Abstract
Plasmid pB1000 is a mobilizable replicon bearing the blaROB-1 β-lactamase gene that we have recently described in Haemophilus parasuis and Pasteurella multocida animal isolates. Here we report the presence of pB1000 and a derivative plasmid, pB1000′, in four Haemophilus influenzae clinical isolates of human origin. Pulsed-field gel electrophoresis showed unrelated patterns in all strains, indicating that the existence of pB1000 in H. influenzae isolates is not the consequence of clonal dissemination. The replicon can be transferred both by transformation and by conjugation into H. influenzae, giving rise to recipients resistant to ampicillin and cefaclor (MICs, ≥64 μg/ml). Stability experiments showed that pB1000 is stable in H. influenzae without antimicrobial pressure for at least 60 generations. Competition experiments between isogenic H. influenzae strains with and without pB1000 revealed a competitive disadvantage of 9% per 10 generations for the transformant versus the recipient. The complete nucleotide sequences of nine pB1000 plasmids from human and animal isolates, as well as the epidemiological data, suggest that animal isolates belonging to the Pasteurellaceae act as an antimicrobial resistance reservoir for H. influenzae. Further, since P. multocida is the only member of this family that can colonize both humans and animals, we propose that P. multocida is the vehicle for the transport of pB1000 between animal- and human-adapted members of the Pasteurellaceae.
Haemophilus influenzae is a Gram-negative rod usually involved in community-acquired respiratory tract infections, acute otitis media, acute sinusitis, exacerbation of chronic bronchitis, and childhood meningitis (4, 28). β-Lactam antibiotics are the drugs of choice for the treatment of infectious processes caused by this pathogen (28). In H. influenzae, high-level β-lactam resistance is mediated by the production of β-lactamases. To date, only two β-lactamases have been described for this pathogen: TEM-1 and ROB-1. Both enzymes belong to the class A serine β-lactamases and confer high-level resistance to penicillin and ampicillin. In addition, ROB-1, but not TEM-1, confers high-level resistance to cefaclor (11, 15). Although there are geographical differences, TEM-1 is largely more prevalent than ROB-1 among β-lactamase-positive H. influenzae strains worldwide (8).
Recently, it was reported that all β-lactam-resistant isolates of Haemophilus parasuis and Pasteurella multocida recovered from animals in Spain carried plasmid pB1000, bearing blaROB-1 (25, 26). pB1000 is a mobilizable plasmid belonging to the ColE1 superfamily and the MOBHEN family (10, 14); it confers high-level resistance to ampicillin and cefaclor (25). It is known that animals can play important roles as reservoirs and sources of antimicrobial-resistant pathogens (20, 21), posing an important concern for public health. Not only the zoonotic circulation of resistant bacteria (6, 20, 21) but also the spread of antimicrobial resistance determinants between animal and human pathogens has been widely described (1, 16). Here we characterized four high-level cefaclor-resistant H. influenzae human clinical isolates recovered between 2000 and 2009 in Spain. Three isolates bore plasmid pB1000, and one carried the nearly identical plasmid pB1000′. To better understand the epidemiology of pB1000, sequencing and transfer analysis were performed for human and animal strains of Pasteurellaceae. In addition, the stability and fitness cost of pB1000 in H. influenzae were determined.
(An initial report of this study was presented at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, September 2009.)
MATERIALS AND METHODS
Bacterial strains, culture conditions, and susceptibility testing.
