Abstract
Bacteria belonging to the “Burkholderia cepacia complex” (Bcc) often cause fatal pulmonary infections in cystic fibrosis patients, yet little is know about the underlying molecular mechanisms. These Gram-negative bacteria can adopt an intracellular lifestyle, although their ability to replicate intracellularly has been difficult to demonstrate. Here we show that Bcc bacteria survive and multiply in macrophages of zebrafish embryos. Local dissemination by nonlytic release from infected cells was followed by bacteremia and extracellular replication. Burkholderia cenocepacia isolates belonging to the epidemic electrophoretic type 12 (ET12) lineage were highly virulent for the embryos; intravenous injection of <10 bacteria of strain K56-2 killed embryos within 3 days. However, small but significant differences between the clonal ET12 isolates K56-2, J2315, and BC7 were evident. In addition, the innate immune response in young embryos was sufficiently developed to control infection with other less virulent Bcc strains, such as Burkholderia vietnamiensis FC441 and Burkholderia stabilis LMG14294. A K56-2 cepR quorum-sensing regulator mutant was highly attenuated, and its ability to replicate and spread to neighboring cells was greatly reduced. Our data indicate that the zebrafish embryo is an excellent vertebrate model to dissect the molecular basis of intracellular replication and the early innate immune responses in this intricate host-pathogen interaction.
In the 1980s, bacteria belonging to the “Burkholderia cepacia complex” (Bcc) emerged as opportunistic pathogens in immunocompromised patients, particularly patients with cystic fibrosis (CF) and chronic granulomatous disease (CGD) (42, 60). Formerly classified into genomovars, Bcc bacteria are now grouped in 17 species (25), all of which have been found in CF patients. Although Pseudomonas aeruginosa is the most common CF pathogen, Bcc infections are associated with poorer clinical prognosis, with Burkholderia cenocepacia and Burkholderia multivorans being the most prevalent species. After an unpredictable period of colonization, the bacteria can cause acute, fatal necrotizing pneumonia and septicemia, known as “cepacia syndrome,” in a some colonized patients (20, 60). The intrinsic multiple resistance of Bcc to antibiotics makes it very difficult to treat. Several epidemic outbreaks have resulted from the rapid spread of certain strains of B. cenocepacia that were highly transmissible between CF patients through both hospital and social contacts (19, 69), and the best-described outbreaks were caused by members of the electrophoretic type 12 (ET12) lineage identified in Canadian and United Kingdom CF populations in the late 1980s (20, 29).
Over the past decade much progress has been made in understanding the epidemiology of clinical infections (42), and information concerning the pathogenesis and genetics of virulence has begun to provide clues about how Bcc causes disease. Although several virulence factors, including hemolysin, proteases, siderophores, lipopolysaccharide, flagella, cable pili, a type III secretion system, catalases, superoxide dismutase, and quorum sensing, have been identified (for a review, see reference 45), the molecular mechanisms of this disease are still largely unknown. Important questions are how the bacteria evade host immune responses, how they persist, what triggers the often fatal septicemia, what the bacterial intracellular survival strategy is, and how the bacteria disseminate.
B. cenocepacia has been detected inside airway and alveolar epithelial cells, macrophages, and neutrophils in the lungs of CF patients (62, 72) and has been documented to be present intracellularly in murine models (8, 63, 72). Using cell culture infection models, it was shown that B. cenocepacia can invade and survive in both professional and nonprofessional phagocytes, as well as in amoebae (4, 46, 47, 61), by actively interfering with phagosome maturation (35) and delaying phagosome acidification (34). Recently, it was shown that the delay in acidification and phagolysosomal fusion is more compromised in cystic fibrosis transmembrane conductance regulator (CFTR)-negative macrophages (36) and that assembly of the NADPH-oxidase complex at the phagosomal membrane is compromised in B. cenocepacia-infected macrophages, which is also further delayed by inhibition of the CFTR (30). The ability of B. cenocepacia to invade and survive intracellularly as well as its ability to resist toxicity by reactive oxygen species in highly inflamed lungs (37) has been proposed to contribute significantly to its pathogenesis. However, the key question of whether B. cenocepacia can replicate intracellularly has been difficult to answer conclusively due to the intrinsic antibiotic resistance of this organism, which complicates the use of classical gentamicin protection assays (35). Colocalization with cellular markers in infected bronchial epithelial cells suggested that B. cenocepacia replicates inside endoplasmic reticulum-derived vacuoles (65). However, studies of the cell biology of Bcc infection are lagging far behind research on intracellular pathogens such as Legionella pneumophila (28) and Brucella (7), and new research tools are indispensable for increasing our knowledge of the cellular biology of infection by this group of important pathogens.
To address the complexity of chronic respiratory infections, a number of mammalian infection models, such as the rat agar bead and mouse cftr−/− models, have been developed (9, 63, 70, 74). Alternative nonvertebrate animal models, such as Caenorhabditis elegans, have been developed to study Bcc virulence (6, 32), and recently larvae of the wax moth were shown to be valuable for examining Bcc virulence (67). Nematode models have been instrumental in identifying bacterial virulence factors and showing fundamental conservation of virulence mechanisms for infection of evolutionarily divergent hosts with, for instance, P. aeruginosa (41). The invertebrate immune system, however, shows many differences from the immune system of humans, and other models are needed to address specific questions related to the innate immune response to a specific pathogen in great detail. The ability to mount an effective adaptive immune response, involvement of the complement system, and development of complex hematopoietic cell lineages are restricted to vertebrates.
Originally established as a powerful model for developmental biology and human genetics, the zebrafish (Danio rerio) has emerged as a remarkably good nonmammalian vertebrate model to study development of the immune system and infectious diseases (48, 76-78, 80). A growing number of both Gram-positive and Gram-negative bacteria, including both natural pathogens of fish and true human pathogens, have been found to infect zebrafish (3, 10, 56, 76, 83). The zebrafish has many useful features as a model system, and the number of available cellular, molecular, and genetic tools, such as forward and reverse genetic screens and antisense techniques using morpholinos, is rapidly increasing. Importantly, proteins with significant homology to major factors in inflammation in humans, such as Toll-like receptors (TLR), the complement system, proinflammatory cytokines, acute-phase response proteins, and counterparts of the mammalian viral and bacterial interferon-dependent defense functions, are present in fish (2, 40, 49, 68, 75, 82). Whereas an adaptive immune system develops at later stages during development, an innate immune system resembling that of mammals is already developing in young embryos (13, 23, 33, 38).
In this study, we exploited the transparency of the zebrafish embryo to visualize Bcc infections in real time. We show here that several Bcc species are highly virulent by establishing an intracellular replication niche in macrophages, followed by dissemination and bacteremia. In addition, a cepR quorum-sensing mutant was attenuated, and we observed differences in virulence between strains obtained from a panel of Bcc clinical isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are described in Table 1. These strains include clinical isolates belonging to several species from a panel of well-characterized Bcc strains (44). Escherichia coli and Bcc strains were cultured at 37°C in Luria-Bertani (LB) broth. Bcc and E. coli strains carrying plasmids pIN25, pIN29, or pIN62 (Table 1; see supplemental text and Fig. S1 in the supplemental material) were grown in the presence of 100 μg ml−1 and 30 μg ml−1 chloramphenicol, respectively. For selection with trimethoprim, concentrations of 250 μg ml−1 and 1,500 μg ml−1 were used, whereas plasmids encoding a tetracycline resistance cassette were selected using 250 μg ml−1 and 10 μg ml−1, respectively.
TABLE 1.
Bcc strains and mutants and plasmids used in this study
| Species or plasmid | Isolate | Descriptiona | Source | Reference |
|---|---|---|---|---|
| Species | ||||
| E. coli | DH5α | φ80lacZΔM15 ΔlacU169 endA1 recA1 hsdR17 supE44 thi-1 gyrA96 relA1 | ||
| B. cenocepacia | K56-2 (= LMG18863) | ET12, Toronto, Canada, CF | C. Mohr | 14 |
| J2315 (= LMG16656) | ET12 index strain, Edinburgh, UK, CF | J. Govan | 19 | |
| BC7 (= LMG18826) | ET12, Toronto, Canada, CF | BCCM/LMG | 64 | |
| J415 (= LMG16654) | Not associated with patient-to-patient spread, CF | BCCM/LMG | 18 | |
| K56-2 R2 | cepR mutant | P. Sokol | 39 | |
| K56-2 R2(pSLR100) | Complemented cepR mutant containing plasmid pSLR100 | P. Sokol | 39 | |
| B. stabilis | LMG14294 | Belgian CF patient, stable condition; detected in one other patient | BCCM/LMG | 58 |
| B. vietnamiensis | FC441 (= LMG18836) | 9-year-old boy with X-linked recessive CGD who survived septicemia | BCCM/LMG | 44 |
| B. cepacia | CEP509 (= LMG18821) | CF patient, Sydney, Australia; recovered from three other patients | BCCM/LMG | 44 |
| Plasmids | ||||
| pIN25 | oripBBR Δmob, Cmr, GFP | This study | ||
| pIN29 | oripBBR Δmob, Cmr, DSRed | This study | ||
| pIN62 | oripBBRmob+, Cmr, DSRed | This study | ||
| pIN63 | oripBBRmob+, Tpr, DSRed | This study | ||
| pIN72 | oripBBRmob+, Tetr, DSRed | This study |
CF, cystic fibrosis patient; CGD, chronic granulomatous disease; Cmr, chloramphenicol resistance; Tpr, trimethoprim resistance; Tetr, tetracycline resistance; Δmob, mobilization deficient.
