Abstract
ExoU, a cytotoxin translocated into host cells via the type III secretion system of Pseudomonas aeruginosa, is associated with increased mortality and disease severity. We previously showed that impairment of recruited phagocytic cells allowed survival of ExoU-secreting P. aeruginosa in the lung. Here we analyzed types of cells injected with ExoU in vivo using translational fusions of ExoU with a β-lactamase reporter (ExoU-Bla). Cells injected with ExoU-Bla were detectable in vitro but not in vivo, presumably due to the rapid cytotoxicity induced by the toxin. Therefore, we used a noncytotoxic ExoU variant, designated ExoU(S142A)-Bla, to analyze injection in vivo. We determined that phagocytic cells in the lung were frequently injected with ExoU(S142A). Early during infection, resident macrophages constituted the majority of cells into which ExoU was injected, but neutrophils and monocytes became the predominant types of cells into which ExoU was injected upon recruitment into the lung. We observed a modest preference for injection into neutrophils over injection into other cell types, but in general the repertoire of injected immune cells reflected the relative abundance of these cells in the lung. Our results indicate that phagocytic cells in the lung are injected with ExoU and support the hypothesis that ExoU-mediated impairment of phagocytes has a role in the pathogenesis of pneumonia caused by P. aeruginosa.
Pseudomonas aeruginosa is a Gram-negative bacterium that is ubiquitous in both natural and manmade environments, including hospitals. In the health care setting, it is a frequent cause of acute infections, such as urinary tract infections, burn and wound infections, sepsis, and severe pneumonia (23). The risk of P. aeruginosa pneumonia is increased in hospitalized patients who are immunosuppressed or require mechanical ventilation. In fact, P. aeruginosa accounts for almost 20% of all pneumonia cases in intensive care units (ICU) and is the leading cause of ventilator-associated pneumonia (VAP) (30). Additionally, as a causative agent of VAP, P. aeruginosa has a higher attributable mortality rate than most other bacteria, making it a particularly dangerous pathogen (4).
Of the many bacterial factors that contribute to the pathogenesis of P. aeruginosa, the type III secretion system is important for bacterial persistence in the presence of elaborate host defense mechanisms and has been associated with poor clinical outcomes in individuals with VAP (13, 32). While the genes encoding the type III secretion apparatus are present in all P. aeruginosa isolates, the repertoire of effector-encoding genes is variable. Approximately 28% of strains obtained from acute infections encode a potent cytotoxin, ExoU (8), which is a marker of highly virulent P. aeruginosa strains isolated from patients with VAP (35). In animal models, ExoU secretion greatly augments the virulence of P. aeruginosa (1, 20, 21, 37). The contribution of ExoU to virulence is attributable to its phospholipase A2 activity (27, 34). Upon injection into host cells, ExoU is activated and targeted to the plasma membrane, where it cleaves membrane phospholipids, resulting in rapid and complete cell lysis (10, 14, 27-29, 34). In addition to ExoU, P. aeruginosa isolates can encode other effectors, including ExoS, ExoT, and ExoY in various combinations (8). ExoS and ExoT are bifunctional enzymes with 75% amino acid identity and similar functional domains, and each of them has GTPase-activating protein (GAP) and ADP-ribosyltransferase (ADPRT) activities (2). ExoY is an adenylate cyclase (41). Interestingly, most strains contain either exoS or exoU, but strains containing both genes are uncommon (8).
Previously, we showed that ExoU-secreting P. aeruginosa strains are capable of persisting in the lungs of infected animals despite the fact that they elicit an exaggerated immune response consisting of infiltrating inflammatory cells. Recruited phagocytic cells were impaired by ExoU during acute lung infection, resulting in a localized paucity of functional phagocytes within the airways, which allowed bacteria to persist and cause severe disease (6). However, whether phagocytes were directly injected with ExoU or were compromised indirectly following intoxication of other cell types was unclear.