The characteristics of the strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description and characteristicsa | Isolation date (day/mo/yr) | Reference or GenBank accession no. |
|---|---|---|---|
| Strains | |||
| H. influenzae | |||
| BB1050 | Ampr clinical isolate (pB1000′); nontypeable | 22/1/2001 | This work |
| BB1051 | Ampr clinical isolate (pB1000); nontypeable | 22/8/2001 | This work |
| BB1052 | Ampr clinical isolate (pB1000); nontypeable | 31/12/2001 | This work |
| BB1053 | Ampr clinical isolate (pB1000); serotype e | 12/04/2007 | This work |
| BB1054 | Amps clinical isolate; serotype e | 16/6/2007 | This work |
| BB1055 | Amps clinical isolate; serotype e | 30/7/2007 | This work |
| BB1056 | Amps clinical isolate; serotype e | 30/11/2007 | This work |
| Rd KW20 | Amps recipient strain for pB1000 in electroporation | NC_000907 | |
| Rd KW20 Tetr | Amps Tetr recipient strain for pB1000 in conjugation (p9956) | This work | |
| P. multocida | |||
| ATCC 43137 | Amps recipient strain for pB1000 in electroporation | Collectionb | |
| BB1057 | Amps Tetr recipient strain for pB1000 in conjugation | This work | |
| BB1034 | Swine clinical isolate (pB1000); farm 1 | 12/3/2003 | 26 |
| BB1037 | Swine clinical isolate (pB1000); farm 2 | 18/12/2003 | 26 |
| BB1038 | Swine clinical isolate (pB1000); farm 2 | 18/12/2003 | 26 |
| BB1044 | Swine clinical isolate (pB1000); farm 3 | 2/1/2004 | 26 |
| BB1045 | Swine clinical isolate (pB1000); farm 4 | 7/1/2004 | 26 |
| H. parasuis BB1021 | Swine clinical isolate (pB1000); farm 4 | 30/12/2003 | 25 |
| E. coli BB1058(pB1000) | K-12 with the conjugation machinery of an IncP plasmid encoded in the chromosome, transformed with pB1000 | 26 | |
| Plasmids | |||
| pB1000 | mobA mobB mobC blaROB-1; ColE1 superfamily | DQ840517 | |
| pB1000′ | mobA mobB mobC blaROB-1; ColE1 superfamily | GU08006 | |
| p9956 | mobA mobB mobC tet(H) tetR; ColE1 superfamily | AY362554 |
Amp, ampicillin; Tet, tetracycline. Superscript letters “r” and “s” indicate resistance and susceptibility, respectively.
Pasteur Institute Collection; P. multocida CIP103286.
Strains BB1050 and BB1051 were obtained from two adult patients in the Hospital Gregorio Marañón, Madrid, Spain. BB1050 was isolated from a bronchial suction sample in January 2001, and BB1051 was isolated from a telescoped catheter sample in August 2001. Strain BB1052 was recovered from a bronchial suction sample from an 86-year-old man with severe chronic obstructive pulmonary disease (COPD) and acute respiratory symptoms in the Hospital Son Dureta, Majorca Island, Eastern Spain, in December 2001. Finally, BB1053 was recovered in April 2007 in the Hospital General Yagüe, in Burgos, Northern Spain, from a sputum sample from a 54-year-old man. Control ampicillin-susceptible serotype e strains BB1054, BB1055, and BB1056 were isolated in 2007 in different hospitals in Spain. Isolates were identified according to standard microbiological methods (3), serotyped, and confirmed by a molecular capsular typing method (7). H. influenzae and P. multocida were cultured as previously described (4, 26). The susceptibilities of H. influenzae clinical isolates to antibiotics were determined by broth microdilution in microtiter plates (Sensititre Enzyme 13; Trek Diagnostics, Inc., Westlake, OH) according to the CLSI guidelines (5). Antibiotic quality control results for H. influenzae ATCC 49247 and ATCC 49766 were within recommended values according to guidelines for MIC tests (5).
DNA analysis, electroporation, and conjugation experiments.
The identification of pB1000 in resistant P. multocida strains was assessed using PCR mapping as previously described (26). Plasmid DNA extraction, automated sequencing, sequence analysis, and pB1000 stability analysis were performed as described by San Millan et al. in 2009 (26). The molecular epidemiology of the H. influenzae isolates was determined by pulsed-field gel electrophoresis (PFGE) as described previously (12). A genetic similarity dendrogram was obtained according to the Dice correlation coefficient with a tolerance level of 1% (Fingerprinting II software; Bio-Rad). Electrocompetent cells were prepared from strains H. influenzae Rd KW20 and P. multocida ATCC 43137 as described previously (22). Electroporation was performed using the following conditions: 2.5 kV/cm, 25 μF, and 200 Ω for H. influenzae; 2.5 kV/cm, 25 μF, and 800 Ω for P. multocida. Conjugation experiments were carried out in duplicate with BB1058 bearing pB1000 as the donor, as previously described (26). The recipient strains for conjugation were P. multocida BB1057 and H. influenzae Rd KW20 Tetr (Table 1). Transconjugants were selected on plates containing oxytetracycline (10 μg/ml) and ampicillin (25 μg/ml).