Zebrafish care and maintenance.
Mixed male and female populations of zebrafish (D. rerio) variety Gold (purchased from Antinea SARL, Montpellier, France) or transgenic Tg(mpx::eGFP)i114 zebrafish (AB strain) (57) were kept in 35-liter tanks containing conditioned water at 27°C (53). The Gold variety lacks strong pigment, and therefore the use of melanization inhibitors was not necessary. The animals were handled according to national regulations for animal welfare. A light cycle consisting of 10 h of darkness and 14 h of light was used. Embryos were obtained by natural spawning in 2-liter breeding tanks (Tecniplast, France) that were placed in the fish tanks the evening before spawning, and they were raised at 28°C in E3 egg water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 0.00005% methylene blue) (53). The ages of the embryos are expressed in hours postfertilization (hpf).
Microinjection of bacteria into zebrafish embryos.
Microinjection was performed basically as described by van der Sar et al. (81), with a few modifications. Briefly, bacteria expressing DSRed or green fluorescent protein (GFP) were grown overnight in LB broth with appropriate antibiotics. Bacteria were collected by centrifugation at 3,000 × g for 2 min and resuspended in phosphate-buffered saline (PBS) (Gibco). Bacterial dilutions (generally 50 bacteria/nl) were prepared in PBS (with 0.05% phenol red to visualize microinjection). Embryos were dechorionated and anesthetized in E3 medium containing 0.02% buffered MS222 (tricaine; ethyl-3-aminobenzoate methanesulfonate salt; Sigma). Microinjection of bacteria into embryos, staged between 28 and 32 hpf, was performed with a Femtojet microinjector (Eppendorf) and a micromanipulator with pulled microcapillary pipettes using a stereo light microscope (Leica MS5). A 0.5- to 1-nl portion of a bacterial suspension was injected directly into the blood circulation of each embryo, either in the blood island or in the axial vein. In initial experiments we determined the inoculum size by direct microinjection onto agar plates. However, we found that plating 5 embryos individually (see below) immediately after microinjection (zero time) resulted in more accurate determination of the numbers of CFU actually injected. To follow infection kinetics, inoculated embryos were incubated individually in E3 medium in 24-well plates at 28°C and sampled at later stages during infection. Five embryos were collected at each time point, generally 2, 24, and 48 h postinfection (hpi), and individually treated for bacterial enumeration. Each embryo was rinsed in 1× PBS, anesthetized in tricaine, and transferred to a 1.5-ml Eppendorf tube in 50 μl (total volume) of tissue-culture-grade trypsin-EDTA. The embryos were disrupted by vigorous pipetting (40 times) with a yellow tip. Next, 50 μl 2% Triton X-100 was added, and the preparations were incubated for 20 min at room temperature, which was followed by additional homogenization by pipetting (20 times). Depending on the expected numbers of bacteria (visualized by fluorescence microscopy), the complete mixture was plated on LB agar plates containing 100 mg/liter chloramphenicol or serial dilutions were prepared in PBS and plated as 10-μl droplets, essentially as described by Miles and Misra (51). The trypsin-EDTA and Triton X-100 treatments did not affect bacterial viability.
For survival assays, embryos were similarly microinjected and maintained individually in 24-well plates in E3 medium at 28°C. At regular time points after infection, the number of dead embryos was determined visually based on the absence of a heartbeat.
Statistical analysis.
The number of CFU per time point for each strain was expressed as the geometric mean ± standard error of the mean for five individually plated embryos. Experiments were performed at least three times, unless stated otherwise. Student's t test (with two-tailed distribution and equal variance) was performed to determine whether the results for two strains were significantly different (P < 0.05).
Microscopic analysis.
A Leica DM IRB inverted microscope equipped for bright-field, differential interference contrast (DIC), and fluorescence imaging was used. GFP and DSRed were excited using a 100-W mercury lamp, and fluorescence was detected using filter sets L5 (band pass [BP] 480/40; beam splitter [BS] 505; emission BP527/30) and N2.1 (515 to 560; BS 580; emission long pass [LP] 590), respectively. For imaging we used a Coolsnap fx (Roper Scientifique) and MetaVue software, and images were processed further using Adobe Photoshop. Embryos were transferred to E3 containing MS222 in glass-bottom dishes (MatTek Corp., Ashland, MA) for direct visualization using 40×, 63× and 100× oil immersion objectives.
FISH with combined immunodetection of B. cenocepacia.
We used the method described by Meijer et al. (50) for combined detection of bacteria and detection of expression of the myeloid-cell-specific marker genes csf1R (macrophages) and mpx (neutrophils) by fluorescence in situ hybridization (FISH), based on protocols developed previously by Clay and Ramakrishnan (12) and Welten et al. (84). Digoxigenin (DIG)-labeled riboprobes for csf1R (CD759443) and mpx (BC056287) (both sense and antisense) were prepared using primers and methods described by Meijer et al. (50) and an in vitro transcription kit obtained from Ambion. Specific details concerning the method and minor differences from previously described protocols are described below.
For FISH, approximately 500 CFU of B. cenocepacia was microinjected into K56-2 embryos. Infected embryos were collected at 5 hpi and 24 hpi and fixed in 4% paraformaldehyde (PFA) in PBS at 4°C overnight. After dehydration of the embryos in methanol and subsequent rehydration, treatment with proteinase K (10 μg ml−1) for 20 min was performed at room temperature. For hybridization we used a probe concentration of 200 to 1,000 ng ml−1 and a hybridization temperature of 66°C. For specific detection we incubated the embryos in a 1:2,000 dilution of horseradish peroxidase-conjugated anti-DIG Fab fragments (Roche) in blocking solution (BS) (1% Western blocking solution [Roche] in PBS with 0.1% Tween 20 [PBST]) overnight at 4°C, which was followed by TSA Plus Cy3 (Perkin Elmer) signal amplification treatment for 20 min. To continue immune detection of bacteria, embryos were incubated twice for 30 min in 6% hydrogen peroxide in PBST before they were incubated for 2 h in BS. Embryos were incubated in B. cenocepacia rabbit antiserum (1:500 dilution in BS), kindly provided by Holger Scholz (Munich, Germany), and this was followed by detection with a 1:1,000 dilution of Alexa Fluor 488 goat anti-rabbit IgG (Molecular Probes) in BS.
RESULTS
Construction of highly fluorescent bacterial strains for real-time analysis.
For optimal visualization of Bcc strains by fluorescence microscopy after infection of translucent zebrafish embryos, we created a series of mobilizable and nonmobilizable pBBR-derived plasmids (Table 1; see supplemental text and Fig. S1 in the supplemental material) with a range of selectable marker genes (chloramphenicol, tetracycline, and trimethoprim resistance genes) that expressed DSred or GFP at high levels under control of a strong constitutive tac promoter sequence. In the absence of antibiotic selection, the plasmids remained stable for more than 3 days after microinjection of Bcc into the embryos (data not shown).
Zebrafish embryos are extremely sensitive to B. cenocepacia ET12 strains, but differences between clonal isolates can be seen.
To determine the levels of virulence of B. cenocepacia in young (28 to 32 hpf) zebrafish embryos, we microinjected red fluorescent bacteria of ET12 lineage strains K56-2 and J2315, which are known to be highly virulent in other infection models (1, 6, 67), directly into the bloodstream (Fig. 1A). The infection was followed by determination of embryo mortality, analysis of the progression of infection in real time by fluorescence microscopy, and determination of the bacterial load by plating infected embryos. Both B. cenocepacia strains were highly virulent for the young embryos. In contrast to E. coli or heat-killed B. cenocepacia, which did not kill embryos (data not shown), live K56-2 or J2315 killed embryos between 2 and 4 days after infection (Fig. 1C). Virulence was confirmed by quantitative plating assays (Fig. 1B). E. coli was not virulent for zebrafish embryos and was phagocytosed and killed within 4 to 5 h after injection (23); however, as few as 5 microinjected CFU of K56-2 were sufficient to establish a virulent infection with growth kinetics similar to those seen in embryos infected with a higher dose (Fig. 1B, compare K56-2 LD and K56-2) and to kill embryos within 3 days (not shown).
FIG. 1.
