Abstract
An Aeromonas allosaccharophila environmental isolate recovered from the Seine River (Paris, France) produced a novel extended-spectrum β-lactamase, PER-6, that shared 92% amino acid identity with the closest ß-lactamase, PER-2. The kinetic properties of PER-6 showed a slightly increased affinity for carbapenems. The blaPER-6 gene was chromosomally located and bracketed by non-transposon-related structures.
Aeromonas species are ubiquitous in aquatic environments. Formerly considered strict fish pathogens, Aeromonas spp. are reported increasingly as emerging human pathogens for both immunocompetent and immunocompromised patients as a source of gastroenteritis, bacteremia, meningitis, and skin and soft tissue infections (10).
Extended-spectrum β-lactamases (ESBLs) have rarely been reported to occur in Aeromonas species, with TEM-24 and CTX-M-1 identified in Aeromonas hydrophila (6, 24) and PER-1 in Aeromonas media (18). Although β-lactamases of the PER type are not the most common ESBLs (15) identified, they have been reported increasingly, with PER-1 being detected mainly in Europe, especially in Turkey (20) and in Italy (17). It has been also identified in Korea in Providencia spp. (12) and more recently in Algeria in Proteus vulgaris and Providencia stuartii (8). β-Lactamase PER-2, which is distantly related to PER-1 (86% amino acid identity), has been reported to occur in Enterobacteriaceae in South America, mostly in Argentina (2, 23). The PER-3, -4, and -5 β-lactamases, which are closely related to PER-1 (99% amino acid identity), have been identified in Aeromonas punctata, P. vulgaris, and Acinetobacter baumannii, according to data available in GenBank (accession no. AY740681, EU748544, and EU687473, respectively).
Aeromonas spp. are Gram-negative, mostly environmental species frequently containing plasmids and integrons with multiple genes for antibiotic resistance (9). Aeromonas spp. have been shown to be the source of plasmid-mediated resistance to quinolones of the Qnr type associated with novel genetic elements (3, 19).
The blaPER-1 gene has been identified mostly as part of a Tn1213 composite transposon (20). The blaPER-2 gene has been identified on a large self-conjugative plasmid downstream of the ISPa12 tnpA gene in Citrobacter freundii (23).
The present study was initiated by isolation of an Aeromonas allosaccharophila isolate from a water sample of the Seine River in Paris in January 2009, as a result of screening for multidrug-resistant isolates. The sampling procedure consisted of filtrating 100 ml of water through nitrocellulose membranes (0.45 μm; Millipore, Molsheim, France), resuspending the filters in 1 ml of sterile water, and plating 100-μl aliquots on ceftazidime (2 μg/ml)-containing MacConkey agar plates. Isolate AL-1 was identified by the API 32GN system (bioMérieux, Marcy-l'Etoile, France) and by sequencing of the 16S rRNA genes. MICs were determined by Etest (AB Biodisk, Solna, Sweden) and by the agar dilution method with Mueller-Hinton agar plates for imipenem and imipenem clavulanate and were interpreted according to the CLSI guidelines (4). A. allosaccharophila AL-1 was resistant or reduced in susceptibility to amoxicillin, ticarcillin, ceftazidime, and cefotaxime (Table 1). A synergy image between aztreonam or ceftazidime and clavulanic acid-containing disks suggested expression of an ESBL gene. Standard PCR amplification experiments performed with primers specific for the genes encoding β-lactamases TEM, SHV, PER-1, VEB, and