Abstract
We characterized 67 Escherichia coli isolates with reduced susceptibility to cefotaxime or ceftiofur obtained from healthy broilers housed in five Italian farms. The blaCTX-M-1, blaCTX-M-32 and blaSHV-12 β-lactamase genes were identified on IncI1, IncN, or IncFIB plasmids. Considerable genetic diversity was detected among the extended-spectrum β-lactamase (ESBL)-producing isolates, and we identified indistinguishable strains in unrelated farms and indistinguishable plasmids in genetically unrelated strains. The detection of highly mobile plasmids suggests a potential animal reservoir for β-lactamase genes.
Escherichia coli resistant to extended-spectrum cephalosporins through the production of extended-spectrum β-lactamases (ESBLs) is isolated at an increasing rate among different hosts and countries, thus representing a potential public health risk (2, 3). Typically, ESBLs are borne on mobile genetic elements and can therefore spread both clonally and horizontally among different lineages of bacteria (5).
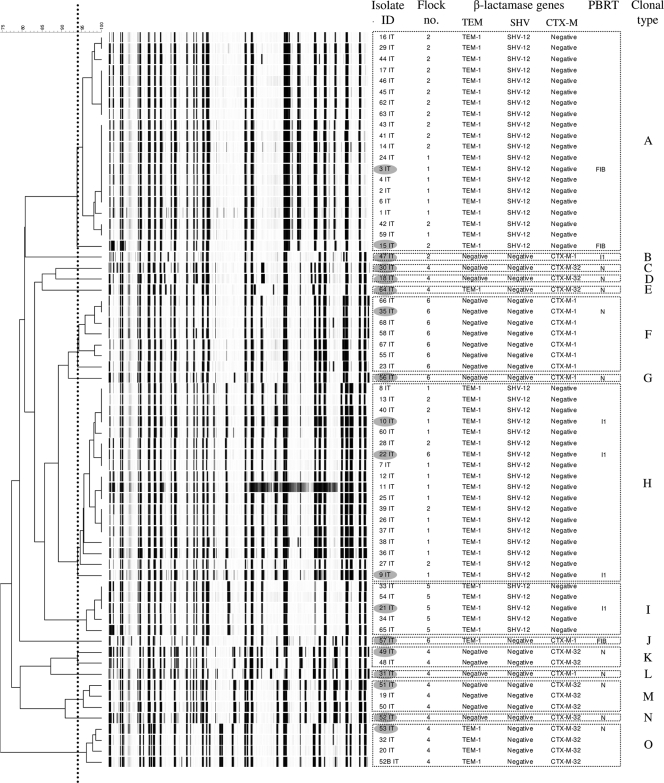
To determine the role of animals as potential reservoirs for ESBL-producing bacteria, there is a need for improved knowledge of the factors that promote dissemination of resistance determinants, including the role of the intestinal flora (4). In this study, we characterized a collection of 67 E. coli isolates showing reduced susceptibility to cefotaxime (MIC ≥ 2 μg/ml) or ceftiofur (MIC ≥ 8 μg/ml). The isolates were obtained during July 2007 by processing fecal samples collected by the “sock method” (1, 22) in a broiler-parent farm and four broiler farms located in northeastern Italy. The farms were ordinary with respect to farm management and biosecurity, thus implying that no contact among the animals reared in different farms could have taken place. One flock (i.e., a group of chickens sharing the same airspace) per farm was sampled (Table 1). All broiler flocks (here identified as flocks 1, 2, 4, and 5) hatched from eggs incubated in the same hatchery, and the parent flock (here identified as flock 6) was among the flocks that provided eggs to that hatchery. Indication of ESBL presence resulted from the ESBL phenotypic detection test performed with the disk method (7, 10). Amplification of the blaTEM, blaSHV, and blaCTX-M genes was done by PCR using primers and conditions previously described (15, 16) on whole-genome bacterial DNA (GenElute bacterial genomic DNA kit; Sigma-Aldrich, Brøndby, Denmark). PCR products were purified (exonuclease I, E. coli, and FastAP thermosensitive alkaline phosphatase; Fermentas, Helsingborg, Sweden) and sequenced (Macrogen Inc., Seoul, South Korea). Comparison of the nucleotide sequences and the derived amino acid sequences with previously described sequences (GenBank database http://www.ncbi.nlm.nih.gov/ and www.lahey.org/studies/webt.html, respectively) showed that the blaTEM-1, blaSHV-12, blaCTX-M-1, and blaCTX-M-32 genes were present in this collection (Fig. 1). The genetic relatedness of the E. coli isolates was determined by amplified fragment length polymorphism (AFLP) analysis as previously described (1, 6). Using a similarity cutoff value of ≥94% (1), 15 different clonal types were identified (A to O in Fig. 1). Eighteen strains were selected for plasmid analysis by selecting one strain per clonal type from each flock. Within one AFLP type, however, two strains from the same flock were examined to determine the intraclonal