Abstract
Pseudomonas aeruginosa is equipped with the Sec and Tat protein secretion systems, which translocate the xenobiotic transporter MexAB-OprM and the pathogenic factor phospholipase C (PlcH), respectively. When the signal sequence of MexA was replaced with that of PlcH, the hybrid protein was successfully expressed and recovered from the periplasmic fraction, suggesting that the hybrid protein had been translocated across the inner membrane. MexA-deficient cells harboring the plasmid carrying the plcH-mexA fusion gene showed antibiotic resistance comparable to that of the wild-type cells. This result suggested that MexA secreted via the Tat machinery was properly assembled and functioned as a subunit of the MexAB-OprM efflux pump. A mutation was introduced into the chromosomal tatC gene encoding an inner membrane component of the Tat protein secretion machinery in mexA-deficient cells, and they were transformed with the plasmid carrying the plcH-mexA fusion gene. The transformants showed antibiotic susceptibility comparable to that of mexA-deficient cells, indicating that the hybrid protein was not transported to the periplasm. Whole-cell lysate of the mexA-tatC double mutant harboring the plcH-mexA plasmid produced mainly unprocessed PlcH-MexA. The periplasmic fraction showed no detectable anti-MexA antibody-reactive material. On the basis of these results, we concluded that MexA could be translocated across the inner membrane through the Tat pathway and assembled with its cognate partners, MexB and OprM, and that this complex machinery was fully functional. This hybrid protein translocation system has the potential to be a powerful screening tool for antimicrobial agents targeting the Tat system, which is not present in mammalian cells.
Infectious diseases caused by bacteria seemed to be well controlled at one time by the use of effective therapeutic agents, but now these are becoming less useful due to resistance. Problems have arisen through the reckless use of antimicrobial agents, causing the emergence of drug-resistant bacteria. Special attention has been paid to the spread of multidrug-resistant bacteria, including Pseudomonas aeruginosa, Mycobacterium tuberculosis, methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant MRSA (28). To combat these multidrug-resistant pathogens, an intensive ongoing search for new compounds active against them has been carried out, yet the discovery of new lead compounds seems to be more and more difficult (12). An alternative approach that differs from the traditional means may be possible (14, 32). If the cell machinery essential for the expression of bacterial virulence but not the life-sustaining processes was the target of in vitro screening systems, it might be possible to discover a compound(s) that attenuates bacterial virulence. Importantly, it is unlikely that such a compound(s) would cause the emergence of resistant cells (2).
A potential antibiotic target appears to be protein secretion machinery that is present in bacterial cells but not in mammalian cells (17). One such candidate may be the twin-arginine translocation (Tat) system that was originally discovered in plant cells (21); more recently, the presence of a similar Tat system has been confirmed in bacterial cells (24). The Tat system is an electrochemical-gradient-driven protein secretion system that transports the proteins across the cytoplasmic membrane (17, 21, 24). Proteins to be translocated by the Tat system have a consensus signal sequence with an SRRXFLK motif, where the presence of consecutive arginine residues is essential and X could be replaced with any polar amino acid (17, 18). Lines of evidence have been accumulating that show the Tat system plays an important role in the secretion of virulence factors in pathogenic bacteria (5, 16, 19). Therefore, a Tat system inhibitor could act as a potent pathogenicity attenuator.
Besides the Tat system, most bacterial cells are equipped with the general protein secretion (Sec) system, which translocates a wide variety of proteins across the cytoplasmic membrane (20). The Sec system secretes proteins having a consensus signal peptide that is similar to, but distinct from, that of the Tat system. The Sec signal sequence lacks an N-terminal consecutive-arginine sequence and has a relatively hydrophobic central region and a relatively short signal sequence compared with that of Tat (17, 18). Thus, whether proteins are secreted via the Sec system or the Tat system largely depends on the characteristic features of their signal sequences.
To search for antimicrobial agents that can knock out the Tat system, it is necessary to develop a high-sensitivity reporter assay system. Our strategy was as follows. If the signal sequence of the protein to be secreted via the Sec system was replaced with that of the protein to be translocated by the Tat system, the Sec system-dependent protein could be translocated via the Tat system. If the candidate protein was a component involved in antibiotic resistance, the hybrid proteins secreted via the Tat system would render the cells resistant to antibiotics. If a compound that blocked or inactivated the Tat system was present, the reporter protein would not be translocated across the inner membrane, and therefore, the cell would become antibiotic susceptible. We employed the MexA subunit of the MexAB-OprM antibiotic efflux pump of P. aeruginosa as a reporter of the Sec system and phospholipase C (PlcH) as a donor of the Tat system signal sequence.