GES (7) failed. Shotgun cloning using EcoRI-restricted genomic DNA and EcoRI-restricted pBKCMV plasmid (Invitrogen, Life Technologies, Cergy- Pontoise, France) was performed as previously described (16). Selection on amoxicillin (100 μg/ml)- and kanamycin (30 μg/ml)-agar plates yielded a recombinant Escherichia coli DH10B(pEco-1) clone expressing an ESBL phenotype. The cloned insert of 4,716 bp was sequenced by using universal primers T3 and T7 and the primers listed in Table 2. Recombinant E. coli(pEco-1) expressed a novel β-lactamase, PER-6 (www.lahey.org/Studies/). PER-6 shares 92% amino acid identity with PER-2, with the blaPER-6 gene sharing 79% nucleotide identity with the blaPER-1 gene, explaining the lack of PCR detection with the use of the blaPER-1-specific primers. The G+C content of the blaPER-6 gene (45.5%) was much lower than that of the A. allosaccharophila genes (56.9%) (5), suggesting that the blaPER-6 gene in A. allosaccharophila Al-1 originated from another bacterial species. Transfer of the β-lactam resistance marker by conjugation or by transformation as described previously (21) from A. allosaccharophila AL-1 to an E. coli reference strain failed. Plasmid extraction performed as described previously (11) identified four plasmids of ca. 5, 12, 22, and 160 kb. None of these plasmids gave a hybridization signal with a blaPER-6-specific probe after Southern transfer (Hybond N+; GE Healthcare), suggesting a chromosomal location for the blaPER-6 gene (data not shown). This location was confirmed by restricting total DNA with the I-CeuI enzyme (New England Biolabs, Saint-Quentin-en-Yvelines, France), followed by pulsed-field gel electrophoresis and hybridization with the blaPER-6 and rRNA probes, as described previously (13) (data not shown).
TABLE 1.
MICs of β-lactams for A. allosaccharophila AL-1 and E. coli DH10B harboring recombinant plasmids pPER-1, pPER-2, and pPER-6 and the E. coli DH10B reference strain
| β-Lactam(s) | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| A. allosaccharophila AL-1(PER-6) |
E. coli DH10B harboring: |
E. coli DH10B | |||
| pPER-6a | pPER-1 | pPER-2 | |||
| Amoxicillin | >256 | >256 | >256 | >256 | 4 |
| Amoxicillin-CLAb | 12 | 4 | 4 | 6 | 4 |
| Ticarcillin | >256 | >256 | >256 | >256 | 4 |
| Ticarcillin-CLA | 256 | 12 | 16 | 16 | 4 |
| Piperacillin | 4 | 256 | 32 | 256 | 1 |
| Cephalothin | 16 | >256 | >256 | >256 | 2 |
| Cefoxitin | 0.5 | 2 | 2 | 2 | 1 |
| Ceftazidime | 128 | >256 | >256 | >256 | 0.5 |
| Cefotaxime | 2 | 64 | 48 | 64 | 0.12 |
| Cefepime | 0.5 | 32 | 16 | 32 | 0.06 |
| Cefpirome | 0.5 | 8 | 16 | 16 | 0.06 |
| Moxalactam | <0.06 | 2 | 1 | 0.5 | 0.12 |
| Aztreonam | 1 | >256 | >256 | >256 | 0.25 |
| Imipenem | 1 | 0.25 | 0.25 | 0.25 | 0.25 |
| Imipenem-CLA | 0.5 | 0.25 | 0.25 | 0.25 | 0.25 |
| Meropenem | 0.06 | 0.0 | 0.03 | 0.03 | 0.06 |
| Ertapenem | 0.5 | <0.03 | <0.03 | <0.03 | <0.03 |
E. coli DH10B(pPER-6) expressed β-lactamase PER-6 from A. allosaccharophila AL-1.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml.
TABLE 2.
Nucleotide sequences of primers used for amplification and sequence analysis
| Primer | Sequence (5′ → 3′) |
|---|---|
| T3.1 | TTAAGTTCATGGGTCGTCTCTG |
| T3.2 | ATTCCGCCGAATCAGCAAGAAC |
| T7.1 | ATCAGAAATGAGCGCCAGTC |
| T7.2 | ATTGCCGATATCACTGATGG |
| PERextS | AAGGACARTCSKATGAATGTCa |
| PER-1extAS | TAGTGTACAACCAGAGTCAGC |
| PER-2extAS | TTGCTCAATCCGGACTCACTGC |
| PER-6extAS | TCGTTTAATCCGGACTTACTGCGG |
R represents A or G, S represents C or G, and K represents G or T.