diversity. Plasmid DNA (GeneJET plasmid miniprep kit; Fermentas, Helsingborg, Sweden) was used to transform electrocompetent Genehog E. coli (Invitrogen, Taastrup, Denmark). Transformants were selected on brain heart infusion (BHI) agar (Merck, Glostrup, Denmark) plates containing 2 μg/ml of cefotaxime and tested by colony PCR (15, 16) to confirm the presence of ESBL-encoding genes. Plasmid DNA (GeneJET plasmid miniprep kit; Fermentas, Helsingborg, Sweden) from the transformants was analyzed by restriction fragment length polymorphism (RFLP) analysis and PCR-based replicon typing (PBRT) (3). IncN (n = 10), IncI1 (n = 5), or IncFIB (n = 3) plasmids were identified (Fig. 1 and Table 2). Twelve different plasmid profiles were visualized on a 0.8% agarose gel after enzymatic digestion with ClaI and PstI (FastDigest; Fermentas, Helsingborg, Sweden). Notably, indistinguishable band patterns were obtained in (i) two blaCTX-M-1 IncN plasmids found in two closely related E. coli strains (with 91.6% similarity according to the analysis of the AFLP patterns) from flock 6, (ii) five blaCTX-M-32 IncN plasmids found in five unrelated E. coli strains from flock 4, and (iii) two blaSHV-12 IncFIB plasmids found in two clonally related E. coli strains from flocks 1 and 2 (Fig. 1 and Table 2). The disk diffusion test (9, 10) showed that all donors were resistant to at least two additional antimicrobial classes (Table 2). Seventeen (94%) isolates were resistant to tetracycline, and 15 (83%) isolates were resistant to nalidixic acid, 8 (44%) of which were ciprofloxacin resistant. Sulfonamide resistance alone or in combination with trimethoprim resistance was detected in 4 (22%) and 10 (55%) donor isolates, respectively. No resistance against amikacin, colistin, and imipenem was detected. Sulfonamide resistance occurred in seven (39%) transformants and was associated with trimethoprim resistance in one (5%) case (Table 2). The cotransfer of sul2 (n = 1), sul3 (n = 5), and sul1 and sul3 (n = 1) genes was determined in such transformants by the use of PCR as previously described (Table 2) (14). As measured by agar dilution (8), the MIC of ciprofloxacin for all the transformants was <0.03 μg/ml, indicating that possible qnr-mediated resistance to this fluoroquinolone was not cotransferred by the ESBL-producing plasmids.
TABLE 1.
Anamnestic information about the flocks sampled
| Flock | Type of animals | Age (days) | Antimicrobial treatment history | Antimicrobial treatment history of the preceding flock | No. of isolates included in the study |
|---|---|---|---|---|---|
| 1 | Broilers | 35 | None | Oxytetracycline, sulfadiazine-trimethoprim | 19 |
| 2 | Broilers | 12 | Amoxicillin | Flumequine, amoxicillin, sulfadiazine-trimethoprim, colistin | 19 |
| 4 | Broilers | 70 | Enrofloxacin, tylosin | None | 14 |
| 5 | Broilers | 35 | Amoxicillin | Amoxicillin | 5 |
| 6 | Parents | 240 | Tylosin, tetracycline, colistin | Tylosin, tetracycline, colistin | 10 |
FIG. 1.
Dendrogram showing the genotypic relatedness of 67 Escherichia coli isolates based on AFLP fingerprints. A Dice coefficient and the unweighted-pair group method using average linkages (UPGMA) algorithm were used with a position tolerance of 1%. The scale bar represents the percentage of similarity, and the vertical, dotted line indicates the cutoff value for identifying clonal types. PBRT, plasmid-based replicon typing as determined with a subset of isolates indicated by gray-shaded ovals.
TABLE 2.