The MexA protein is a component of the MexAB-OprM efflux pump and functions as a molecular clamp in the pump assembly (1, 35). Though the MexAB-OprM pump confers multiantibiotic resistance on cells, the absence of MexA totally inactivates the pump function, thus rendering the cells hypersusceptible to antibiotics (35). MexA anchors the inner membrane via fatty acids attached to the N-terminal cysteine residue (33). When the N-terminal signal sequence plus an adjacent cysteine residue of MexA was replaced with the signal sequence of periplasmic azurin, this hybrid protein was properly translocated, processed, and folded in the periplasmic space as the soluble form (33). In addition, the cells producing the hybrid protein but lacking the chromosomal mexA gene restored antibiotic resistance, indicating that membrane anchoring is not essential for the function of MexA (33).
The PlcH protein, an important virulence factor of P. aeruginosa, has the consensus signal sequence RRRTFLK and appears to be secreted into the extracellular medium via the Tat system (27). Accordingly, we designed the experiment to replace the signal sequence of MexA with that of PlcH and assessed whether MexA could be translocated via the Tat machinery.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Cells were grown aerobically in L broth containing 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter (pH 7.2) at 37°C. For the pyoverdine assay, cells were grown in medium containing 5 g per liter of Casamino Acids, 5 mM K2HPO4, and 1 mM MgSO4 with shaking at 37°C overnight (16). The MICs of aztreonam (Sigma) and chloramphenicol (Wako Pure Chemical, Japan) were determined by the agar dilution method using Mueller-Hinton agar (35).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17(rK−rK+) e14− (mcrA−) supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | Laboratory strain |
| E. coli S17-1 | RP4-2-Tc::Mu-Km::Tn7 pro res−mod+ | 26 |
| P. aeruginosa PA04290 | leu-10 argF10 aph-9004 FP− | Matsumoto collection |
| P. aeruginosa TNP070 | ΔmexA derivative of PAO4290 | 35 |
| P. aeruginosa TNP080 | ΔtatC derivative of TNP070 | This study |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Apr | Stratagene |
| pUC18 | Cloning vector; Apr | Takara |
| pTH18ks1 | pSC101 derivative; repAts1 Kmr | 9 |
| pK18mob | pK18 derivative carrying RP4 mob region; Kmr | 25 |
| pMMB67HE | Broad-host-range vector; Apr IncQ | 8 |
| pMEXA1 | pMMB67HE derivative carrying wild-type mexA gene; Apr | 34 |
| pBlueMexA | pBluescript II SK(+) derivative carrying mexA gene; Apr | This study |
| pTH18-PlcMexA | pTH18ks1 derivative carrying chimeric mexA gene with plcH signal sequence; Kmr | This study |
| pPlcMexA | pMMB67HE derivative carrying chimeric mexA gene with plcH signal sequence; Apr | This study |
| pK18ΔTat | pK18mob derivative carrying internal tatC gene and mob gene; Kmr | This study |
| pTH18-TatPlcMexA | pTH18ks1 derivative carrying entire tatC gene and the chimeric mexA gene; Kmr | This study |
| pTatPlcMexA | pMMB67HE derivative carrying entire tatC gene and the chimeric mexA gene; Apr | This study |
Ap, ampicillin; Km, kanamycin.
Replacement of the signal peptide of mexA with that of plcH.
The modified overlap extension method (10) was used to construct recombinant mexA having the signal sequence of the plcH gene (27). The sequence including the plcH signal sequence was amplified by ExTaq polymerase (Takara, Japan) using forward (PlcH-F) and reverse (PlcH-R) primers (Table 2) from the P. aeruginosa PAO1 chromosomal DNA as a template. The gene encoding mature MexA was amplified by ExTaq using forward (MexA-F) and reverse (MexA-R) primers from a derivative of pBluescript II SK(+) (Stratagene) carrying the MexA gene as a template. The PCR products were fused by a second DNA amplification using primers PlcH-F and MexA-R, followed by cloning onto pTH18ks1 (9) digested with BamHI and KpnI to yield pTH18-PlcMexA. We next amplified the fused gene using the PlcH-F and MexA-R primers and pTH18-PlcMexA as a template, and then the fragment was cloned onto a shuttle vector, pMMB67HE (8), pretreated with BamHI and KpnI to yield pPlcMexA. The nucleotide sequence of the fused gene was determined by the dideoxy chain termination method (23). P. aeruginosa TNP070 and TNP080 were transformed with the plasmid pPlcMexA by electroporation with a GenePulser II (Bio-Rad) at 2.5 kV, 200 Ω, and 25 μF.