Growth curve and growth competition experiments.
Growth kinetics were determined for H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000). Volumes of 25 ml Haemophilus test medium (HTM) and 25 ml HTM with 25 μg/ml of ampicillin were inoculated independently with 107 CFU of each strain. Cultures were grown for 12 h at 100 rpm and 37°C under 5% CO2, and absorbance at 600 nm was measured every hour.
The fitness cost of plasmid pB1000 was determined by four independent competition experiments between H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000), as previously described (9, 18). Strains were grown for 16 h at 37°C, under 5% CO2, in HTM broth. Then 106 CFU of H. influenzae Rd KW20 was mixed with 106 CFU of H. influenzae Rd KW20(pB1000) in 10 ml of antibiotic-free HTM. The mixture was grown at 37°C under 5% CO2 at 100 rpm. A total of 2 × 106 CFU was transferred to 10 ml of fresh HTM every 24 h. Samples were taken every hour during the first 12 h, at 24 h, and then every 24 h for 9 days. For each sample, aliquots were plated onto nonselective chocolate agar, and the proportion of resistant colonies was deduced by replica plating of 100 colonies on chocolate agar plates containing 100 μg/ml of ampicillin. Relative fitness is expressed as the competition index (CI), calculated as the ratio of the mean CFU for four independent competition experiments between the resistant and susceptible strains at a given timepoint (t1) divided by the same ratio at t0 (2). The selection coefficient, s, was calculated as the slope of the linear regression model: s = ln(CI)/ln(d), where d is the dilution factor (18). The selection coefficient estimates the difference between the relative fitnesses of the two competitors over the entire competition experiment.
Nucleotide sequence accession numbers.
The nucleotide sequences of the following plasmids, determined in this study, have been deposited in GenBank under the accession numbers given in parentheses: pB1000 from BB1034 (GU080062), BB1037 (GU080070), BB1038 (GU080069), BB1044 (GU080068), BB1045 (GU080067), BB1051 (GU080063), BB1052 (GU080064), and BB1053 (GU080065) and pB1000′ from BB1050 (GU080066).
RESULTS AND DISCUSSION
Characterization of high-level cefaclor-resistant H. influenzae clinical isolates.
The collection of the Spanish Reference Centre for Haemophilus was surveyed for ROB-1-positive H. influenzae strains isolated between 2000 and 2009. Strains were selected based on their high-level resistance to cefaclor (MIC, ≥8 μg/ml) and were then analyzed by PCR for the presence of blaROB-1 (13). Four H. influenzae blaROB-1-positive clinical isolates were identified; of these, three were nontypeable (BB1050 to BB1052) and one belonged to serotype e (BB1053) (Table 1). The β-lactam resistance profiles of the blaROB-1-positive strains were determined (Table 2). The clinical isolates showed high-level resistance to ampicillin and cefaclor and susceptibility to cefotaxime (Table 2).
TABLE 2.
β-Lactam resistance profiles of H. influenzae strains
| Strain | MIC (μg/ml)a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMX | AMC | AMP | CEC | FUR | CTX | FIX | IMI | AZT | |
| Rd KW20 | 0.5 | ≤0.5 | 0.5 | ≤0.25 | 1 | 0.5 | ≤0.03 | ≤0.03 | 0.5 | ≤0.12 |
| Rd KW20(pB1000) | >256 | >64 | 2 | >128 | 64 | 4 | ≤0.03 | 0.06 | 1 | ≤0.12 |
| BB1050 | >256 | >64 | 4 | >128 | 64 | 4 | 0.06 | 0.06 | 2 | ≤0.12 |
| BB1051 | >256 | >64 | 2 | >128 | 64 | 4 | ≤0.03 | 0.06 | 0.5 | ≤0.12 |
| BB1052 | >256 | >64 | 2 | >128 | 64 | 2 | ≤0.03 | ≤0.03 | 1 | ≤0.12 |
| BB1053 | >256 | >64 | 1 | >128 | 64 | 4 | ≤0.03 | 0.06 | 0.5 | ≤0.12 |
AMC, amoxicillin-clavulanic acid (2:1 ratio); AMP, ampicillin; AMX, amoxicillin; AZT, aztreonam; CEC, cefaclor; FIX, cefixime; CTX, cefotaxime; FUR, cefuroxime; IMI, imipenem; PEN, penicillin.