B. cenocepacia ET12 lineage strains, but not J415, are highly virulent for zebrafish embryos. (A) Drawing of zebrafish embryo 30 hpf. The site of microinjection in the blood island is indicated. Scale bar, 100 μm. (B) Bacterial multiplication in embryos microinjected with K56-2 and J2315. The data are geometric means ± standard errors of the means (n = 5). The mean sizes of the inoculum for each embryo were 143 CFU (K56-2) and 4 CFU (K56-2 LD) for K56-2 and 117 CFU for J2315. J2315 and K56-2 differed significantly from each other in growth kinetics at 24 and 48 hpi (P = 0.0081 and P = 0.0067, Student's t test). The standard errors of the means at 24 hpi and 48 hpi were reproducibly less for K56-2 than for J2315 (for K56-2, 4.06 ± 0.03 and 5.54 ± 0.13 log CFU; for J2315, 2.94 ± 0.32 and 4.20 ± 0.27 log CFU). (C and D) Results of experiments in which embryos were inoculated with B. cenocepacia strains BC7 (45 CFU), J415 (72 CFU), J2315 (60 CFU), and K56-2 (126 CFU). (C) Survival following injection of different strains (n = 20 for each strain). (D) Bacterial multiplication within embryos. The data are geometric means ± standard errors of the means (n = 5). In this experiment the results for BC7 differed significantly from the results for K56-2 and J415 at 24 hpi (P = 0.00076 and P = 0.0051, respectively) and at 48 hpi (P = 0.008 and P = 1.5 × 10−5, respectively), and the results for J2315 and K56-2 differed significantly at 48 hpi (P = 0.003) but not at 24 hpi (P = 0.14). (E and F) Fluorescent images of embryos microinjected with K56-2 (E) and J2315 (F) expressing DSRed at 46 hpi and 68 hpi, respectively. The images show the final stages of infection. The arrowhead and arrow in panel F indicate bacterial aggregate formation in the intersegmental vessels in the tail. Scale bar, 300 μm.
Interestingly, the bacterial growth kinetics (Fig. 1B) and the time of onset of mortality (Fig. 1C) suggested that K56-2 was more virulent than J2315, even though these are clonal isolates. In a total of 8 experiments, we showed that K56-2 was significantly more virulent than J2315 (see Fig. S2 in the supplemental material). Between 2 and 48 hpi, the average increase in the number of CFU for K56-2 was 4,000-fold, while the increase for J2315 was only 250-fold (see Fig. S2C in the supplemental material). The animal-to-animal variation for K56-2-infected fish was very small both within an experiment (Fig. 1B) and between experiments (see Fig. S2A in the supplemental material); however, much greater variation was seen for J2315 (see Fig. S2B in the supplemental material). In addition, a more variable time to death for J2315-infected embryos was observed (data not shown), suggesting that the outcome of infection with J2315 is more unpredictable. The greater virulence of K56-2 than of J2315 was also evident when fish were inoculated with very low doses (8 and 6 CFU, respectively [see Fig. S2A and B in the supplemental material]), as the disease caused by a few bacteria was more severe when K56-2 was used.
After all bacteria had been phagocytosed in the hours following microinjection, both strains slowly reentered the blood circulation at about 18 hpi (see below for details). During later stages of the infection (>40 hpi) we observed formation of extracellular bacterial aggregates, possibly biofilms, in the blood vessels of about 50% of the J2315-infected embryos by fluorescence microscopy, and these aggregates often clearly marked large parts of the vascular system in the final stages of infection (Fig. 1F). K56-2 was still circulating freely in the bloodstream with only minor formation of bacterial aggregates in the vessels at the time of death (Fig. 1E), but it killed the embryos much more rapidly than J2315 killed the embryos (48 hpi versus 72 hpi), possibly by inducing a greater inflammatory response.
Zebrafish embryos can control infection with a less virulent B. cenocepacia strain.
Next we analyzed whether the virulence observed with K56-2 and J2315 was specific for ET12 lineage strains (Bcc subgroup IIIA) or whether the young embryos were highly sensitive to infection with any B. cenocepacia strain. Therefore, we tested the virulence of another ET12 lineage strain, BC7, and of B. cenocepacia strain J415 (subgroup IIIB strain [Eshwar Mahenthiralingam, personal communication]) that was isolated from a single patient and was not associated with patient-to-patient spread. In infection assays, BC7 differed significantly from K56-2, but it was not significantly more or less virulent than J2315 (Fig. 1C and D). Interestingly, during later stages of infection, BC7 formed bacterial aggregates in the veins similar to those observed with J2315 (Fig. 1F and data not shown), showing that clonal isolates BC7 and J2315 behave similarly and have pathogenic traits distinct from those of K56-2. J415 was not very virulent for zebrafish embryos with the infection doses used in this study, and although injected bacteria could resist killing by the host and persist in the fish for longer time periods, most embryos could clear the infection (Fig. 1C and D). Differences in the in vitro growth rates did not match the observed differences between the strains in the embryo model (data not shown), indicating that differences in virulence between the strains are due to variation in virulence factors and not the growth rate. The findings obtained with BC7 confirm that B. cenocepacia strains belonging to the ET12 lineage are highly virulent for young zebrafish embryos. However, the innate immune system of these embryos was sufficiently developed to control infection with B. cenocepacia J415.
Different Bcc strains show distinct levels of virulence in zebrafish embryos.
To investigate whether the zebrafish embryo model also reflects differences in virulence between different Bcc species, we compared B. cepacia strain CEP509, Burkholderia stabilis LMG14294, and Burkholderia vietnamiensis FC441 with B. cenocepacia K56-2 in bacterial replication and survival assays. As shown in Fig. 2A, K56-2 was still the most virulent strain, producing significantly greater bacterial loads. However, CEP509 was also highly virulent and multiplied to produce levels significantly higher than those of FC441 or LMG14294. The difference was less pronounced when very small inocula were used for CEP509 (<15 CFU) (data not shown). LMG14294 and FC411 did not differ significantly (P < 0.05) in several independent experiments, and although these strains were not very virulent, they were not eradicated by the host's immune system and were able to survive within the fish for longer periods of time.
FIG. 2.
Different Bcc strains show distinct levels of virulence in zebrafish embryos, reflecting clinical information. Embryos were microinjected with B. cenocepacia K56-2 (134 CFU), B. cepacia CEP509 (114 CFU), B. stabilis LMG14294 (62 CFU), and B. vietnamiensis FC441 (234 CFU). One subgroup was used to determine bacterial multiplication, and another subgroup was used to determine mortality rates. (A) Bacterial multiplication within embryos. The data are geometric means ± standard errors of the means (n = 5). The bacterial loads of K56-2 at 24 hpi and 48 hpi were significantly greater than those of CEP509 (P = 0.002 and P = 0.09), LMG14294 (P = 0.001 and P = 0.005), and FC441 (P = 1.0 × 10−5 and 1.5 × 10−6). CEP509 was significantly more virulent than LMG14294 and FC441 at 24 hpi (P = 0.01 and 0.0002, respectively) and at 48 hpi (P = 0.002 and 8.3 × 10−5, respectively). (B) Embryo survival following infection with Bcc strains (n = 20 for each strain).
The differences in bacterial growth kinetics were reflected in the results of survival assays; embryos infected with K56-2 died reproducibly earlier than embryos infected with CEP509 (Fig. 2B), whereas LMG14294 and FC441 were still unable to cause a lethal infection at 8 days postinfection (dpi), the last time point analyzed. After infection with LMG14294 and FC441, at 5 days postinfection about 20% of the embryos had completely cleared the invading pathogen, whereas 20% and 5% of the embryos, respectively, were heavily infected (but still alive). Microscopic analysis showed that the other infected embryos all contained between 1 and 10 infected cells at this stage. Interestingly, the heavily infected embryos were still alive at 8 dpi, and even though in some embryos bacteria were able to replicate intracellularly and spread (see below for details of infection at the cellular level), the bacteria had not reentered the blood circulation, after they were phagocytosed rapidly after microinjection, to cause a bacteremic infection (not shown).
B. cenocepacia replicates intracellularly, which is followed by dissemination and bacteremia.
To dissect the course of the infection of zebrafish embryos by B. cenocepacia, we first examined the kinetics of K56-2 infection in more detail during the early stages of infection. Embryos were sampled in one experiment at 0, 1, 2, 4, 5, 6, 18, 24, 30, 42, and 48 hpi and in another experiment at 0, 2, 5, 7, 8, 10, 12, and 24 hpi. Enumeration of the bacteria showed that K56-2 was not killed after microinjection into the blood circulation of the embryos; rather, all bacteria survived in the host (Fig. 3). K56-2 began to replicate 6 to 8 h after infection, and the numbers of bacteria at the time of death of the embryos were high.
FIG. 3.
K56-2 survives in zebrafish embryos and starts to replicate between 6 and 8 hpi. Embryos were microinjected with B. cenocepacia K56-2 (61 CFU in experiment 1 [exp1] and 25 CFU in experiment 2 [exp2]). The data are geometric means ± standard errors of the means (n = 5).