TABLE 2.
Primers
| Primer | Sequencea | Tag |
|---|---|---|
| PlcH-F | 5′-GAGGATCCTCGCTGTATTCCCTGCC-3′ | BamHI |
| PlcH-R | 5′-CGGCGCCTCGCTTTTTCCCTCGACGGCCAGGGCGCG-3′ | |
| MexA-F | 5′-CGCGCCCTGGCCGTCGAGGGAAAAAGCGAGGCGCCG-3′ | |
| MexA-R | 5′-AGGGTACCCTTGATCAGCCCTTGC-3′ | KpnI |
| TatC-F1 | 5′-GTGGATCCACCTGACCGAACTGCGTACG-3′ | BamHI |
| TatC-R1 | 5′-ACGGATCCGTCAGGACGAAGTCCTAGGTAG-3′ | BamHI |
| TatC-F2 | 5′-GTGGATCCACCTGACCGAACTGCGTACG-3′ | BamHI |
| TatC-R2 | 5′-ACGGATCCGTCAGGACGAAGTCCAGGTAG-3′ | BamHI |
Restriction site tags are underlined.
Construction of a tatC-deficient mutant.
To inactivate the tatC gene in the mexA-deficient strain, TNP070, we first amplified an internal region of tatC (446 bp) with ExTaq using forward (TatC-F1) and reverse (TatC-R1) primers and the PAO1 chromosomal DNA as a template. The DNA fragment produced was cloned into the BamHI site of pK18mob (25) to generate pK18ΔTat. Escherichia coli S17-1 was transformed with pK18ΔTat (36), and the chromosomal tatC of P. aeruginosa TNP070 was disrupted by integration of the plasmid by conjugation with selection on an agar plate containing 12.5 μg/ml of kanamycin (Wako Pure Chemical, Japan) and 50 μg/ml of tetracycline (Wako Pure Chemical, Japan), yielding TNP080. Mutation in the tatC gene was confirmed by Southern blot analysis as described previously (36).
Construction of the tatC expression vector.
The tatC gene was amplified with PrimeStar (Takara) using forward (TatC-F2) and reverse (TatC-R2) primers, with the PAO1 chromosomal DNA as a template. The amplified fragment was cloned into the BamHI site of pUC18, and the presence of the cloned tatC was verified by nucleotide sequencing. The BamHI fragment excised from the resulting plasmid was cloned into the BamHI site of pTH18-PlcMexA, yielding pTH18-TatPlcMexA, which carried the intact tatC gene in front of the plcH-mexA fusion. After digestion of pHT18-TatPlcMexA with HindIII and KpnI, the fragment encompassing tatC and the mexA gene fusion was purified using the QIAEXII gel extraction kit (Qiagen). Next, the isolated fragment was cloned into pMMB67HE pretreated with HindIII and KpnI to generate pTatPlcMexA. TNP080 lacking chromosomal mexA and tatC was transformed with pTatPlcMexA.
Fractionation of the periplasmic proteins.
Crude periplasmic materials were obtained by cold shock treatment (15). Briefly, overnight-grown cells in 20 ml of L broth containing 12.5 μg/ml of kanamycin and 50 μg/ml of sulbenicillin (Takeda, Japan), if needed, were diluted with 200 ml of prewarmed (37°C) L broth containing 2 mM isopropyl-1-thio-β-d-galactopyranoside and incubated at 37°C until late log phase. The cells were harvested by centrifugation (9,000 × g; 10 min; 23°C) and suspended in 50 mM Tris-HCl, 0.2 M MgCl2 (pH 7.3) at the wet-cell-weight/buffer ratio of 1 to 5. The cell suspension was alternatively immersed in a 37°C water bath for 10 min and an ice water bath for 15 min for two cycles. After centrifugation at 17,500 × g for 15 min at 10°C, the supernatant was centrifuged at 108,000 × g for 60 min at 10°C (Beckman rotor TLA-120.1). The supernatant was saved as the periplasmic protein fraction.