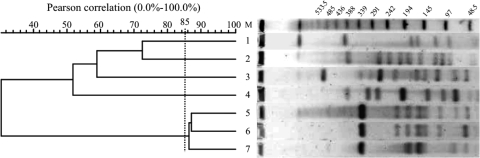
The molecular relatedness of the H. influenzae isolates was determined by PFGE (Fig. 1). The resulting profiles showed that the blaROB-1-carrying strains were not genetically related. Interestingly, BB1053 was highly related to two ampicillin-susceptible clinical isolates of serotype e, BB1054 and BB1056.
FIG. 1.
PFGE fingerprint patterns and genetic similarity dendrogram of H. influenzae clinical isolates. Lane 1, BB1051; lane 2, BB1050; lane 3, BB1054; lane 4, BB1052; lane 5, BB1053; lane 6, BB1056; lane 7, BB1055; lane M, molecular size marker. Numbers above the bands are molecular sizes (in kilobases). PFGE patterns with coefficients of similarity greater than 85% were considered to define a particular clone. Three ampicillin-susceptible clinical H. influenzae strains of serotype e (BB1054 to BB1056), isolated from three different hospitals in Spain in the same year as BB1053, were included for comparison (Table 1).
Plasmids pB1000 and pB1000′ confer β-lactam resistance on H. influenzae.
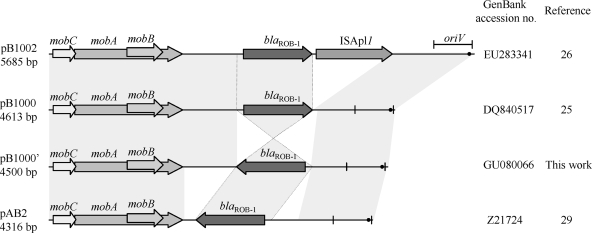
To identify the genetic platforms bearing blaROB-1, the plasmid contents of the four H. influenzae strains were analyzed. Plasmid preparation and plasmid digestion profiles with the PstI and XbaI endonucleases, as well as blaROB-1 inverted-PCR analysis (26) of BB1050 to BB1053, were compatible with the presence of plasmid pB1000 (data not shown). PCR mapping of pB1000 was performed as previously described (26) and confirmed the presence of pB1000 in isolates BB1051 to BB1053. In contrast, for BB1050, plasmid mapping was not conclusive at first, but further PCR analysis showed that the plasmid carried by this strain, pB1000′, had the same structure as pB1000 but an inverted blaROB-1 gene (Fig. 2). All plasmids were completely sequenced. In BB1051 to BB1053, the pB1000 plasmids were 4,613 bp long, with a nucleotide identity of ≥99.9% among them and a nucleotide identity of ≥99.8% with the first pB1000 sequence from H. parasuis (25) (Table 1). Plasmid pB1000′ from BB1050 has 4,500 bp and a backbone identical to that of pB1000 (Fig. 2), although blaROB-1 is inverted compared to that from pB1000. The nucleotide sequences of blaROB-1 were identical in the four plasmids, as were the sequences of the remaining genes, mobA, mobB, and mobC. pB1000 and pB1000′ were compared with plasmid pB1002 from P. multocida and plasmid pAB2 from Mannheimia haemolytica (29), both bearing blaROB-1 β-lactamase (Fig. 2). These four replicons, reported for different members of the family Pasteurellaceae, are very similar, sharing the same backbone, with the origin of replication and the mobABC genes corresponding to those of the MOBHEN family. The variable region of each plasmid bears the blaROB-1 gene. Although blaROB-1 is arranged in different orientations in the plasmids, the genetic environments upstream and downstream of this gene are identical in all of them (Fig. 2). In pB1002, the insertion sequence ISApl1 is located downstream of blaROB-1. This gene was recently reported for the first time in Actinobacillus pleuropneumoniae (27). Whether ISApl1 is involved in the mobilization of this β-lactamase is currently under study.