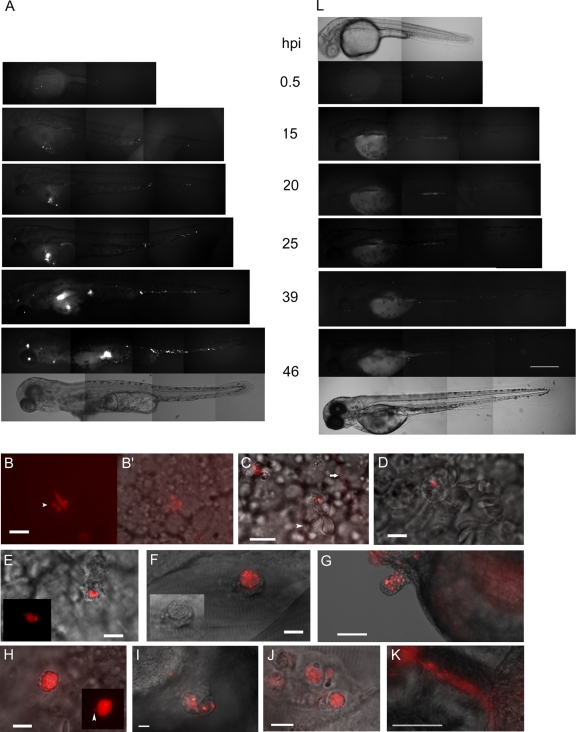
We also followed the fate of microinjected bacteria in real time by fluorescence microscopy. The progression of the infection was visualized by examining individual embryos at different time points after infection (a representative embryo is shown in Fig. 4A). At the cellular level, K56-2 was taken up by phagocytic cells circulating in the blood, anchored to the wall of the blood vessels, or wandering in the yolk sac circulation valley (Fig. 4C). Most circulating cells settled very quickly after uptake of bacteria, and they often seemed to attract other cells. Within 4 to 5 h the majority of the bacteria circulating freely in the blood (even with higher infection doses up to 500 to 1,000 CFU) had been taken up by phagocytic cells. Unlike for E. coli, which was phagocytosed and killed within 4 to 5 h, as indicated by the presence of diffuse red fluorescence without clear bacterial morphology (Fig. 4B), microscopic analysis of K56-2-infected embryos confirmed the data obtained in enumeration assays (Fig. 3) showing that bacterial multiplication began between 6 and 8 hpi (Fig. 4D and E). In these experiments we used very small inocula (<10 CFU), which ensured that cells originally phagocytosed only one or two bacteria. The bacteria seemed to replicate in membrane-bound vacuoles. By 15 hpi, some cells were packed with bacteria, often clearly visible in vacuoles (Fig. 4F), and generally at this stage no freely circulating bacteria were detected in the blood. The very small inoculum ruled out the possibility that these infected cells were the result of engulfment of large numbers of intravenously injected bacteria, and thus the results clearly demonstrated that there was intracellular replication within phagocytes.
FIG. 4.
Real-time visualization of K56-2 infection showing intracellular replication, dissemination, and bacteremia. (A and C to L) Real-time analysis by fluorescence microscopy of embryos infected with K56-2 (A and C to K) and a cepR mutant (L) expressing DSRed. (A and L) Fluorescence images taken at 30 min to 46 hpi of two embryos (lateral view, anterior side to the left) infected with ∼175 CFU of K56-2 (A) or ∼130 CFU of cepR (L). For 0.5 hpi and 46 hpi corresponding bright-field images are included. The embryo infected with K56-2 was dead at 46 hpi. Scale bar, 300 μm. (B and B′) Fluorescence image (B) and fluorescence and bright-field overlay image (B′) of an embryo inoculated with 500 CFU E. coli, at 2 hpi. A macrophage in the yolk sac valley containing bacteria is shown. Three vacuoles with degraded E. coli are distinguished by the diffuse red signal and the round small vacuole, in contrast to the large E. coli bacteria (the arrowhead indicates one of the three phagolysosomes). Scale bar, 10 μm. (C) Zebrafish embryo inoculated intravenously with ∼500 CFU K56-2: overlay of fluorescence and DIC images showing a phagocytic cell 30 min after infection, anchored to the side of a blood vessel that has a single K56-2 bacterium adhered to it or taken up. Two erythrocytes (arrowhead) are attached to the macrophage. The two red lines (one indicated by a horizontal arrow) are bacteria moving in the blood circulation at the time of image acquisition. In the left corner is an out-of-focus phagocytic cell with engulfed bacteria. Scale bar, 10 μm. (D and E) Embryos were inoculated with 5 to 10 CFU of K56-2. The DIC and fluorescence overlay images obtained at 9 hpi show B. cenocepacia-containing phagocytic cells that initially took up one or two bacteria. The inset in panel E shows the fluorescence image. Scale bars, 10 μm. (F) Fluorescence and DIC overlay image taken at 23 hpi, showing a highly infected cell in the tail region (rostral side to the right, dorsal side up) in which bacteria are clearly present in a membrane-bound vacuole. Scale bar, 10 μm. The inset shows a DIC image. (G) Fluorescence and bright-field overlay image of an infected embryo (rostral side to the right, dorsal side up) at 26 hpi, showing budding of infected cells near the pericardium. Scale bar, 100 μm. (H) Representative DIC and fluorescence overlay image after embryos were inoculated with 5 to 10 CFU of K56-2, showing a cell at 24 hpi that is packed with bacteria and a single bacterium apparently “escaping” from the vacuole. The inset shows the fluorescence image. Scale bar, 10 μm. (I) DIC and fluorescence overlay image of a cell aggregate with Burkholderia-containing cells that formed in an embryo infected with 50 CFU K56-2, at 24 hpi. Scale bar, 50 μm. (J) DIC and fluorescence overlay image of a cell aggregate that formed in an embryo inoculated with 5 to 10 CFU of K56-2, at 28 hpi. At this point in infection bacteria had already spread from the initially infected macrophage to neighboring cells in the aggregate. Scale bar, 10 μm. (K) Bright-field and fluorescence overlay image of an intersegmental blood vessel in the tail, showing formation of extracellular bacterial aggregates. Scale bar, 50 μm. (L) Fluorescence and bright-field images of embryos inoculated with cepR mutant R2 (see above).
Single infected cells were observed to be moving in the bloodstream at different times during infection, showing occasional spread to other locations. Interestingly, infected cells often (but not always) attracted other, noninfected host cells and formed cell aggregates (Fig. 4G, I, and J). From 15 to 20 hpi on, local spreading was observed, initially mainly within the cell aggregates that formed. Although occasionally bacteria had reentered the blood circulation at his point, the highly infected cells did not lyse. Currently, we do not know the precise mechanism of spreading; however, from the onset of dissemination, single bacteria were often observed to be close to infected cells, also in embryos infected with only a few bacteria, and at the periphery of cells (Fig. 4H). We found no evidence for formation of apoptotic blebs, like those recently described for Leptospira interrogans infection (15), suggesting that the bacteria had apparently actively escaped from vacuoles of the infected cells. From this time on, dissemination of bacteria to neighboring cells in the aggregates was followed by intracellular replication (Fig. 4I and J). Sometimes the aggregates were shed from the embryo (Fig. 4G). At this time, infection also became bacteremic with extracellular multiplication. Occasionally, K56-2 formed extracellular aggregates in the vessels (Fig. 4K), as described above. By 24 hpi, host tissue deformation started to become visible, and at later stages host cells were massively shed from the embryos (Fig. 1E and 4A, 46 hpi), which may have been induced by a high inflammatory response and cell death. By 48 hpi (depending on the infection dose) the embryos had finally succumbed to the invading pathogen.
B. cenocepacia survives and replicates inside macrophages.
In CF patients, bacterial infections coincide with massive neutrophil infiltration in the inflamed lungs, and B. cenocepacia has been detected inside neutrophils, as well as inside alveolar macrophages in the lungs of patients (62, 72). In zebrafish, primitive macrophages are the first leukocytes that appear, invading embryonic tissues after their differentiation. They have been shown to be actively involved in phagocytosis of microinjected bacteria as early as 28 h postfertilization (hpf) (23, 81). Neutrophilic granulocytes start appearing at 33 to 35 hpf and have been shown to phagocytose P. aeruginosa and Staphylococcus aureus (3, 56); at 2 days postfertilization (dpf) neutrophilic granulocytes are mature, and they are the major leukocyte type (38). In this study, the morphology of the cells (large cells with vacuolated cytoplasm) that engulfed Bcc, their amoeba-like movements, and the presence of vacuoles containing methylene blue taken up from the medium (38) suggested that the majority of Burkholderia-engulfing cells were macrophages (data not shown).
To determine more precisely whether B. cenocepacia is indeed phagocytosed mainly by macrophages, we first performed coinfection studies with high doses (500 CFU) of two bacteria that were labeled differently: E. coli (GFP), which was shown to be ingested primarily by macrophages by several groups of workers (23, 50, 81), and B. cenocepacia (DSRed). B. cenocepacia was taken up by the same phagocytic cells as E. coli (Fig. 5A to C).
FIG. 5.