Other methods.
Visualization of the chromosomal tatC region was carried out using the DIG labeling and detection system (Roche) according to the manufacturer's instructions. Pyoverdine was detected by measuring the absorption spectra of the culture supernatants from 380 to 500 nm (30). For Western blotting, cellular extracts were subjected to electrophoresis and blotted to a polyvinylidene fluoride membrane (Millipore), and the protein band was probed with the anti-MexA antibody using alkaline phosphatase-conjugated secondary antibody (35). SDS-PAGE was carried out as described by Laemmli (11). The protein concentration was quantified by the method of Lowry et al. (13).
RESULTS AND DISCUSSION
The function of MexA encoded by the plcH-mexA fusion gene.
The hybrid protein encoded by the fused gene of the azurin signal sequence and the mature mexA sequence had been found to be properly secreted into the periplasmic space and fully functional (33). We assumed, therefore, that the signal sequence of MexA might be replaced with that of phospholipase C (PlcH) and that the hybrid PlcH-MexA might be transported via the Tat secretion machinery. TNP070 cells lacking the chromosomal mexA gene were transformed with a plasmid carrying the fused gene of the plcH signal peptide and the mexA sequence encoding the mature form of MexA. The MICs of aztreonam and chloramphenicol in the transformant TNP070(pPlcMexA) were 2 and 32 μg/ml, respectively (Table 3). These values were 8 and 4 times, respectively, higher than the MICs of aztreonam and chloramphenicol in cells lacking MexA (TNP070) and were fully comparable to the MICs in cells producing a full amount of MexA, TNP070(pMEXA1). These results suggested that the hybrid MexA had been expressed and translocated across the inner membrane via the Tat protein secretion machinery.
TABLE 3.
MICs of aztreonam and chloramphenicol for transformants expressing chimeric MexA
| Strain | Plasmid | MIC (μg/ml) |
|
|---|---|---|---|
| Aztreonam | Chloramphenicol | ||
| PAO4290 | −a | 4.0 | 64 |
| TNP070 | − | 0.25 | 8.0 |
| TNP070 | pMEXA1 | 2.0 | 32 |
| TNP070 | pPlcMexA | 2.0 | 32 |
| TNP080 | − | 0.13 | 4.0 |
| TNP080 | pMexA1 | 2.0 | 32 |
| TNP080 | pPlcMexA | 0.25 | 4.0 |
| TNP080 | pTatPlcMexA | 2.0 | 32 |
−, no plasmid.
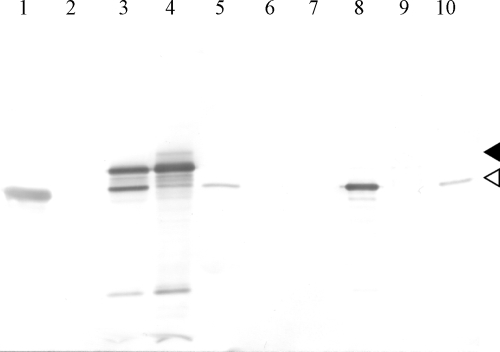
We analyzed the whole-cell lysate of the transformant harboring pPlcMexA by the Western blotting method, probing with anti-MexA antibody, and found that there were two anti-MexA antibody-reactive bands. (Fig. 1, lane 3). The lower band corresponded to the mature form of MexA, and therefore, the upper band was most likely the unprocessed form of MexA. If this was the case, it could be expected that at least the mature form of MexA would be localized in the periplasmic aqueous phase. To confirm this prediction, the periplasmic materials were released from the cells into the aqueous phase by the cold osmotic shock method, and then the extracts were subjected to Western blotting and probed with the anti-MexA antibody. The electrophoretogram showed only one major band reactive with the anti-MexA antibody and corresponding to the location of the mature form of MexA (Fig. 1, lane 8). This result suggests that hybrid MexA was translocated across the inner membrane, most likely through the Tat secretion machinery. As a control experiment, the wild-type cell, PAO4290, which produces fatty acid-modified MexA and hence contains MexA, which anchors the inner membrane, was subjected to the osmotic shock experiment. The result showed that MexA was undetectable in the periplasmic materials, confirming the previous results (33).