Genetic and phenotypic traits of ESBL-producing Escherichia coli from broiler flocks
| Flock | Isolate | AFLP clonal type (no. of strains within clonal type) | β-Lactamase gene(s)a | ClaI plasmid RFLP profile | PstI plasmid RFLP profile | Replicon typing | Additional resistancesb | sul genesc |
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | A (20) | TEM-1, SHV-12 | 9 | l | FIB | TET, SUL, TMP, FFC, CHL, NAL, CIP, STR | sul1, sul3 |
| 1 | 9 | H (18) | TEM-1, SHV-12 | 3 | c | I1 | TET, SUL, TMP, FFC, CHL, NAL, STR | sul2, sul3 |
| 1 | 10 | H (18) | TEM-1, SHV-12 | 11 | n | I1 | TET, SUL, TMP, NAL, STR | sul1, sul2 |
| 2 | 15 | A (20) | TEM-1, SHV-12 | 9 | l | FIB | TET, SUL, TMP, FFC, CHL, NAL, CIP, STR | sul1, sul3 |
| 2 | 47 | B (1) | CTX-M-1 | 8 | i | I1 | TET, SUL, TMP, NAL | sul1, sul3 |
| 4 | 18 | D (1) | CTX-M-32 | 5 | f | N | TET, NAL | |
| 4 | 30 | C (1) | CTX-M-32 | 5 | f | N | TET, NAL | |
| 4 | 31 | L (1) | CTX-M-1 | 2 | b | N | SUL, NAL, CIP, STR | sul1 |
| 4 | 49 | K (2) | CTX-M-32 | 6 | g | N | TET, SUL, NAL, CIP, SPT | sul2 |
| 4 | 51 | M (3) | CTX-M-32 | 4 | d | N | TET, SUL, TMP, CHL, NAL, CIP, STR | sul2 |
| 4 | 52 | N (1) | CTX-M-32 | 5 | f | N | TET, SUL, TMP, CHL, NAL, CIP, STR | sul1, sul2 |
| 4 | 53 | O (4) | TEM-1, CTX-M-32 | 5 | f | N | TET, SUL, TMP, CHL, NAL, CIP, STR, GEN | sul1, sul2, sul3 |
| 4 | 64 | E (1) | TEM-1, CTX-M-32 | 5 | f | N | TET, SUL, CHL, NAL, CIP | sul3 |
| 5 | 21 | I (5) | TEM-1, SHV-12 | 1 | a | I1 | TET, SUL, TMP, FFC, CHL, STR | sul1, sul2, sul3 |
| 6 | 22 | H (18) | TEM-1, SHV-12 | 7 | h | I1 | TET, SUL, TMP, FFC, CHL, NAL, STR | sul1, sul2, sul3 |
| 6 | 35 | F (7) | CTX-M-1 | 5 | e | N | TET | |
| 6 | 56 | G (1) | CTX-M-1 | 5 | e | N | TET | |
| 6 | 57 | J (1) | TEM-1, CTX-M-1 | 10 | m | FIB | TET, SUL, CHL, NAL | sul1, sul3 |
β-Lactamase genes were determined by PCR and by sequencing in the donors. Genes transferred by transformation, as determined by PCR but not by sequencing, are underlined.
CHL, chloramphenicol; CIP, ciprofloxacin; FFC, florfenicol; GEN, gentamicin; NAL, nalidixic acid; STR, streptomycin; SUL, sulfonamides; TET, tetracycline; TMP, trimethoprim. Patterns transferred by transformation are underlined.
Genes transferred by transformation are underlined.
We isolated ESBL-producing E. coli in flocks that did not receive treatments with cephalosporins (Table 1). Treatments with other antimicrobials (Table 1) could have coselected for the bacteria described. Although a limited number of flocks and bacterial isolates was included in the study, the genetic environment for the ESBL-encoding genes was highly variable. The blaSHV-12 gene was associated with three major clones (A, H, and I) detected in four flocks. These strains carried IncI1 (H and I) or IncFIB (A) plasmids. The blaCTX-M-1 gene and the closely related blaCTX-M-32 gene were mainly associated with IncN plasmids found in 10 different clonal types in three flocks. As clearly shown in flock 4 (Table 2), IncN plasmids were broadly distributed among genetically unrelated strains. The horizontal mobility of IncN plasmids has already been reported by Moodley and Guardabassi, who described the spread of a blaCTX-M-1 IncN plasmid among multiple E. coli lineages of porcine and human origin (18). The finding of IncN plasmids that could disseminate by horizontal transmission is worrisome. Our findings concerning the association of ESBL-encoding genes and plasmid replicons are similar to previous findings in humans, chickens, pigs, and dogs (5, 11, 12, 13, 17, 18, 19, 21, 23). The similarity between the ESBL variants we detected and those present in humans in Italy (20) indicates a possible role of poultry in the dissemination of these resistance determinants.
Acknowledgments
This study was supported by a grant from the Danish Poultry Council, the Graduate School FOOD, and the University of Copenhagen.