Recently, a fluorogenic β-lactam substrate, CCF2-AM, has been used to detect translocation of bacterial proteins into host cells in vitro and in vivo (3, 11, 17, 24, 25). Upon diffusion into host cells, intact CCF2-AM exhibits fluorescence resonance energy transfer (FRET) resulting in green fluorescence. Cleavage of this substrate by a β-lactamase molecule disrupts FRET, resulting in a shift to blue fluorescence (3, 42). Fusion of a bacterial protein (in this study ExoU) with a β-lactamase tag allows detection of protein translocation into cells by virtue of the change in fluorescence emission. Here we used CCF2-AM to identify cell types intoxicated with ExoU in a mouse model of acute pneumonia. We found that phagocytic cells were injected with ExoU in the lung. Resident alveolar macrophages were injected with ExoU in the earliest stages of infection, but as neutrophils and monocytes were rapidly recruited to the lung, they became the predominant cell types injected with ExoU. In comparison, only a small number of lymphocytes and type II alveolar epithelial cells were injected in vivo. Cells injected with ExoU were detected as early as 3 h after infection, and then the level increased steadily to a maximum of approximately 10% of all recovered cells at 18 h postinfection. These findings are consistent with a model in which extensive injection of ExoU into phagocytic cells incapacitates these cells, which facilitates persistence of P. aeruginosa in the lung and progression to severe disease.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains, mutants, and plasmids used in this study are listed in Table 1. PA99 is a P. aeruginosa clinical isolate that naturally contains the exoU, exoS, and exoT genes but lacks the exoY gene (8). PA99null and PA99secr− (PA99 ΔpscJ) were generated from PA99 by in-frame deletion as previously described (37). Complementation restored the levels of virulence of the mutants to expected levels, and neither mutant displayed a growth defect on laboratory medium (37). Chemically competent Escherichia coli TOP10 cells (Invitrogen) were used for cloning.
TABLE 1.
P. aeruginosa strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference and/or source |
|---|---|---|
| P. aeruginosa strains | ||
| PA99 | Clinical isolate; naturally secretes ExoS, ExoT, and ExoU | 8 |
| PA99null | Secretes no effectors; PA99 ΔexoS ΔexoT ΔexoU; gentamicin resistant | 37 |
| PA99secr− | Secretes no effectors; does not make secretion apparatus; PA99 ΔpscJ | 37 |
| PA99S | Secretes only ExoS | 36 |
| PA99ST | Secretes ExoS and ExoT | 36 |
| PA99null+ExoU | PA99null complemented with exoU in the attB locus | This study |
| PA99null+ExoU(S142A) | PA99null complemented with exoU(S142A) in the attB locus | 6 |
| PA99null+ExoU-Bla | PA99null complemented with exoU-bla in the attB locus | This study |
| PA99null+ExoU(S142A)-Bla | PA99null complemented with exoU(S142A)-bla in the attB locus | This study |
| PA99null+GST-Bla | PA99null complemented with gst-bla under the control of native exoU promoter in the attB locus | This study |
| PA99secr−+ExoU-Bla | PA99secr− complemented with exoU-bla in the attB locus | This study |
| PA99secr−+ExoU(S142A)-Bla | PA99secr− complemented with exoU(S142A)-bla in the attB locus | This study |
| PA99S+ExoU(S142A)-Bla | PA99S complemented with exoU(S142A)-bla in the attB locus | This study |
| PA99ST+ExoU(S142A)-Bla | PA99ST complemented with exoU(S142A)-bla in the attB locus | This study |
| Plasmids | ||
| mini-CTX-1 | Plasmid for chromosomal gene integration into attB locus; Tetr | H. Schweizer (16) |
| mini-CTXexoU | exoU gene ligated into mini-CTX-1; Tetr | 29 |
| mini-CTXexoU-bla | mini-CTXexoU with C-terminal bla tag; Tetr | This study |
| mini-CTXexoU(S142A)-bla | mini-CTXexoU(S142A) with C-terminal bla tag; Tetr | This study |
| mini-CTXgst-bla | gst-bla replaces exoU coding sequence in mini-CTXexoU-bla; Tetr | This study |
Bacterial strains were streaked from frozen cultures onto Luria-Bertani (LB) agar or Vogel-Bonner minimal (VBM) agar (39). For infection, overnight cultures of P. aeruginosa grown in 5 ml MINS medium (26) at 37°C were diluted into fresh medium and regrown to exponential phase.
Generation of effector-Bla fusions.