FIG. 1.
Western blotting of periplasmic materials extracted from cells expressing hybrid MexA proteins. Whole-cell lysate (4 μg protein) (lanes 1 to 5) and periplasmic materials (1 μg protein) (lanes 6 to 10) were subjected to SDS-PAGE (12% gel), blotted onto a polyvinylidene fluoride membrane, and visualized with anti-MexA antibody. Lanes 1 and 6, PAO4290; lanes 2 and 7, TNP070; lanes 3 and 8, TNP070(pPlcMexA); lanes 4 and 9, TNP080(pPlcMexA); lanes 5 and 10, TNP080(pTatPlcMexA). The white and black arrowheads indicate mature and unprocessed MexA.
Translocation of the hybrid MexA through the Tat pathway.
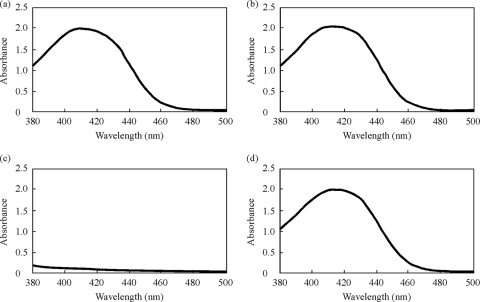
The results described above could not distinguish whether the hybrid protein was translocated via the Tat pathway or the Sec pathway. If the hybrid protein was secreted via the Tat pathway, introduction of a mutation into the subunit protein of the Tat machinery should abolish the secretion. Therefore, we disrupted the tatC gene by single-crossover mutagenesis in the mexA-deficient mutant (TNP080). Disruption was confirmed by Southern blot analysis (data not shown). The function of the Tat system in the mutant was assessed by measuring the pyoverdine in the medium, the production of which is Tat dependent (31). Though the culture supernatant of the wild-type cells, PAO4290, and the MexA-deficient cells, TNP070, showed the typical absorption spectra of pyoverdine (Fig. 2), the cells carrying the mutation in the tatC gene secreted an undetectable level of pyoverdine, suggesting that the Tat secretion machinery was not functional. This result was consistent with what had been seen before (31). We also noted that the growth rate of the TNP080 cells was slowed down compared with that of the parent strain, TNP070 (data not shown), consistent with the characteristic feature of the tatC-deficient mutant of P. aruginosa (16). When the tatC-deficient TNP080 cells were transformed with the plasmid carrying unimpaired tatC, pTatPlcMexA, the pyoverdine-secreting capability of the cells was restored (Fig. 2).
FIG. 2.
Absorption spectra of culture supernatants. Cells were grown in Casamino Acid medium overnight at 37°C under aeration by reciprocal shaking, and the culture was centrifuged at 13,700 × g for 10 min. The clear supernatant was scanned with a spectrophotometer (V-630 Bio; Jasco Co., Japan). (a) PAO4290. (b) TNP070. (c) TNP080. (d) TNP080(pTatPlcMexA).
We then investigated whether inactivation of the Tat machinery interfered with the function of the MexAB-OprM efflux pump by determining the MICs of the antibiotics. The TNP080 cells with double mutations in mexA and tatC and harboring pPlcMexA showed aztreonam and chloramphenicol MICs of 0.25 μg/ml and 4.0 μg/ml, respectively, and these values were comparable with those in MexA-deficient TNP070 cells (Table 3). This result suggested that the hybrid MexA protein was not transported to the periplasmic space, implying that the hybrid protein was not the substrate of the Sec system. It is likely that the unprocessed form of the hybrid MexA remained in the cytoplasmic space or stacked at the inner membrane. In fact, a significant amount of unprocessed MexA was recovered from the whole-cell lysate of TNP080 harboring pPlcMexA (Fig. 1, lane 4), whereas nearly no detectable processed form of MexA was recovered from the periplasmic fraction (Fig. 1, lane 9). These results clearly indicated that the hybrid MexA was translocated across the inner membrane exclusively through the Tat pathway. In contrast, TNP080 harboring pMEXA1, and thus expressing wild-type MexA, restored resistance to aztreonam (2.0 μg/ml) and chloramphenicol (32 μg/ml) to levels similar to those of TNP070 harboring pMEXA1, confirming that MexA was translocated through the Sec system.