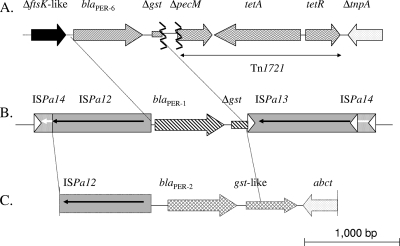
Analysis of the surrounding sequences did not identify composite transposon Tn1213 (20, 23). Instead, part of an open reading frame of 550 bp was identified 80 bp upstream of the blaPER-6 gene, encoding a putative protein that shared weak identity (<28% amino acid identity) with a DNA translocase domain-containing protein (FtsK-like) (Fig. 1). Downstream of the blaPER-6 gene, the 5′ extremity (only 180 bp) of a gene encoding a glutathione S-transferase was identified, exhibiting 86% nucleotide identity with a gene identified downstream of the blaPER-1 and blaPER-2 genes (23). The location of part of an identical gst gene downstream of all those blaPER-like genes indicates a common origin for these β-lactamase genes. That gst gene was truncated by a genetic structure containing a gene encoding a PecM-like protein, together with the tetA and tetR genes, usually part of the Tn1721 transposon (22) (Fig. 1).
FIG. 1.
Schematic map of the blaPER-6-containing structure identified in A. allosaccharophila (this study) (A), the blaPER-1-containing structure identified in Pseudomonas aeruginosa (20) (B), and the blaPER-2-containing structure identified in C. freundii (23) (C).
In order to compare the catalytic properties of PER-1, PER-2, and PER-6 and their contributions to β-lactam resistance, the corresponding genes were cloned and expressed in an isogenic E. coli background under the control of the same promoter (E. coli DH10B; Invitrogen, Cergy-Pontoise, France). Cloning experiments were performed with the pCR-BluntII-TOPO vector (Invitrogen) by following the manufacturer's instructions, using external primer PERextS and specific primers PER-1extAS, PER-2extAS, and PER-6extAS (Table 2), encompassing the entire group of blaPER genes. It gave rise to recombinant strains E. coli DH10B(pPER-1), E. coli DH10B(pPER-2), and E. coli DH10B(pPER-6), expressing β-lactamases PER-1, PER-2, and PER-6, respectively. The MIC values of β-lactams showed no significant differences between those three different E. coli recombinant strains (Table 1).
In order to characterize more precisely whether PER-6 might possess specific catalytic properties, a kinetic study was initiated. E. coli DH10B(pPER-6) produced a β-lactamase with a pI value of 6.4 according to isoelectric focusing analysis performed as described previously (14). This pI value was different from those of PER-1 (16) and PER-2 (23) (both with a pI value of 5.4). PER-6 was purified (>90% as estimated by SDS-PAGE analysis; data not shown) from the E. coli DH10B(pPER-6) crude extract by using a two-step chromatography process (a cation exchange at pH 6.8 using an S-Sepharose column, followed by an anion exchange at pH 8 using a Q-Sepharose column). This protocol allowed 5 mg of purified PER-6 β-lactamase to be obtained (specific activity of 36,800 nmol/min·mg of protein with the use of 100 μM cephalothin as a substrate). β-Lactamase PER-6 had a broad-spectrum hydrolysis profile, including penicillins, broad-spectrum cephalosporins, and, surprisingly, carbapenems but excluding cephamycins (Table 3). The activity of PER-6 was less susceptible to inhibitions by clavulanic acid or tazobactam than that of PER-2. The fifty percent inhibitory concentrations (IC50) for clavulanic acid were 0.3 and 0.07 μM for PER-6 and PER-2 (23), respectively, and those for tazobactam were 1 and 0.1 μM for PER-6 and PER-2 (23), respectively. ß-Lactamase PER-6 was weakly inhibited by sulbactam, with an IC50 of 4 μM. In comparison to PER-2, β-lactamase PER-6 showed overall low catalytic efficiencies (kcat/Km) for most β-lactams (Table 3). This was due to a significant alteration of the Km values for all substrates, particularly for cephalosporins, which were from 20-fold (cefotaxime) to 120-fold (cefepime) higher than those observed for PER-2 (Table 3) (23). A significant hydrolytic activity against carbapenems was detected with PER-6. The kcat values for PER-6 for imipenem, meropenem, and ertapenem were low, but the Km values were also very low, showing a good affinity of PER-6 for those molecules. To better assess whether PER-6 may possess specific properties in comparison to other PER derivatives, specific activities for PER-1, PER-2, and PER-6 were determined with imipenem as a substrate, using culture extracts of isogenic E. coli recipient strains. No significant difference could be observed between those different extracts (data not shown), and no difference in the MICs of carbapenems could be observed (Table 1). However, among the 20 amino acid changes identified between PER-6 and PER-2, two were located in the Ω loop: alanine 161 was replaced by an aspartic acid residue, and glutamine 178 was replaced by a lysine residue in PER-6. These changes may be involved in the peculiar catalytic properties of PER-6.