The stop codon of exoU was altered, and an AgeI site was engineered at the 3′ end of exoU by site-directed mutagenesis using plasmid mini-CTXexoU (29) as the template to generate mini-CTXexoUΔstopAgeI. The mature fragment of TEM-1 β-lactamase was amplified from pBR322 with engineered AgeI sites using primers BlacAgeUp (5′-AAAACCGGTACACCCAGAAACGCTGGTGA-3′) and BlacAgeDwn (3′-AAAACCGGTATTACCAATGCTTAATCAGTGA-5′) and ligated into mini-CTXexoUΔstopAgeI. A Bla fusion of a noncytotoxic ExoU variant [ExoU(S142A)] was created by removing the stop codon of exoU(S142A) by site-directed mutagenesis using mini-CTXexoU(S142A) (6) as the template, followed by amplification of the resulting exoU(S142A)Δstop construct with existing NsiI and engineered AgeI sites at its 5′ and 3′ ends using primers exoU5′ (5′-AAAATGCATATCCAATCGTTGG-3′) and exoUAgeI (3′-AAAACCGGTATCGGCCATGTGAACTCCTTATTCC-5′). The amplified fragment was ligated into mini-CTXexoUΔstopAgeI digested with NsiI and AgeI to generate mini-CTXexoU(S142A)ΔstopAgeI. In a second reaction, the bla fragment was ligated into mini-CTXexoU(S142A)ΔstopAgeI digested with AgeI to create mini-CTXexoU(S142A)-bla.
A glutathione S-transferase (GST)-Bla fusion that is expressed under the control of the native exoU promoter was generated by amplifying the open reading frame of glutathione S-transferase from pGEX-5X1 using primers GSTNsiI (5′-AAAATGCATATGTCCCCTATACTAGGTTATTGG-3′) and GSTAgeI (3′-AAAACCGGTTCAGTCAGTCACGATGAATTCCCG-5′) and was ligated into mini-CTXexoUΔstopAgeI digested with NsiI and AgeI to generate mini-CTXgstAgeI. The stop codon of gst was altered by site-directed mutagenesis. In a subsequent reaction, the bla fragment was ligated into AgeI-digested mini-CTXgstAgeI to create mini-CTXgst-bla.
All plasmids were confirmed by sequence analysis. In each case, the resulting construct [mini-CTXexoU-bla, mini-CTXexoU(S142A)-bla, or mini-CTXgst-bla] was transformed into E. coli strain S17.1 (38) and, following conjugation, was introduced into a neutral site in the PA99null chromosome via integrase-mediated recombination at the attB site using previously described approaches (16) to generate PA99null+ExoU-Bla, PA99null+ExoU(S142A)-Bla, and PA99null+GST-Bla, respectively. mini-CTXexoU-bla and mini-CTXexoU(S142A)-bla were also introduced into the chromosome of PA99secr− to create strains PA99secr−+ExoU-Bla and PA99secr−+ExoU(S142A)-Bla. Integration was confirmed by PCR amplification of each construct from intact bacterial colonies.
Immunoblot analysis.
Immunoblot analysis for secretion of ExoU-Bla fusion proteins was performed using polyclonal ExoU antiserum as previously described (35).
Fluorescence microscopy.
J774 macrophage-like cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and grown on glass coverslips in a 24-well plate overnight. Cells were incubated with P. aeruginosa at a multiplicity of infection (MOI) of 10 for 3 h. Cells were washed once with Hanks' balanced salt solution (HBSS) and incubated with CCF2-AM (Invitrogen) at a final concentration of 1× (prepared according to the manufacturer's instructions) for 1 h at room temperature protected from light. Cells were washed with HBSS to eliminate excess CCF2-AM and fixed with 3.7% formaldehyde. Coverslips were placed on slides with 1 drop of Fluoromount-G (Southern Biotechnology Associates, Inc.) and sealed with nail polish. Cells were viewed with a Leica DMR microscope with a 100-W mercury lamp and equipped with an Axiocam MR3 color camera. Pictures were taken using the AxioVision release 4.6.3 software.
Mouse model of acute pneumonia.
Studies of acute pneumonia were conducted using the aspiration mouse model described by Comolli et al. (5). Briefly, bacteria were collected by centrifugation and resuspended to the appropriate concentration in phosphate-buffered saline (PBS). Six- to 8-week-old female BALB/c mice were anesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/ml) and xylazine (20 mg/ml). Mice were inoculated intranasally with 1.2 × 107 CFU of bacteria in 50 μl PBS, as determined by optical density. Inoculum sizes were confirmed by plating serial dilutions.
Animals were purchased from Harlan and housed in the containment ward of the Center for Comparative Medicine at Northwestern University. All experiments were performed in accordance with the guidelines of the Northwestern University Animal Care and Use Committee.
Analysis of cells into which ExoU was injected by flow cytometry.