B. cenocepacia is taken up mainly by macrophages, and there is minimal phagocytosis by neutrophils. (A to C) Simultaneous microinjection of high doses (∼500 CFU) of E. coli expressing GFP and B. cenocepacia K56-2 expressing DSRed was followed by engulfment of both bacterial species by the same cell type, at 0.5 hpi. (A) Fluorescence image overlay showing overview of colocalization of E. coli (green) and K56-2 (red) in the yolk sac circulation valley, at 0.5 hpi. Scale bar, 50 μm. (B and B′) DIC close-up image of phagocytic cell at 1 hpi (B) and DIC and fluorescence overlay image of the same cell (B′) with E. coli (green) and K56-2 (red) adhering to or engulfed by the cell. Scale bar, 10 μm. (C) Fluorescence image of a phagocytic cell with E. coli (green) and K56-2 (red). Scale bar, 10 μm. (D and E) Colocalization of FISH signal (red) with macrophage-specific riboprobe cfs1R and an immunolabeling signal for B. cenocepacia (green), at 5 hpi. Scale bar, 10 μm. The inset in panel E is an enlarged fluorescence image (green filter). (F and G) FISH using a neutrophil-specific mpx riboprobe (red signal) and an immunolabeling signal for B. cenocepacia (green), at 24 hpi. The images are two images of the same embryo (rostral side to left, dorsal side up) but in different focal planes. (F) The microscope was focused on the B. cenocepacia-containing cells, and mpx-stained cells are not in focus. (G) The microscope was focused on mpx-positive cells, and B. cenocepacia containing cells are not in focus. Scale bar, 100 μm. (H to J). K56-2 (∼500 CFU) infection of Tg(mpx::eGFP)i114 embryos at 30 hpf. (H) From left to right, images of the embryo sac taken with the red channel, the green channel, green and red overlay, and fluorescence-DIC overlay. Cells in the embryo sac valley at 3 hpi that have taken up many DSRed-expressing bacteria do not colocalize with mpx-GFP-expressing neutrophils. Scale bar, 50 μm. (I) Close-up image of individual fluorescence and overlay (combined fluorescence and DIC) images showing mpx-GFP-expressing neutrophils and K56-2 at 3 hpi. The inset is an enlargement of a neutrophil that has taken up a DSRed-expressing bacterium, indicated by an arrowhead. The arrows indicate non-GFP-fluorescing cells containing multiple DSRed-expressing bacteria. Scale bar, 10 μm. (J) Fluorescent overlay images showing six individual GFP-expressing neutrophils, at 18 hpi. About 20 to 30% of the cells with GFP fluorescence contained one or a few bacteria (arrowheads). The arrows indicate nonfluorescent cells containing bacteria. Scale bar, 10 μm.
Next, we performed fluorescence in situ hybridizations (FISH) with macrophage-specific colony-stimulating factor 1 receptor (csf1R) and neutrophil-specific myeloperoxidase (mpx) riboprobes (22, 50) in K56-2-infected embryos. At 5 hpi, B. cenocepacia, which was detected with B. cenocepacia-specific antibodies, was colocalized with csf1R-positive cells (Fig. 5D and E). Although mpx-positive cells have been shown to ingest bacteria as early as 32 hpf (3, 56), using FISH we found no evidence that neutrophils were actively involved in phagocytosis of B. cenocepacia in this model (data not shown). FISH colocalization studies at 24 hpi, just prior to bacterial dissemination, also produced no evidence that there was colocalization of K56-2 with neutrophils; in fact, neutrophils seemed to be present in a different tissue layer than the infected cells (Fig. 5F and G).
To more accurately determine the role of neutrophils in phagocytosis, we used a transgenic reporter fish line, Tg(mpx::eGFP)i114, that specifically expresses GFP in neutrophils (57). After microinjection of high doses (∼500 CFU) of DSRed-expressing K56-2 into 30-hpf embryos, we monitored the bacteria in live embryos using fluorescence microscopy. Shortly after microinjection, most bacteria were found to concentrate in GFP-negative cells (Fig. 5H and I), in line with our previous findings showing that macrophages are the major phagocytosing population. However, use of the transgenic zebrafish revealed that K56-2 bacteria were sometimes also ingested by GFP-expressing cells (Fig. 5I), although the number of ingested bacteria was low (1 to 5 bacteria) compared to the number of bacteria taken up by macrophages. Although neutrophils were recruited to wounded tissue at the site of microinjection, they did not seem to phagocytose more bacteria at these sites. We did not observe degradation of bacteria like that observed for E. coli (Fig. 4B), which was visualized as a diffuse red signal in GFP-expressing cells, indicating that Bcc bacteria survive killing by zebrafish neutrophils. As granulocytic neutrophils start appearing around 33 hpf and are not fully mature at this time, we infected older (50 hpf) embryos containing many granulocytic neutrophils in a similar way. The neutrophils in these older embryos behaved like those in infected 30-hpf embryos (data not shown). At 18 hpi, some of the GFP-expressing cells (20 to 30%) were observed to contain one or a few bacteria, suggesting that Bcc cells survive inside neutrophils (Fig. 5J) without replicating.
Together, our data show that macrophages form the main phagocytosing population of cells after intravenous infection of young zebrafish embryos with Bcc and that macrophages are the site of intracellular replication.
Quorum sensing is important for virulence in zebrafish embryos.
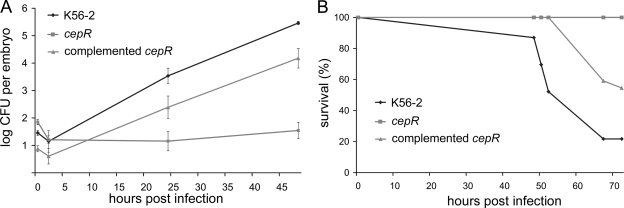
The expression of many virulence factors has been found to be controlled by the cepIR-encoded N-acyl homoserine lactone-dependent quorum-sensing system. A functional CepIR quorum-sensing system has also been shown to contribute to the severity of B. cenocepacia infections in rat and cftr−/− mouse models (71). We found that a B. cenocepacia K56-2 cepR mutant, R2 (39), was significantly attenuated compared to its parent (Fig. 6A). The bacterial loads in infected embryos either increased slightly or remained constant. Survival assays also showed that the cepR mutant was highly attenuated and generally did not kill embryos (Fig. 6B). Occasionally, an embryo infected with the cepR mutant succumbed to infection, but only at very late times (>7 days) (data not shown). The complemented mutant showed restoration of virulence to the cepR mutant in both assays (Fig. 6A and B).
FIG. 6.
K56-2 cepR mutant is highly attenuated in virulence: infection kinetics (A) and survival assay results (B) for wild-type strain K56-2, cepR mutant R2, and the cepR mutant complemented with pSLR100. (A) The average numbers of microinjected bacteria were 39 CFU (K56-2), 77 CFU (cepR), and 8 CFU (cepR mutant complemented with pSLR100). The data are geometric means ± standard errors of the means (n = 5). The growth of the mutant differed significantly from the growth of the wild-type parent at 24 hpi (P = 0.0015) and at 48 hpi (P = 1.3 × 10−6) and from the growth of the complemented mutant at 48 hpi (P = 0.0007). (B) Embryo survival following infection with Bcc strain K56-2 (31 CFU), strain R2 (83 CFU), and complemented strain R2 (16 CFU) (n = 20 for each strain).
Real-time visualization of infected embryos clearly showed the attenuated phenotype (compare Fig. 4L to Fig. 4A). In contrast to the findings for K56-2, some of the cepR mutant bacteria were unable to resist killing by macrophages, while others were able to survive intracellularly for longer time periods (more than 24 h). Occasionally, infected cells in which the mutant replicated to wild-type levels were observed; however, the bacteria did not disseminate efficiently (data not shown). In transgenic mpx-GFP embryos infected with the cepR mutant, the phagocytic behavior of macrophages and neutrophils was similar to the phagocytic behavior observed with wild-type bacteria (data not shown).
Our findings with zebrafish embryos suggest that the factors regulated by the CepIR system are required for efficient intracellular replication and dissemination.
DISCUSSION
Zebrafish as a novel model to study the involvement of host and pathogen determinants in B. cepacia complex infections.
This study demonstrates that the zebrafish embryo is an exciting new animal model to study virulence mechanisms of Bcc species. In this model, virulent B. cenocepacia bacteria are taken up primarily by host macrophages. Based on results obtained with very small intravenous inocula, we concluded that individual bacteria, after internalization and an initial 6- to 8-h adaptation phase, replicated in vacuolar compartments of macrophages and then disseminated by a mechanism that involved nonlytic release, leading to systemic infection and death of the embryos. B. cenocepacia strains belonging to the epidemic ET12 lineage were highly virulent. However, other clinical Bcc isolates and a K56-2 cepR mutant were far less virulent, showing that the innate immune system of the embryos is sufficiently developed to control infection with some Bcc strains.
