. De Corte, R. W. Kavash, C. Y. Ho, H. He, M. A. Lichtenstein, K. Andries, R. Pauwels, M. P. De Bethune, P. L. Boyer, P. Clark, S. H. Hughes, P. A. Janssen, and E. Arnold.2004. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 47:2550-2560. [DOI] [PubMed] [Google Scholar]
- 5.Duan, Y., C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. Kollman.2003. A point-charged force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24:1999-2012. [DOI] [PubMed] [Google Scholar]
- 6.Gatanaga, H., D. Das, Y. Suzuki, D. D. Yeh, K. A. Hussain, A. K. Ghosh, and H. Mitsuya.2006. Altered HIV-1 Gag protein interactions with cyclophilin A (CypA) on the acquisition of H219Q and H219P substitutions in the CypA binding loop. J. Biol. Chem. 281:1241-1250. [DOI] [PubMed] [Google Scholar]
- 7.Gatanaga, H., A. Hachiya, S. Kimura, and S. Oka.2006. Mutations other than 103N in human immunodeficiency virus type 1 reverse transcriptase (RT) emerge from K103R polymorphism under non-nucleoside RT inhibitor pressure. Virology 344:354-362. [DOI] [PubMed] [Google Scholar]
- 8.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya.2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 9.Hachiya, A., S. Aizawa-Matsuoka, M. Tanaka, Y. Takahashi, S. Ida, H. Gatanaga, Y. Hirabayashi, A. Kojima, M. Tatsumi, and S. Oka.2001. Rapid and simple phenotype assay for drug susceptibility of human immunodeficiency virus type 1 using CCR5-expressing HeLa/CD4(+) cell clone 1-10 (MAGIC-5). Antimicrob. Agents Chemother. 45:495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hachiya, A., H. Gatanaga, E. Kodama, M. Ikeuchi, M. Matsuoka, S. Harada, H. Mitsuya, S. Kimura, and S. Oka.2004. Novel patterns of nevirapine resistance-associated mutations of human immunodeficiency virus type 1 in treatment-naïve patients. Virology 327:215-224. [DOI] [PubMed] [Google Scholar]
- 11.Hachiya, A., E. N. Kodama, S. G. Sarafianos, M. M. Schuckmann, Y. Sakagami, M. Matsuoka, M. Takiguchi, H. Gatanaga, and S. Oka.2008. Amino acid mutation N348I in connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 82:3261-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachiya, A., S. Matsuoka-Aizawa, K. Tsuchiya, H. Gatanaga, S. Kimura, M. Tatsumi, and S. Oka.2003. “All-in-One Assay,” a direct phenotypic anti-human immunodeficiency virus type 1 drug resistance assay for three-drug combination therapies that takes into consideration in vivo drug concentrations. J. Virol. Methods 111:43-53. [DOI] [PubMed] [Google Scholar]
- 13.Holguín, A., and V. Soriano.2002. Resistance to antiretroviral agents in individuals with HIV-1 non-B subtypes. HIV Clin. Trials 3:403-411. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Scapiro, and D. D. Richman.2009. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 17:138-145. [PubMed] [Google Scholar]
- 15.Kantor, R.2006. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr. Opin. Infect. Dis. 19:594-606. [DOI] [PubMed] [Google Scholar]
- 16.Kodama, E. I., S. Kohgo, K. Kitano, H. Machida, H. Gatanaga, S. Shigeta, M. Matsuoka, H. Ohrui, and H. Mitsuya.2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob. Agents Chemother. 45:1539-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kosalaraksa, P., M. F. Kavlick, V. Maroun, R. Le, and H. Mitsuya.1999. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J. Virol. 73:5356-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazzarin, A., T. Campbell, B. Clotet, M. Johnson, C. Katlama, A. Moll, W. Towner, B. Trottier, M. Peeters, J. Vingerhoets, G. De Smedt, B. Baeten, G. Beets, R. Sinha, B. Woodfall, and the DUET-2 Study Group.2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomized, double-blind, placebo-controlled trial. Lancet 370:39-48. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. C., and Y. Duan.2004. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized Born solvent model. Proteins 55:620-634. [DOI] [PubMed] [Google Scholar]
- 20.Lindberg, J., S. Sigurdsson, S. Lowgren, H. O. Andersson, C. Sahlberg, R. Noreen, K. Fridborg, H. Zhang, and T. Unge.2002. Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant. Eur. J. Biochem. 269:1670-1677. [DOI] [PubMed] [Google Scholar]
- 21.Madruga, J. V., P. Cahn, B. Grinsztejn, R. Haubrich, J. Lalezari, A. Mills, G. Pialoux, T. Wilkin, M. Peeters, J. Vingerhoets, G. De Smedt, L. Leopold, R. Trefiglio, B. Woodfall, and the DUET-1 Study Group.2007. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomized, double-blind, placebo-controlled trial. Lancet 370:29-38. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Cajas, J. L., N. Pant-Pai, M. B. Klein, and M. A. Wainberg.2008. Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: a systemic review of virologic and biochemical evidence. AIDS Rev. 10:212-223. [PubMed] [Google Scholar]
- 23.McBurney, S. P., and T. M. Ross.2008. Viral sequence diversity: challenges for AIDS vaccine designs. Expert Rev. Vaccines 7:1405-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onufriev, A., D. Bashford, and D. A. Case.2004. Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins 55:383-394. [DOI] [PubMed] [Google Scholar]
- 25.Quan, Y., B. G. Brenner, R. G. Marlink, M. Essex, T. Kurimura, and M. A. Wainberg.2003. Drug resistance profiles of recombinant reverse transcriptase from human immunodeficiency virus type 1 subtypes A/E, B, and C. AIDS Res. Hum. Retroviruses 19:743-753. [DOI] [PubMed] [Google Scholar]
- 26.Ren, J., R. Esnouf, E. Garman, D. Somers, C. Ross, I. Kerby, J. Keeling, G. Darby, Y. Jones, D. Stuart, and D. Stammers.1995. High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat. Struct. Biol. 2:293-302. [DOI] [PubMed] [Google Scholar]
- 27.Shafer, R. W., and J. M. Schapiro.2008. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 10:67-84. [PMC free article] [PubMed] [Google Scholar]
- 28.Tebit, D. M., I. Nankya, E. J. Arts, and Y. Gao.2007. HIV diversity, recombination and disease progression: how does fitness “fit” into the puzzle? AIDS Rev. 9:75-87. [PubMed] [Google Scholar]
- 29.Tee, K. K., A. Kamarulzaman, and K. P. Ng.2006. Prevalence and pattern of drug resistance mutations among antiretroviral-treated HIV-1 patients with suboptimal virological response in Malaysia. Med. Microbiol. Immunol. 195:107-112. [DOI] [PubMed] [Google Scholar]
- 30.Wang, J., R. M. Wolf, J. W. Caldwell, P. A. Kollman, and D. A. Case.2004. Development and testing of a general amber force field. J. Comput. Chem. 25:1157-1174. [DOI] [PubMed] [Google Scholar]
- 31.Yoshimura, K., R. Feldman, E. Kodama, M. F. Kavlick, Y. L. Qiu, J. Zemlicka, and H. Mitsuya.1999. In vitro induction of human immunodeficiency virus type 1 variants resistant to phosphoralaninate prodrugs of Z-methylenecyclopropane nucleoside analogues. Antimicrob. Agents Chemother. 43:2479-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.