FIG. 2.
Schematic representation of blaROB-1-carrying plasmids described for members of the family Pasteurellaceae. The reading frames for genes are shown as arrows, with arrowheads pointing in the direction of transcription. Regions with more than 99% identity are shaded. Two vertical bars bracket the region containing the putative origin of replication (oriV). A black dot indicates the putative transfer origin (oriT). Dotted lines connect the blaROB-1 cassettes in the different plasmids.
Fitness cost of pB1000 in H. influenzae.
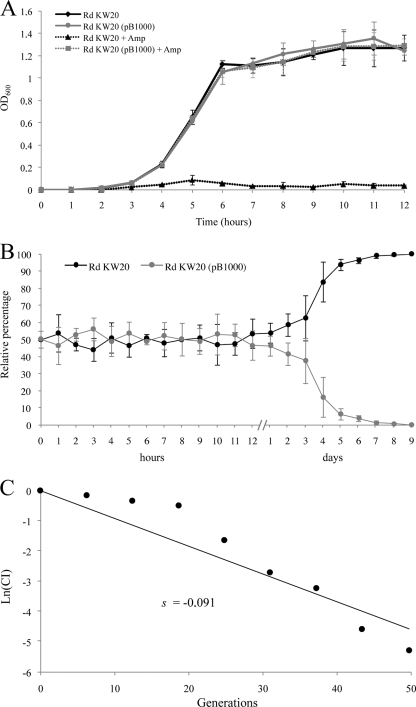
The growth kinetics of H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000) in the absence and in the presence of ampicillin (25 μg/ml) were determined (Fig. 3A). Using these isogenic strains, the biological cost of pB1000 was assessed. Growth curves showed no significant difference for the strains in the absence of selective pressure. In the presence of ampicillin, as expected, the strain lacking pB1000 was not able to grow, whereas the growth of H. influenzae Rd KW20(pB1000) showed no significant difference at any point of the assay from that of the strains growing in the absence of ampicillin. Hence, the growth kinetics obtained over 12 h showed no evidence of any fitness cost exacted by pB1000.
FIG. 3.
Fitness cost of pB1000 in H. influenzae. (A) Comparison of the growth kinetics of strains H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000) in the absence and presence of ampicillin. Error bars represent the standard deviations of the means. Growth curves were constructed with the average results from three independent experiments. (B) Growth competition between H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000). The initial ratio was 1:1. The relative percentage of each strain at different time points is shown. Competition curves were constructed with the average results of four independent experiments. (C) The selection coefficient (s) was calculated from the competition experiment. s is the slope of the linear regression model, ln(CI)/ln(d), where CI is the ratio between the CFU counts of the resistant and susceptible populations at t1 divided by the same ratio at t0, and d is the dilution factor.
Competition experiments offer a more discriminative and precise measurement of fitness, since they reflect the competitive disadvantage during all the phases of the growth cycle and in several consecutive cycles (9). A volume of 10 ml of HTM was inoculated with H. influenzae Rd KW20 and H. influenzae Rd KW20(pB1000), and 2 × 106 CFU from this mixture was transferred to 10 ml fresh HTM every 24 h for 9 days. The proportion of bacteria carrying pB1000 was monitored every hour for the first 12 h. For this period, consistent with the growth curve, the ratio was almost 1:1 at every point (Fig. 3B). Measurements were made at the 24-h point and then every 24 h until day 9. From day 2 on, a constant decrease in the proportion of pB1000-carrying strains was observed. At day 9, all the colonies tested were pB1000 free (Fig. 3B). The selection coefficient (s) was established to estimate the difference in relative fitness between the strains in competition over the entire experiment. H. influenzae Rd KW20 bearing pB1000 presented a competitive disadvantage of ca. 9% per 10 generations relative to H. influenzae Rd KW20. This fitness cost could be responsible for the low frequency of ROB-1 relative to TEM-1 in H. influenzae, since blaROB-1 is associated with pB1000-like plasmids. Other factors may be involved, such as the fitness of TEM-1, or the probable coselection of blaTEM-1 by other antimicrobial resistance determinants present in the large genetic platforms where it is encoded (17, 28). Further reports on blaROB-1, or even on pB1000, will be of great interest for determining the importance of this plasmid in β-lactam resistance in H. influenzae.