Recent data from several laboratories indicate that both the macrophage and neutrophil responses in young zebrafish embryos are pathogen specific and parallel those in human infection (3), illustrating the benefit of using the zebrafish model to study the role of immune cells in infection. P. aeruginosa and S. aureus are phagocytosed by both primitive neutrophils and macrophages (3, 56), whereas primitive neutrophils, although highly attracted to sites of infection, were shown to be only marginally involved in phagocytosis of E. coli, Salmonella enterica serovar Typhimurium, or Mycobacterium marinum (11, 38, 50). Using three different experimental approaches, coinjection of E. coli and B. cenocepacia labeled with different colors, FISH, and zebrafish embryos displaying neutrophil-specific expression of GFP, we showed that macrophages are the major cell population that phagocytoses Bcc. Bcc isolates have been shown to be able to survive intracellularly within macrophages and amoebae in vitro and to replicate in respiratory epithelial cells, although the situation in human infection is unclear (for a review, see reference 66). Here we visualized Bcc survival and efficient replication in macrophages of zebrafish embryos, which provide a novel animal model for studying the capacity of intracellular Bcc to redirect maturation of the phagosome and create an intracellular replication niche. It will be interesting to learn how Bcc bacteria are able to replicate efficiently in this model, compared to their survival or minimal replication in in vitro cell culture models.
Real-time visualization of K56-2 infection in mpx-GFP transgenic embryos revealed that a small percentage of neutrophils also phagocytosed intravenously introduced K56-2 but that the number of ingested bacteria was low compared to the number of bacteria ingested by macrophages. We found no evidence of bacterial degradation, suggesting that K56-2 can evade killing by neutrophils after phagocytosis in this model. This finding contrasts with results obtained using neutrophils in vitro and gp91phox−/− mice (a mouse model of chronic granulomatous disease [CGD]) and results obtained for CGD patients, which showed that neutrophils play an important role in the host defense against Bcc requiring NADPH phagocyte oxidase (5, 17, 72, 73). Bcc, however, appears to be able to resist the massive influx of neutrophils in CF lungs, one possible explanation for which is compromised production of reactive oxygen species (ROS) (72), and in the mouse cftr−/− model recruited neutrophils exhibited minimal evidence of activation (63). In young zebrafish embryos, it is possible that the immature granulocytic neutrophils lack specific defense mechanisms that are needed to counteract K56-2. ROS production, for instance, was not detected in embryos up to 48 hpf but was detected in older embryos (24), and this could explain the ability of K56-2 to survive in such neutrophils. Also, at this point we cannot exclude the possibility that there are strain-dependent effects on the efficiency of phagocytosis and neutrophil killing. Although we observed a minimal role for neutrophils in phagocytosis of intravenously introduced K56-2 during early stages of infection, our preliminary data obtained with mpx-GFP reporter fish show that once bacterial dissemination begins and tissue inflammation is apparent, neutrophils are recruited to infected sites. The massive neutrophil infiltration seen in the lungs of CF patients is not mimicked in zebrafish; however, in future studies it will be of great interest to determine how K56-2 is able to avoid neutrophil phagocytosis and killing in this model and to employ zebrafish embryos and older larvae to study the role and competence of neutrophils at different developmental stages during Bcc infection.
Interestingly, the role of phagocytes after infection of zebrafish embryos with P. aeruginosa, the most common CF pathogen, seems to differ from their role described here after infection with Bcc. Unlike Bcc, P. aeruginosa is phagocytosed and rapidly killed by both macrophages and neutrophils, and zebrafish embryos are relatively resistant to intravenously injected P. aeruginosa (3, 10). These findings are not inconsistent with the possibility that in the neutrophils of very young zebrafish the oxidative response may be insufficiently developed to kill Bcc, as it has been shown that neutrophils from CGD patients can kill P. aeruginosa by nonoxidative defense mechanisms, whereas Bcc can resist nonoxidative stress (73). The differences between the two CF pathogens may play a role in the increased severity of the disease due to infection with Bcc compared to the disease due to infection with P. aeruginosa in the zebrafish model.
Revolutionary work using the zebrafish-Mycobacterium model provided evidence that tuberculous granulomas, which were long considered to be host-protective structures, actually contribute to early multiplication and spread of bacteria (16). We found that B. cenocepacia-infected macrophages attracted other, noninfected cells and formed cell aggregates. However, individual infected macrophages not in cell aggregates were also observed. Currently, we do not know which cells are present in the aggregates or whether the cell aggregates are specific for the zebrafish-Burkholderia interaction. Our microscopic observations showed that the infected cells, either individually or in aggregates, did not lyse when they were heavily infected with bacteria; rather, bacteria seemed to actively escape from the replicative vacuoles into neighboring cells. Entry and exit are important steps in the life cycle of intracellular bacteria, yet little is known about the mechanisms used by intracellular bacteria to escape from host cells once their intracellular replication cycle is finished. There is recent evidence indicating that the escape of microbes is directed by the pathogen itself and that in many cases the host cell remains intact (27). Unlike Listeria monocytogenes and Burkholderia pseudomallei, two species that use actin-based protrusions as an exit strategy, B. cenocepacia does not escape rapidly from phagosomes to replicate in the cytoplasm. Several exit strategies have been described for intracellular bacteria that replicate in a protected vacuole. L. pneumophila uses pore formation-mediated cytolysis to egress from mammalian and protozoan cells (52), whereas Chlamydia escapes by a mechanism that was called “extrusion” and was shown to require actin polymerization, neuronal Wiskott-Aldrich syndrome protein, myosin II, and Rho GTPase (26). Very recent evidence showed that Mycobacterium uses a mechanism involving nonlytic ejection from infected amoebae, using an actin-based structure which was called the ejectosome (21). It will be interesting to study in more detail whether bacterial exit and dissemination contribute to the fatal infections resulting from septicemia seen in some CF patients and whether they contribute to the severity of the disease.
In addition to being used to study the intracellular stages of Bcc and the involvement of host factors, the zebrafish can be used to study the genetics of Bcc virulence. Genes encoding many Bcc virulence factors are regulated by quorum sensing via the CepIR proteins. A cepR mutant was attenuated in nematodes (32) and in mice, in which the cepIR system was suggested to contribute to virulence, probably by regulating production of virulence factors involved in invasion (71). In zebrafish embryos a cepR mutant was strongly attenuated; its ability to create an intracellular replication niche and disseminate was highly impaired. Our data show that virulence factors regulated by CepI/R quorum sensing are also essential for full virulence in zebrafish embryos. It will be interesting to identify the specific quorum-sensing-regulated virulence factors that are involved in virulence in this model (79).
Investigating the molecular basis of the highly variable virulence of Bcc.
Significant differences were seen between isolates that were described as clonal isolates (44); K56-2 was significantly more virulent than J2315 and BC7. For all of the ET12 lineage, a major difference between the strains is that K56-2 has smooth lipopolysaccharide (LPS), whereas J2315 has been shown to have an insertion element in the wbxE gene of the O-side chain polysaccharide biosynthesis gene cluster (54). A similar difference in virulence between K56-2 and J2315 was described previously using the nematode model. Although in nematodes LPS did not seem to contribute to this difference (6), whether the difference in LPS contributes to the observed differences in virulence in the fish model remains to be determined. The differences between clonal isolates were also detectable in the rat chronic infection model and the alfalfa model (1).
Bcc strains are ubiquitous in the environment, and, for a clinician, knowing the pathogenic potential of a strain isolated from a CF patient would be a great help in the management of the disease. Unfortunately, the assignment of environmental or clinical strains to a species does not predict the potential pathogenicity (6). We analyzed the virulence of a panel of Bcc clinical isolates. B. cenocepacia J415, which was associated with the first United Kingdom report of cepacia syndrome but was not involved in patient-to-patient spread (18), was much less virulent in the zebrafish model than the highly transmissible ET12 strains. J415 (= LMG16654) also has reduced virulence in rats, in which it infected a relatively small number of animals compared to ET12 strains (55). B. cepacia CEP509, which was transmitted among 4 patients in an Australian clinic (43), was virulent in the zebrafish model. This strain was also highly virulent in alfalfa and rats, although it was not highly virulent in nematodes (1, 6, 9). Two other strains, B. stabilis LMG14294 and B. vietnamiensis FC441, were significantly less virulent in zebrafish embryos than ET12 strains and CEP509. LMG14294 was isolated from a Belgian CF patient, and, although an identical isolate was recovered from a second patient, neither patient developed a lethal infection; similarly, FC441 was isolated from a 9-year-old CGD patient who survived septicemia (44). FC441 was described previously as an isolate that is not invasive in a cell infection model (9), although its virulence was relatively high in the nematode, alfalfa, and rat models. B. stabilis LMG14294, in contrast, was highly attenuated in the nematode, alfalfa, and rat models.
Visualization of LMG14294 and FC441 in zebrafish embryos clearly showed that the abilities of these bacteria to replicate and spread were highly impaired, suggesting that virulence is correlated with the ability to survive host killing mechanisms, create intracellular replication vacuoles, and then disseminate. This is in agreement with previous suggestions that factors such as intracellular survival and the capacity to be highly invasive contribute to the severity of the disease (59, 62, 63). The finding that these strains persisted for longer periods of time in some of the infected embryos at a controllable level suggests the development of a chronic infection, and we will analyze this by performing infection experiments over longer periods of time. Some embryos, however, developed a severe infection with the same strains, showing that any Bcc strain has the potential to produce a serious infection.