Restoration of hybrid MexA translocation by TatC expressed from the plasmid-borne tatC gene.
We next designed an experiment to recover the antibiotic resistance in TNP080 cells by restoring the Tat pathway-dependent translocation of the hybrid MexA. The pTatPlcMexA plasmid carrying unimpaired tatC was constructed, and TNP080 was transformed with the plasmid (Table 1). The transformants harboring pTatPlcMexA excreted pyoverdine into the culture medium, as assayed by measurement of the absorption spectrum (Fig. 2), indicating that the Tat machinery was reconstituted from the plasmid expressing the TatC subunit and functioned properly. In addition, it should be noted that the lowered growth rate of the tatC mutant fully recovered, as the plasmid-borne TatC was expressed (data not shown). Functional restoration of the Tat machinery was assessed by determination of antibiotic susceptibility. The transformants harboring pTatPlcMexA showed the MIC of aztreonam and chloramphenicol to be 2.0 and 32 μg/ml, respectively, and these values were comparable to those in PAO4290, TNP070(pMEXA1), TNP070(pPlcMexA), and TNP080(pMexA) (Table 1), indicating again that the hybrid MexA had been translocated through the reconstituted Tat pathway, processed, and properly assembled into a functional efflux pump.
On the basis of these results, we concluded that the hybrid MexA with the Tat-directed signal sequence is exclusively translocated across the inner membrane via the Tat pathway. Thus, this system, which is a combination of the plasmid carrying the plcH-mexA fusion gene and the mexA-deficient host cell of P. aeruginosa, provides a high-sensitivity screening strategy for Tat pathway inhibitors. If the Tat pathway inhibitors were to be discovered, the compounds could be used as pathogenicity attenuators of P. aeruginosa, because the Tat machinery is involved in several aspects of virulence and pathogenic factors (16).
There are growing lines of evidence that the Tat pathway recognizes and translocates the folded proteins across the cytoplasmic membrane, in marked contrast to the Sec pathway, which translocates unfolded substrate proteins cotranslationally. Thus, the system may function as a protein-folding sensor or a folding quality control system (4). This implies that the Tat system senses the folding status of its substrate proteins, as unfolded substrate proteins are generally rejected (4, 18). It is intriguing to note that the MexA protein could be translocated in either the unfolded state via the Sec system or the folded state via the Tat system. This feature of the Tat system-mediated translocation of folded proteins has been well studied by the replacement of the Sec system-directed signal sequence of alkaline phosphatase and cytochrome c with the Tat system-directed signal sequence (4, 22). Another interesting reporter protein would be the green fluorescent protein (GFP) attached to the Tat-directing signal sequence (29), which is transported solely through the Tat pathway, leading to proper fluorescence of the secreted protein. GFP transported through the Sec pathway does not fluoresce (7). The translocation pathway-dependent fluorescence of the exported GFP is most likely due to the fact that GFP may be folded only in a cytoplasmic environment with reduced conditions (7). The maltose binding protein was transported by either the Sec system or the Tat system, depending upon the signal sequence attached to it (3). MexA is the first example, to our knowledge, of an RND family protein (6) that could be exported by the Tat machinery while retaining full activity. Accordingly, the hybrid MexA should prove to be a useful tool for the study of the molecular mechanism of protein translocation of the P. aeruginosa Tat machinery.
Acknowledgments
We are grateful to Keietsu Abe of Tohoku University and Hideaki Maseda of Tokushima University for helpful discussion.
This study was supported in part by a grant from the Japan Society for the Promotion of Science (KAKENHI 20659066).