We thank Henrik Hasman and Arshnee Moodley for assistance with ESBL and plasmid characterization.
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Bortolaia, V., M. Bisgaard, and A. M. Bojesen.2009. Distribution and possible transmission of ampicillin- and nalidixic acid-resistant Escherichia coli within the broiler industry. Vet. Microbiol. doi: 10.1016/j.vetmic. 2009.10.024. [DOI] [PubMed]
- 2.Cantón, R., A. Novais, A. Valverde, E. Machado, L. Peixe, F. Baquero, and T. M. Coque.2008. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14(Suppl. 1):144-153. [DOI] [PubMed] [Google Scholar]
- 3.Carattoli, A., A. Bertini, L. Villa, V. Falbo, K. L. Hopkins, and E. J. Threlfall.2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219-228. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli, A.2008. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 14(Suppl. 1):117-123. [DOI] [PubMed] [Google Scholar]
- 5.Carattoli, A.2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, H., M. Bisgaard, A. M. Bojesen, R. Mutters, and J. E. Olsen.2003. Genetic relationships among avian isolates classified as Pasteurella haemolytica, ′Actinobacillus salpingitidis' or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 53:275-287. [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute.2006. Performance standards for antimicrobial disk susceptibility tests; approved standard, 9th ed. CLSI M2-A9. Clinical and Laboratory Standards Institute, Wayne, PA.
- 8.Clinical and Laboratory Standards Institute.2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed. CLSI M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute.2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 3rd ed. CLSI M31-A3. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Clinical and Laboratory Standards Institute.2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Diestra, K., C. Juan, T. Curiao, B. Moyá, E. Miró, J. Oteo, T. M. Coque, M. Pérez-Vázquez, J. Campos, R. Cantón, A. Oliver, and F. Navarro.2009. Characterization of plasmids encoding blaESBL and surrounding genes in Spanish clinical isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 63:60-66. [DOI] [PubMed] [Google Scholar]
- 12.García-Fernández, A., G. Chiaretto, A. Bertini, L. Villa, D. Fortini, A. Ricci, and A. Carattoli.2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61:1229-1233. [DOI] [PubMed] [Google Scholar]
- 13.Girlich, D., L. Poirel, A. Carattoli, I. Kempf, M.-F. Lartigue, A. Bertini, and P. Nordmann.2007. Extended-spectrum β-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl. Environ. Microbiol. 73:4681-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grape, M., L. Sundström, and G. Kronvall.2003. Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 52:1022-1024. [DOI] [PubMed] [Google Scholar]
- 15.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup.2005. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, X., Y. Ni, Y. Jiang, F. Yuan, L. Han, M. Li, H. Liu, L. Yang, and Y. Lu.2005. Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 beta-lactamase in China. J. Clin. Microbiol. 43:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcadé, G., C. Deschamps, A. Boyd, V. Gautier, B. Picard, C. Branger, E. Denamur, and G. Arlet.2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum β-lactamases. J. Antimicrob. Chemother. 63:67-71. [DOI] [PubMed] [Google Scholar]
- 18.Moodley, A., and L. Guardabassi.2009. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob. Agents Chemother. 53:1709-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mugnaioli, C., F. De Luca, A. Carattoli, and G. M. Rossolini.2006. Characterisation of conjugative plasmids encoding CTX-M-type extended-spectrum beta-lactamases in Italian clinical isolates of Escherichia coli. Clin. Microbiol. Infect. 12(Suppl. 4):24-25.16445721 [Google Scholar]
- 20.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, M. Perilli, G. Amicosante, S. Stefani, A. Toniolo, and G. M. Rossolini.2006. CTX-M-type extended-spectrum β-lactamases in Italy: molecular epidemiology of an emerging countrywide problem. Antimicrob. Agents Chemother. 50:2700-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novais, Â., R. Cantón, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque.2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, -32) plasmids. Antimicrob. Agents Chemother. 51:796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skov, M. N., B. Carstensen, N. Tornøe, and M. Madsen.1999. Evaluation of sampling methods for the detection of Salmonella in broiler flocks. J. Appl. Microbiol. 86:695-700. [DOI] [PubMed] [Google Scholar]
- 23.Vinué, L., A. García-Fernández, D. Fortini, P. Poeta, M. A. Moreno, C. Torres, and A. Carattoli.2009. Escherichia coli clones and plasmid-mediated later transfer disseminate CTX-M1 and CTX-M-32 among animals and humans, abstr. O308. Abstr. 19th Eur. Cong. Clin. Microbiol. Infect. Dis., Helsinki, Finland.