Epidemicity and interspecies mobilization of plasmid pB1000.
Plasmid pB1000 has been reported in clinical samples from three different species belonging to the family Pasteurellaceae (25, 26). However, pB1000 was shown to be largely unstable when hosted in Escherichia coli (26). Hence, pB1000 seems to be adapted to the family Pasteurellaceae and to be able to spread within it. To analyze the dissemination of this plasmid, transfer analysis of pB1000 by transformation and conjugation to P. multocida and H. influenzae was performed. Transformation by electroporation was successful for both H. influenzae Rd KW20 and P. multocida ATCC 43137 (Table 1). For conjugation, E. coli BB1058 with pB1000, maintained with selective pressure, was used as the donor strain. Transconjugants were obtained at similar frequencies, between 10−3 and 10−4 colonies per donor CFU, for both recipients, H. influenzae and P. multocida (Table 1). Transformants and transconjugants were checked for the presence of pB1000 by PCR mapping, plasmid preparation, and plasmid digestion profiles with the PstI and XbaI endonucleases. pB1000 was present after 60 generations without selective pressure in the 100 colonies of P. multocida and H. influenzae tested. These results showed that pB1000 could be acquired by transformation and conjugation and could exhibit high stability in the family Pasteurellaceae.
pB1000 in the family Pasteurellaceae.
To date, pB1000 has been reported in three different members of the Pasteurellaceae between 2001 and 2007 (Table 1). The H. influenzae isolates carrying this replicon are unrelated, as shown by pulsotyping and serotyping, and no link has been found between them and animal isolates of Pasteurellaceae carrying pB1000. On the other hand, for P. multocida and H. parasuis, a link has been found in a swine farm where pB1000 transmission could have occurred, since strains of both species carrying pB1000 were isolated during the same week (Table 1). To elucidate the recent evolution of pB1000 within the family Pasteurellaceae, this plasmid has been completely sequenced from a total of nine strains, a representative sample of the three species. The pB1000 plasmids analyzed were those of H. influenzae isolates BB1051, BB1052, and BB1053, five P. multocida strains (26), and the H. parasuis clone (25) (Table 1). The nucleotide identity of the pB1000 plasmids in the different isolates was 98.8% to 100%. For P. multocida, four out of five strains were clonally related, and the plasmids showed identical nucleotide sequences. In one P. multocida strain, pB1000 showed the most divergent sequence from the eight others. Thus, the highest degree of variability in the pB1000 sequence appeared within P. multocida. In H. parasuis, this plasmid presented only one nucleotide difference from the sequences of the plasmids in the majority of P. multocida strains. Interestingly, in H. influenzae, the three plasmids sequenced showed 99.9% nucleotide identity, even if the strains were isolated within a 7-year interval. These results point to a parallel evolution of pB1000 in animal and human isolates of the Pasteurellaceae in recent years, after the transmission of this plasmid between them.
In vivo and in vitro data confirm the interspecies spread of pB1000 within the family Pasteurellaceae. For animal pathogens, this spread could be easily understood, since multiple species of this family cohabit in the respiratory tracts of animals (19). However, transfer of pB1000 between animal isolates of Pasteurellaceae and H. influenzae is less evident. Interestingly, the only species from the family Pasteurellaceae with a broad host range, producing both animal diseases and human respiratory tract infections, is P. multocida (24). In addition, pB1000 sequence analysis and epidemiological data indicate that this plasmid could be native to P. multocida. Hence, we propose that P. multocida acts as the hinge conveying pB1000 between animal isolates of Pasteurellaceae and H. influenzae. These results support the previous suggestion (23, 24) that animal isolates of Pasteurellaceae could act as an antimicrobial resistance reservoir for H. influenzae.