Concluding remarks.
The zebrafish is a genetically tractable system and should be instrumental in answering several fundamental questions concerning the molecular mechanisms of Bcc virulence, specifically questions concerning the intracellular stages and early innate immune responses. The tools available for the zebrafish model should allow us to study the effects of host factors, including CFTR, on infection and the innate immune response in the absence of an adaptive system in the embryos. Bacterial mutants already defined using other systems can be easily analyzed in this model, and the use of a signature-tagged mutagenesis strategy in zebrafish should allow identification of specific bacterial virulence factors (31). There is a pressing need to develop novel microbiological tools and infection models to advance research on this bacterium to elucidate, in combination with clinical studies, the pathogenic mechanisms and to develop effective therapies.
Supplementary Material
Acknowledgments
We are grateful to Saskia Rueb (IBL, Leiden, the Netherlands) for the initial suggestion to develop the zebrafish model for Bcc. We are indebted to Nicolas Cubedo and Alain Ghysen (INSERM U881, Montpellier, France) for providing fish tanks, fish eggs, needle-pulling facilities, and many helpful suggestions. We thank Christine Felix for taking care of the fish and Mark Thomas (University of Sheffield, United Kingdom) for critically reading the manuscript. We also thank Astrid van der Sar (Vrije Universiteit, Amsterdam, the Netherlands) for kindly providing pRZT3, John Govan (University of Edinburgh, United Kingdom) for providing J2315, Chris Mohr (University of Minnesota) for providing K56-2 and BC7, Pam Sokol (University of Calgary, Canada) for providing cepR mutant R2, Howard Ceri (University of Calgary, Canada) for providing pHKT4, and Holger Scholz (Institut für Mikrobiologie der Bundeswehr, München, Germany) for providing the B. cenocepacia antibodies.
This work was financed by INSERM, Université de Montpellier 1, Vaincre la Mucoviscidose, the Région Languedoc-Rousillion, and the Fondation pour la Recherche Médicale.
Editor: F. C. Fang
Footnotes
Published ahead of print on 19 January 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bernier, S. P., L. Silo-Suh, D. E. Woods, D. E. Ohman, and P. A. Sokol. 2003. Comparative analysis of plant and animal models for characterization of Burkholderia cepacia virulence. Infect. Immun. 71:5306-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boshra, H., J. Li, and J. O. Sunyer. 2006. Recent advances on the complement system of teleost fish. Fish Shellfish Immunol. 20:239-262. [DOI] [PubMed] [Google Scholar]
- 3.Brannon, M. K., J. M. Davis, J. R. Mathias, C. J. Hall, J. C. Emerson, P. S. Crosier, A. Huttenlocher, L. Ramakrishnan, and S. M. Moskowitz. 2009. Pseudomonas aeruginosa type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell. Microbiol. 11:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bylund, J., L. A. Burgess, P. Cescutti, R. K. Ernst, and D. P. Speert. 2006. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. J. Biol. Chem. 281:2526-2532. [DOI] [PubMed] [Google Scholar]
- 6.Cardona, S. T., J. Wopperer, L. Eberl, and M. A. Valvano. 2005. Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol. Lett. 250:97-104. [DOI] [PubMed] [Google Scholar]
- 7.Celli, J. 2006. Surviving inside a macrophage: the many ways of Brucella. Res. Microbiol. 157:93-98. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, C. H., A. Ostry, and D. P. Speert. 2001. Invasion of murine respiratory epithelial cells in vivo by Burkholderia cepacia. J. Med. Microbiol. 50:594-601. [DOI] [PubMed] [Google Scholar]
- 9.Cieri, M. V., N. Mayer-Hamblett, A. Griffith, and J. L. Burns. 2002. Correlation between an in vitro invasion assay and a murine model of Burkholderia cepacia lung infection. Infect. Immun. 70:1081-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clatworthy, A. E., J. S. Lee, M. Leibman, Z. Kostun, A. J. Davidson, and D. T. Hung. 2009. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect. Immun. 77:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clay, H., J. M. Davis, D. Beery, A. Huttenlocher, S. E. Lyons, and L. Ramakrishnan. 2007. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe 2:29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clay, H., and L. Ramakrishnan. 2005. Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish 2:105-111. [DOI] [PubMed] [Google Scholar]
- 13.Crowhurst, M. O., J. E. Layton, and G. J. Lieschke. 2002. Developmental biology of zebrafish myeloid cells. Int. J. Dev. Biol. 46:483-492. [PubMed] [Google Scholar]
- 14.Darling, P., M. Chan, A. D. Cox, and P. A. Sokol. 1998. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect. Immun. 66:874-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, J. M., D. A. Haake, and L. Ramakrishnan. 2009. Leptospira interrogans stably infects zebrafish embryos, altering phagocyte behavior and homing to specific tissues. PLoS Negl. Trop. Dis. 3:e463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis, J. M., and L. Ramakrishnan. 2009. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136:37-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinauer, M. C., M. A. Gifford, N. Pech, L. L. Li, and P. Emshwiller. 2001. Variable correction of host defense following gene transfer and bone marrow transplantation in murine X-linked chronic granulomatous disease. Blood 97:3738-3745. [DOI] [PubMed] [Google Scholar]
- 18.Glass, S., and J. R. Govan. 1986. Pseudomonas cepacia—fatal pulmonary infection in a patient with cystic fibrosis. J. Infect. 13:157-158. [DOI] [PubMed] [Google Scholar]
- 19.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 20.Govan, J. R., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 21.Hagedorn, M., K. H. Rohde, D. G. Russell, and T. Soldati. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbomel, P., B. Thisse, and C. Thisse. 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238:274-288. [DOI] [PubMed] [Google Scholar]
- 23.Herbomel, P., B. Thisse, and C. Thisse. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735-3745. [DOI] [PubMed] [Google Scholar]
- 24.Hermann, A. C., P. J. Millard, S. L. Blake, and C. H. Kim. 2004. Development of a respiratory burst assay using zebrafish kidneys and embryos. J. Immunol. Methods 292:119-129. [DOI] [PubMed] [Google Scholar]
- 25.Holden, M. T., H. M. Seth-Smith, L. C. Crossman, M. Sebaihia, S. D. Bentley, A. M. Cerdeno-Tarraga, N. R. Thomson, N. Bason, M. A. Quail, S. Sharp, I. Cherevach, C. Churcher, I. Goodhead, H. Hauser, N. Holroyd, K. Mungall, P. Scott, D. Walker, B. White, H. Rose, P. Iversen, D. Mil-Homens, E. P. Rocha, A. M. Fialho, A. Baldwin, C. Dowson, B. G. Barrell, J. R. Govan, P. Vandamme, C. A. Hart, E. Mahenthiralingam, and J. Parkhill. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hybiske, K., and R. S. Stephens. 2008. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 6:99-110. [DOI] [PubMed] [Google Scholar]
- 28.Isberg, R. R., T. J. O'Connor, and M. Heidtman. 2009. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keith, K. E., D. W. Hynes, J. E. Sholdice, and M. A. Valvano. 2009. Delayed association of the NADPH oxidase complex with macrophage vacuoles containing the opportunistic pathogen Burkholderia cenocepacia. Microbiology 155:1004-1015. [DOI] [PubMed] [Google Scholar]
- 31.Kizy, A. E., and M. N. Neely. 2009. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect. Immun. 77:1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kothe, M., M. Antl, B. Huber, K. Stoecker, D. Ebrecht, I. Steinmetz, and L. Eberl. 2003. Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol. 5:343-351. [DOI] [PubMed] [Google Scholar]
- 33.Lam, S. H., H. L. Chua, Z. Gong, T. J. Lam, and Y. M. Sin. 2004. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28:9-28. [DOI] [PubMed] [Google Scholar]
- 34.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 9:40-53. [DOI] [PubMed] [Google Scholar]
- 35.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6:1127-1138. [DOI] [PubMed] [Google Scholar]
- 36.Lamothe, J., and M. A. Valvano. 2008. Burkholderia cenocepacia-induced delay of acidification and phagolysosomal fusion in cystic fibrosis transmembrane conductance regulator (CFTR)-defective macrophages. Microbiology 154:3825-3834. [DOI] [PubMed] [Google Scholar]
- 37.Lefebre, M., and M. Valvano. 2001. In vitro resistance of Burkholderia cepacia complex isolates to reactive oxygen species in relation to catalase and superoxide dismutase production. Microbiology 147:97-109. [DOI] [PubMed] [Google Scholar]
- 38.Le Guyader, D., M. J. Redd, E. Colucci-Guyon, E. Murayama, K. Kissa, V. Briolat, E. Mordelet, A. Zapata, H. Shinomiya, and P. Herbomel. 2008. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood 111:132-141. [DOI] [PubMed] [Google Scholar]
- 39.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, B., S. Chen, Z. Cao, Y. Lin, D. Mo, H. Zhang, J. Gu, M. Dong, Z. Liu, and A. Xu. 2007. Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol. Immunol. 44:295-301. [DOI] [PubMed] [Google Scholar]
- 41.Mahajan-Miklos, S., L. G. Rahme, and F. M. Ausubel. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981-988. [DOI] [PubMed] [Google Scholar]
- 42.Mahenthiralingam, E., A. Baldwin, and C. G. Dowson. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539-1551. [DOI] [PubMed] [Google Scholar]
- 43.Mahenthiralingam, E., A. Baldwin, and P. Vandamme. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533-538. [DOI] [PubMed] [Google Scholar]
- 44.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 46.Marolda, C. L., B. Hauroder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 47.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meeker, N. D., and N. S. Trede. 2008. Immunology and zebrafish: spawning new models of human disease. Dev. Comp. Immunol. 32:745-757. [DOI] [PubMed] [Google Scholar]
- 49.Meijer, A. H., S. F. G. Krens, L. A. Medina, I. Rodriguez, S. He, W. Bitter, B. E. Snaar-Jagalska, and H. P. Spaink. 2004. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 40:773-783. [DOI] [PubMed] [Google Scholar]
- 50.Meijer, A. H., A. M. van der Sar, C. Cunha, G. E. Lamers, M. A. Laplante, H. Kikuta, W. Bitter, T. S. Becker, and H. P. Spaink. 2008. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev. Comp. Immunol. 32:36-49. [DOI] [PubMed] [Google Scholar]
- 51.Miles, A. A., and S. S. Misra. 1938. The estimation of the bactericidal power of the blood. J. Hyg. 38:732-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molmeret, M., O. A. Alli, S. Zink, A. Flieger, N. P. Cianciotto, and Y. A. Kwaik. 2002. icmT is essential for pore formation-mediated egress of Legionella pneumophila from mammalian and protozoan cells. Infect. Immun. 70:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nuesslein-Volhard, C., and R. Dahm (ed.). 2002. Zebrafish: a practical approach. Oxford University Press, New York, NY.