TABLE 3.
Steady-state kinetic parameters of the β-lactamase PER-6 and comparison of parameter values obtained for β-lactamase PER-2 (23)a
| β-Lactam | PER-6 |
PER-2 |
||
|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (mM−1·s−1) | kcat/Km (mM−1·s−1) | |
| Benzylpenicillin | 5 | 200 | 25 | 120 |
| Ampicillin | 1 | 20 | 50 | 330 |
| Ticarcillin | 0.4 | 9 | 50 | ND |
| Piperacillin | 0.1 | 4 | 25 | 200 |
| Cephalothin | 8 | 55 | 145 | 670 |
| Ceftazidime | 12 | 1,000 | 10 | 430 |
| Cefotaxime | 40 | 900 | 45 | 760 |
| Cefepime | 10 | 2,000 | 5 | 20 |
| Cefpirome | 12 | 1,500 | 8 | NA |
| Cefoxitin | ND | ND | ND | <10 |
| Aztreonam | 3 | 40 | 75 | 120 |
| Moxalactam | ND | ND | ND | NA |
| Imipenem | 0.006 | 1.5 | 4 | <10 |
| Meropenem | 0.004 | 10 | 0.4 | NA |
| Ertapenem | 0.002 | 7 | 0.3 | NA |
Data are means of results from three independent experiments. Standard deviations were within 10% of the means. ND, not determinable or no detectable hydrolysis (<0.01 s−1); NA, not available.
Conclusion.
This study emphasizes the spread of PER-type ESBLs in Aeromonas species. A novel PER-type β-lactamase was identified, the corresponding gene being associated with a novel genetic environment. However, the fact that identical structures were found immediately downstream of all blaPER genes indicates a likely common origin.
Finally, the recent identification of PER-1 in A. media from a Swiss alpine lake, together with the present identification of PER-6 in A. allosaccharophila from the Seine river, emphasizes the spread of these PER-type determinants in the environment, at least on the European continent, with reservoirs likely being waterborne bacterial species.
Nucleotide sequence accession number.
The nucleotide and protein sequences of the PER-6 ß-lactamase have been registered in GenBank under accession no. GQ396303.
Acknowledgments
This work was funded mostly by a grant from the INSERM (U914) and by grants from the European Community (TROCAR, HEALTH-F3-2008-223031) and from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, France.
Footnotes
Published ahead of print on 9 February 2010.
REFERENCES
- 1.Reference deleted.