Acknowledgments
N. Montero and M. Arroyo are acknowledged for excellent technical assistance. P. Courvalin, M. C. Foucault, and N. Garcia are acknowledged for helpful discussions. We thank O. Cortes for excellent assistance in genetic analysis and Juncal Garmendia for strain RdKW20. We are very grateful to clinical microbiologists of the Spanish hospitals Gregorio Marañón (Madrid), Son Dureta (Mallorca), and General Yagüe (Burgos) for kindly providing isolates.
A.S.M., J.A.E., and L.H. acknowledge the Ministry of Science and Innovation (MICINN), the Universidad Complutense de Madrid, and the Comunidad de Madrid for their respective fellowships. S.G.-C. is a recipient of a fellowship from the Instituto de Salud Carlos III (reference 05/0033). B.G.-Z. acknowledges WP29 of the Med-Vet-Net Network of Excellence (FOOD-CT-2004-506122) and the MICINN (GEN2006-27767-E/PAT). J.C. acknowledges research grants from the REIPI Network (reference RD06/0008/0023) and the Network of Excellence GRACE (PL 518226).
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Ajiboye, R. M., O. D. Solberg, B. M. Lee, E. Raphael, C. Debroy, and L. W. Riley.2009. Global spread of mobile antimicrobial drug resistance determinants in human and animal Escherichia coli and Salmonella strains causing community-acquired infections. Clin. Infect. Dis. 49:365-371. [DOI] [PubMed] [Google Scholar]
- 2.Björkman, J., and D. I. Andersson.2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist. Updat. 3:237-245. [DOI] [PubMed] [Google Scholar]
- 3.Campos, J., and J. A. Sáez-Nieto.2001. Gram-negative infections: Haemophilus and other clinically relevant gram-negative coccobacilli, p. 557 to 580. In N. Cimolai (ed.), Laboratory diagnosis of bacterial infections. Marcel Dekker, Inc., New York, NY.
- 4.Campos, J., M. Hernando, F. Roman, M. Perez-Vazquez, B. Aracil, J. Oteo, E. Lázaro, F. de Abajo, and Group of Invasive Haemophilus Infections of the Autonomous Community of Madrid, Spain.2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute.2009. Performance standards for antimicrobial susceptibility testing, 19th ed. Approved standard M100-S19. CLSI, Wayne, PA.
- 6.Escudero, J. A., A. San Millan, A. Catalan, A. G. de la Campa, E. Rivero, G. Lopez, L. Dominguez, M. A. Moreno, and B. Gonzalez-Zorn.2007. First characterization of fluoroquinolone resistance in Streptococcus suis. Antimicrob. Agents Chemother. 51:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falla, T. J., D. W. Crook, L. N. Brophy, D. Maskell, J. S. Kroll, and E. R. Moxon.1994. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 32:2382-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham.2005. Global distribution of TEM-1 and ROB-1 β-lactamases in Haemophilus influenzae. J. Antimicrob. Chemother. 56:773-776. [DOI] [PubMed] [Google Scholar]
- 9.Foucault, M. L., P. Courvalin, and C. Grillot-Courvalin.2009. Fitness cost of VanA-type vancomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francia, M. V., A. Varsaki, M. P. Garcillan-Barcia, A. Latorre, A. Drainas, and F. de la Cruz.2004. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28:79-100. [DOI] [PubMed] [Google Scholar]
- 11.Galán, J. C., M. I. Morosini, M. R. Baquero, M. Reig, and F. Baquero.2003. Haemophilus influenzae blaROB-1 mutations in hypermutagenic ΔampC Escherichia coli conferring resistance to cefotaxime and beta-lactamase inhibitors and increased susceptibility to cefaclor. Antimicrob. Agents Chemother. 47:2551-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Cobos, S., J. Campos, E. Lázaro, F. Román, E. Cercenado, C. García-Rey, M. Pérez-Vázquez, J. Oteo, and F. de Abajo.2007. Ampicillin-resistant non-β-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob. Agents Chemother. 