- 54.Ortega, X., T. A. Hunt, S. Loutet, A. D. Vinion-Dubiel, A. Datta, B. Choudhury, J. B. Goldberg, R. Carlson, and M. A. Valvano. 2005. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pirone, L., A. Bragonzi, A. Farcomeni, M. Paroni, C. Auriche, M. Conese, L. Chiarini, C. Dalmastri, A. Bevivino, and F. Ascenzioni. 2008. Burkholderia cenocepacia strains isolated from cystic fibrosis patients are apparently more invasive and more virulent than rhizosphere strains. Environ. Microbiol. 10:2773-2784. [DOI] [PubMed] [Google Scholar]
- 56.Prajsnar, T. K., V. T. Cunliffe, S. J. Foster, and S. A. Renshaw. 2008. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell. Microbiol. 10:2312-2325. [DOI] [PubMed] [Google Scholar]
- 57.Renshaw, S. A., C. A. Loynes, D. M. Trushell, S. Elworthy, P. W. Ingham, and M. K. Whyte. 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108:3976-3978. [DOI] [PubMed] [Google Scholar]
- 58.Revets, H., P. Vandamme, Z. A. Van, B. K. De, M. J. Struelens, J. Verhaegen, J. P. Ursi, G. Verschraegen, H. Franckx, A. Malfroot, I. Dab, and S. Lauwers. 1996. Burkholderia (Pseudomonas) cepacia and cystic fibrosis: the epidemiology in Belgium. Acta Clin. Belg. 51:222-230. [DOI] [PubMed] [Google Scholar]
- 59.Ryley, H. 2004. Burkholderia cepacia complex infection in cystic fibrosis patients: mechanisms of pathogenesis. Rev. Med. Microbiol. 15:93-101. [Google Scholar]
- 60.Saiman, L., and J. Siegel. 2004. Infection control in cystic fibrosis. Clin. Microbiol. Rev. 17:57-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 62.Sajjan, U., M. Corey, A. Humar, E. Tullis, E. Cutz, C. Ackerley, and J. Forstner. 2001. Immunolocalisation of Burkholderia cepacia in the lungs of cystic fibrosis patients. J. Med. Microbiol. 50:535-546. [DOI] [PubMed] [Google Scholar]
- 63.Sajjan, U., G. Thanassoulis, V. Cherapanov, A. Lu, C. Sjolin, B. Steer, Y. J. Wu, O. D. Rotstein, G. Kent, C. McKerlie, J. Forstner, and G. P. Downey. 2001. Enhanced susceptibility to pulmonary infection with Burkholderia cepacia in Cftr−/− mice. Infect. Immun. 69:5138-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sajjan, U. S., L. Sun, R. Goldstein, and J. F. Forstner. 1995. Cable (cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J. Bacteriol. 177:1030-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sajjan, U. S., J. H. Yang, M. B. Hershenson, and J. J. LiPuma. 2006. Intracellular trafficking and replication of Burkholderia cenocepacia in human cystic fibrosis airway epithelial cells. Cell. Microbiol. 8:1456-1466. [DOI] [PubMed] [Google Scholar]
- 66.Saldias, M. S., and M. A. Valvano. 2009. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology 155:2809-2817. [DOI] [PubMed] [Google Scholar]
- 67.Seed, K. D., and J. J. Dennis. 2008. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 76:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sieger, D., C. Stein, D. Neifer, A. M. van der Sar, and M. Leptin. 2009. The role of gamma interferon in innate immunity in the zebrafish embryo. Dis. Model. Mech. 2:571-581. [DOI] [PubMed] [Google Scholar]
- 69.Smith, D. L., L. B. Gumery, E. G. Smith, D. E. Stableforth, M. E. Kaufmann, and T. L. Pitt. 1993. Epidemic of Pseudomonas cepacia in an adult cystic fibrosis unit: evidence of person-to-person transmission. J. Clin. Microbiol. 31:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N(5)-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 72.Sousa, S. A., M. Ulrich, A. Bragonzi, M. Burke, D. Worlitzsch, J. H. Leitao, C. Meisner, L. Eberl, I. Sa-Correia, and G. Doring. 2007. Virulence of Burkholderia cepacia complex strains in gp91phox−/− mice. Cell. Microbiol. 9:2817-2825. [DOI] [PubMed] [Google Scholar]
- 73.Speert, D. P., M. Bond, R. C. Woodman, and J. T. Curnutte. 1994. Infection with Pseudomonas cepacia in chronic granulomatous disease: role of nonoxidative killing by neutrophils in host defense. J. Infect. Dis. 170:1524-1531. [DOI] [PubMed] [Google Scholar]
- 74.Speert, D. P., B. Steen, K. Halsey, and E. Kwan. 1999. A murine model for infection with Burkholderia cepacia with sustained persistence in the spleen. Infect. Immun. 67:4027-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stein, C., M. Caccamo, G. Laird, and M. Leptin. 2007. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 8:R251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sullivan, C., and C. H. Kim. 2008. Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 25:341-350. [DOI] [PubMed] [Google Scholar]
- 77.Traver, D., P. Herbomel, E. E. Patton, R. D. Murphey, J. A. Yoder, G. W. Litman, A. Catic, C. T. Amemiya, L. I. Zon, and N. S. Trede. 2003. The zebrafish as a model organism to study development of the immune system. Adv. Immunol. 81:253-330. [PubMed] [Google Scholar]
- 78.Trede, N., A. H. Meijer, B. E. Snaar-Jagalska, and H. P. Spaink. 2007. Model systems for infectious disease and cancer in zebrafish: a report on an EMBO workshop held at the Lorentz Center, Leiden, The Netherlands, July 16-18, 2007. Zebrafish 4:287-292. [DOI] [PubMed] [Google Scholar]
- 79.Uehlinger, S., S. Schwager, S. P. Bernier, K. Riedel, D. T. Nguyen, P. A. Sokol, and L. Eberl. 2009. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect. Immun. 77:4102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Sar, A. M., B. J. Appelmelk, C. M. Vandenbroucke-Grauls, and W. Bitter. 2004. A star with stripes: zebrafish as an infection model. Trends Microbiol. 12:451-457. [DOI] [PubMed] [Google Scholar]
- 81.van der Sar, A. M., R. J. Musters, F. J. van Eeden, B. J. Appelmelk, C. M. Vandenbroucke-Grauls, and W. Bitter. 2003. Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 5:601-611. [DOI] [PubMed] [Google Scholar]
- 82.van der Sar, A. M., O. W. Stockhammer, C. van der Laan, H. P. Spaink, W. Bitter, and A. H. Meijer. 2006. MyD88 innate immune function in a zebrafish embryo infection model. Infect. Immun. 74:2436-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vojtech, L. N., G. E. Sanders, C. Conway, V. Ostland, and J. D. Hansen. 2009. Host immune response and acute disease in a zebrafish model of Francisella pathogenesis. Infect. Immun. 77:914-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Welten, M. C., S. B. de Haan, N. van den Boogert, J. N. Noordermeer, G. E. Lamers, H. P. Spaink, A. H. Meijer, and F. J. Verbeek. 2006. ZebraFISH: fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish 3:465-476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.