- 2.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. An, C. Bal, and J. M. Casellas.1996. Characterization of ß-lactamase gene blaPER-2, which encodes an extended-spectrum class A ß-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cattoir, V., L. Poirel, C. Aubert, C. J. Soussy, and P. Nordmann.2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg. Infect. Dis. 14:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute.2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Esteve, C., M. C. Gutiérrez, and A. Ventosa.1995. DNA relatedness among Aeromonas allosaccharophila strains and DNA hybridization groups of the genus Aeromonas. Int. J. Syst. Bacteriol. 45:390-391. [DOI] [PubMed] [Google Scholar]
- 6.Fosse, T., C. Giraud-Morin, I. Madinier, F. Mantoux, J. P. Lacour, and J. P. Ortonne.2004. Aeromonas hydrophila with plasmid-borne class A extended-spectrum ß-lactamase TEM-24 and three chromosomal class B, C, and D ß-lactamases, isolated from a patient with necrotizing fasciitis. Antimicrob. Agents Chemother. 48:2342-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girlich, D., L. Poirel, A. Leelaporn, A. Karim, C. Tribuddharat, M. Fennewald, and P. Nordmann.2001. Molecular epidemiology of the integron-located VEB-1 extended-spectrum ß-lactamase in nosocomial enterobacterial isolates in Bangkok, Thailand. J. Clin. Microbiol. 39:175-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iabadene, H., C. Dallenne, Y. Messai, D. Geneste, R. Bakour, and G. Arlet.2009. Emergence of extended-spectrum ß-lactamase PER-1 in Proteus vulgaris and Providencia stuartii isolates from Algiers, Algeria. Antimicrob. Agents Chemother. 53:4043-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs, L., and H. Y. Chenia.2007. Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 114:295-306. [DOI] [PubMed] [Google Scholar]
- 10.Jones, B. L., and M. H. Wilcox.1995. Aeromonas infections and their treatment. J. Antimicrob. Chemother. 35:453-461. [DOI] [PubMed] [Google Scholar]
- 11.Kieser, T.1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19-36. [DOI] [PubMed] [Google Scholar]
- 12.Lee, H. W., H. Y. Kang, K. S. Shin, and J. Kim.2007. Multidrug-resistant Providencia isolates carrying blaPER-1, blaVIM-2, and armA. J. Microbiol. 45:272-274. [PubMed] [Google Scholar]
- 13.Liu, S. L., A. Hessel, and K. E. Sanderson.1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mammeri, H., L. Poirel, and P. Nordmann.2003. In vivo selection of a chromosomally encoded β-lactamase variant conferring ceftazidime resistance in Klebsiella oxytoca. Antimicrob. Agents Chemother. 47:3739-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naas, T., L. Poirel, and P. Nordmann.2008. Minor extended-spectrum ß-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):42-52. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann, P., E. Ronco, T. Naas, C. Duport, T. Michel-Briand, and R. Labia.1993. Characterization of a novel extended-spectrum ß-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perilli, M., F. De Santis, C. Mugnaioli, G. M. Rossolini, F. Luzzaro, S. Stefani, M. L. Mezzatesta, A. Toniolo, and G. Amicosante.2007. Spread of Enterobacteriaceae carrying the PER-1 extended-spectrum ß-lactamase gene as a chromosomal insert: a report from Italy. J. Antimicrob. Chemother. 59:323-324. [DOI] [PubMed] [Google Scholar]
- 18.Picão, R. C., L. Poirel, A. Demarta, O. Petrini, A. R. Corvaglia, and P. Nordmann.2008. Expanded-spectrum ß-lactamase PER-1 in an environmental Aeromonas media isolate from Switzerland. Antimicrob. Agents Chemother. 52:3461-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Picão, R. C., L. Poirel, A. Demarta, C. S. Silva, A. R. Corvaglia, O. Petrini, and P. Nordmann.2008. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J. Antimicrob. Chemother. 62:948-950. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., L. Cabanne, H. Vahaboglu, and P. Nordmann.2005. Genetic environment and expression of the extended-spectrum ß-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob. Agents Chemother. 49:1708-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann.1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Post, V., and R. M. Hall.2009. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:2667-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Power, P., J. Di Conza, M. M. Rodríguez, B. Ghiglione, J. A. Ayala, J. M. Casellas, M. Radice, and G. Gutkind.2007. Biochemical characterization of PER-2 and genetic environment of blaPER-2. Antimicrob. Agents Chemother. 51:2359-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radice, M., P. Power, J. Di Conza, and G. Gutkind.2002. Early dissemination of CTX-M-derived enzymes in South America. Antimicrob. Agents Chemother. 46:602-604. [DOI] [PMC free article] [PubMed] [Google Scholar]