51:2564-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Cobos, S., J. Campos, E. Cercenado, F. Román, E. Lázaro, M. Pérez-Vázquez, F. de Abajo, and J. Oteo.2008. Antibiotic resistance in Haemophilus influenzae decreased, except for beta-lactamase-negative amoxicillin-resistant isolates, in parallel with community antibiotic consumption in Spain from 1997 to 2007. Antimicrob. Agents Chemother. 52:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcillán-Barcia, M. P., M. V. Francia, and F. de la Cruz.2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 15.Karlowsky, J. A., G. Verma, G. G. Zhanel, and D. J. Hoban.2000. Presence of ROB-1 beta-lactamase correlates with cefaclor resistance among recent isolates of Haemophilus influenzae. J. Antimicrob. Chemother. 45:871-875. [DOI] [PubMed] [Google Scholar]
- 16.Kruse, H., and H. Sørum.1994. Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leaves, N. I., I. Dimopoulou, I. Hayes, S. Kerridge, T. Falla, O. Secka, R. A. Adegbola, M. P. Slack, T. E. Peto, and D. W. Crook.2000. Epidemiological studies of large resistance plasmids in Haemophilus. J. Antimicrob. Chemother. 45:599-604. [DOI] [PubMed] [Google Scholar]
- 18.Lenski, R. E.1991. Quantifying fitness and gene stability in microorganisms. Biotechnology 15:173-192. [DOI] [PubMed] [Google Scholar]
- 19.Livrelli, V. O., A. Darfeuille-Richaud, C. D. Rich, B. H. Joly, and J. L. Martel.1988. Genetic determinant of the ROB-1 beta-lactamase in bovine and porcine Pasteurella strains. Antimicrob. Agents Chemother. 32:1282-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd, D. H.2007. Reservoirs of antimicrobial resistance in pet animals. Clin. Infect. Dis. 45:148-152. [DOI] [PubMed] [Google Scholar]
- 21.Maidhof, H., B. Guerra, S. Abbas, H. M. Elsheikha, T. S. Whittam, and L. Beutin.2002. A multiresistant clone of Shiga toxin-producing Escherichia coli O118:[H16] is spread in cattle and humans over different European countries. Appl. Environ. Microbiol. 68:5834-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz.2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71:3454-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medeiros, A. A., R. Levesque, and G. A. Jacoby.1986. An animal source for the ROB-1 beta-lactamase of Haemophilus influenzae type b. Antimicrob. Agents Chemother. 29:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenau, A., A. Labigne, F. Escande, P. Courcoux, and A. Philippon.1991. Plasmid-mediated ROB-1 beta-lactamase in Pasteurella multocida from a human specimen. Antimicrob. Agents Chemother. 35:2419-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Millan, A., J. A. Escudero, A. Catalán, S. Nieto, F. Farelo, M. Gibert, M. A. Moreno, L. Dominguez, and B. Gonzalez-Zorn.2007. β-Lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob. Agents Chemother. 51:2260-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.San Millan, A., J. A. Escudero, B. Gutierrez, L. Hidalgo, N. Garcia, M. Llagostera, L. Dominguez, and B. Gonzalez-Zorn.2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob. Agents Chemother. 53:3399-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tegetmeyer, H. E., S. C. Jones, P. R. Langford, and N. Baltes.2008. ISApl1, a novel insertion element of Actinobacillus pleuropneumoniae, prevents ApxIV-based serological detection of serotype 7 strain AP76. Vet. Microbiol. 128:342-353. [DOI] [PubMed] [Google Scholar]
- 28.Tristram, S., M. R. Jacobs, and P. C. Appelbaum.2007. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 20:368-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood, A. R., F. A. Lainson, F. Wright, G. D. Baird, and W. Donachie.1995. A native plasmid of Pasteurella haemolytica serotype A1: DNA sequence analysis and investigation of its potential as a vector. Res. Vet. Sci. 58:163-168. [DOI] [PubMed] [Google Scholar]