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Akama, H., T. Matsuura, S. Kashiwagi, H. Yoneyama, S. Narita, T. Tsukihara, A. Nakagawa, and T. Nakae.2004. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 279:25939-25942. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy.2004. Targeting virulence to prevent infection: to kill or not to kill? Drug Discov. Today Ther. Strateg. 1:483-489. [Google Scholar]
- 3.Blaudeck, N., P. Kreutzenbeck, R. Freudl, and G. A. Sprenger.2003. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J. Bacteriol. 185:2811-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLisa, M. P., D. Tullman, and G. Georgiou.2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. U. S. A. 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding, Z., and P. J. Christie.2003. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 185:760-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinh, T., I. T. Paulsen, and M. H. Saier, Jr.1994. A family of extracytoplasmic proteins that allow transport of large molecules across the outer membrane of gram-negative bacteria. J. Bacteriol. 176:3825-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips.2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furste, J. P., W. Pansegrau, W. Frank, R. Blocker, H. Scholz, P. Bagdasarian, and E. Lanka.1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 9.Gotoh, T. H., M. Yamaguchi, K. Yasojima, A. Tsujimura, Y. Wakabayashi, and Y. Watanabe.2000. A set of temperature sensitive-replication/segregation and temperature resistant plasmid vectors with different copy numbers and in an isogenic background (chloramphenicol, kanamycin, lacZ, repA, par, polA). Gene 241:185-191. [DOI] [PubMed] [Google Scholar]
- 10.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease.1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction, Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 11.Laemmli, U. K.1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 272:680-685. [DOI] [PubMed] [Google Scholar]
- 12.Leeb, M.2004. A shot in the arm. Nature 431:892-893. [DOI] [PubMed] [Google Scholar]
- 13.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall.1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 14.McDevitt, D., and M. Rosenberg.2001. Exploiting genomics to discover new antibiotics. Trends Microbiol. 9:611-617. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima, A., Y. Sugimoto, H. Yoneyama, and T. Nakae.2000. Localization of the outer membrane subunit OprM of resistance-nodulation-cell division family multicomponent efflux pump in Pseudomonas aeruginosa. J. Biol. Chem. 275:30064-30068. [DOI] [PubMed] [Google Scholar]
- 16.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil.2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer, T., and B. C. Berks.2003. Moving folded proteins across the bacterial cell membrane. Microbiology 149:547-556. [DOI] [PubMed] [Google Scholar]
- 18.Palmer, T., F. Sargent, and B. C. Berk.2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13:175-180. [DOI] [PubMed] [Google Scholar]
- 19.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L.-F. Wu.2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugsley, A. P.1993. The complete general-secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson, C., D. Cai, A. Hulford, I. W. Brock, D. Michl, L. Hazell, I. Schmidt, R. G. Herrmann, and R. B. Klosgen.1994. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO J. 13:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders, C., N. Wethkamp, and H. Lill.2001. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 41:241-246. [DOI] [PubMed] [Google Scholar]
- 23.Sanger, F., S. Nicklen, and A. R. Coulson.1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sargent, F., E. G. Bogsh, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer.1998. Overlapping functions of components of a bacterial Sec-independent export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler.1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 26.Simon, R., U. Priefer, and A. Puhler.1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 27.Snyder, A., A. I. Vasil, S. L. Zajdowicz, Z. R. Wilson, and M. L. Vasil.2006. Role of the Pseudomonas aeruginosa PlcH Tat signal peptide in protein secretion, transcription, and cross-species Tat secretion system compatibility. J. Bacteriol. 288:1762-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taubes, G.2008. The bacteria fight back. Science 321:356-361. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson.2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 30.Vandenende, C. S., M. Vlasschaert, and S. Y. K. Seah.2004. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:5596-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voulhoux, R., A. Filloux, and I. J. Schalk.2006. Pyoverdine-mediated iron uptake in Pseudomonas aeruginosa: the Tat system is required for PvdN but not for FpvA transport. J. Bacteriol. 188:3317-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh, C.2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 33.Yoneyama, H., H. Maseda, H. Kamiguchi, and T. Nakae.2000. Function of the membrane fusion protein, MexA, of the MexA, B-OprM efflux pump in Pseudomonas aeruginosa without an anchoring membrane. J. Biol. Chem. 275:4628-4634. [DOI] [PubMed] [Google Scholar]
- 34.Yoneyama, H., A. Ocaktan, N. Gotoh, T. Nishino, and T. Nakae.1998. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 244:898-902. [DOI] [PubMed] [Google Scholar]
- 35.Yoneyama, H., A. Ocaktan, M. Tsuda, and T. Nakae.1997. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 233:611-618. [DOI] [PubMed] [Google Scholar]
- 36.Yoneyama, H., Y. Yamano, and T. Nakae.1995. Role of porins in the antibiotic susceptibility of Pseudomonas aeruginosa: construction of mutants with deletions in the multiple porin genes. Biochem. Biophys. Res. Commun. 213:88-95. [DOI] [PubMed] [Google Scholar]