At the appropriate time points, mice were anesthetized and sacrificed by cervical dislocation and thoracotomy. Lungs were perfused and flushed by injection of 2 ml PBS into the right side of the heart to remove circulating blood cells. Lungs were excised and pressed through 40-μm filters (Falcon). The filters were rinsed repeatedly with PBS. For bronchoalveolar lavage (BAL) experiments, mouse lungs were lavaged by instilling and withdrawing 1 ml of PBS three times. The recovered cells were gently pelleted by centrifugation at 500 × g for 5 min. The supernatant was removed, and red blood cells were lysed by addition of 3 ml cold sterile H2O and incubation for 30 s. Three milliliters of 2× normal saline (1.8% NaCl) was quickly added to prevent additional cell lysis. The remaining cells were pelleted and resuspended in PBS, and trypan blue-excluding cells were quantified using a hemacytometer.
A total of 1 × 105 or 2 × 105 cells were placed in each well of a V-bottom 96-well plate (Nunc), resuspended in 100 μl PBS with CCF2-AM (Invitrogen) at a final concentration of 1×, and incubated at room temperature protected from light for 1 h. Cells were pelleted, and surface Fc receptors and nonspecific binding sites were blocked by incubation in 10% rat serum (Sigma) and anti-CD16/32 in fluorescence-activated cell sorting (FACS) buffer (1% bovine serum albumin, 0.1% NaN3 in PBS) for 5 to 15 min on ice. Cell-discriminating antibodies at the appropriate dilutions in FACS buffer were added, and the final volume in each well was adjusted to 125 μl. Antibodies and cells were incubated for 15 to 30 min on ice. Cells were pelleted by centrifugation at 500 × g for 5 min and resuspended in 2% paraformaldehyde in PBS for 2 min. An equal volume of FACS buffer was added, and cells were pelleted again. Cells were resuspended in FACS buffer and transferred into Falcon 2052 tubes after passage through 70-μm Nitex filters. Antibodies were used at the following final dilutions: anti-CD16/32, 1:50; anti-CD45, 1:1,500; anti-Gr1, 1:1,500; anti-F4/80, 1:50; anti-CD4, 1:500; anti-CD8, 1:500; anti-CD19, 1:500; anti-CD49, 1:500; anti-CD11b, 1:2,500; anti-CD11c, 1:500; and isotype controls, 1:100 each. All antibodies were purchased from eBioscience.
Cells were analyzed by flow cytometry with a Becton Dickinson FACSCantoII. Instrument settings were determined each day using the appropriate positive- and negative-control samples. After data collection, cell debris was removed from the analysis by gating for forward scatter and side scatter. The remaining cells were considered to represent the total number of cells extracted from the lungs. Innate immune cells were defined as follows: CD11b+ Gr1hi cells were defined as neutrophils, CD11b+ Gr1int cells were defined as recruited monocytes, Gr1− F4/80+ cells were defined as resident macrophages, and CD11cint Gr1− cells were defined as dendritic cells. (In separate samples, CD11cint cells were shown to be F480−, indicating that they were dendritic cells rather than CD11c+ alveolar macrophages.) Recruited mononuclear cells that had migrated into tissue were considered monocytes and were distinguished from resident macrophages by expression of Ly6C. Recruited monocytes express intermediate levels of Ly6C (recognized by anti-Gr1 antibody) on the surface, while mature macrophages do not (12). Lymphocyte subtypes were identified as follows: CD4+ helper T cells, CD8+ cytotoxic T cells, CD19+ B cells, and CD49+ NK cells. The total number of viable inflammatory cells per mouse lung was determined by equating the number of total cells with normal scatter characteristics measured by flow cytometry to the number of trypan blue-negative cells counted with the hemacytometer.
Isolation of type II pneumocytes from mouse lungs.
Murine type II pneumocytes were isolated from lungs using a modification of the method described by Ridge et al. (31). Briefly, lungs were perfused via the pulmonary artery, lavaged six times, and digested with elastase for 20 min (30 U/ml; Worthington Biochemical). Cells were isolated from collected lavage fluid for parallel analysis. Digested lung tissue was minced in the presence of fetal bovine serum (FBS) and DNase, filtered through 70-μm cell strainers, and centrifuged at 500 × g for 10 min. Contaminating leukocytes were removed by an indirect magnetic separation technique. Cells were incubated at 37°C for 30 min with biotinylated antibodies at the following dilutions: anti-CD16/32, 1:1,500; anti-TER119, 1:500; anti-CD45, 1:700; and anti-CD90, 1:700. Cells were pelleted, resuspended in DMEM with 10% FBS, and incubated with streptavidin magnetic beads for 30 min at room temperature with rotation; then the preparations were taped to a magnet for 15 min to remove all biotinylated cells. The supernatant containing the alveolar epithelial cells was removed and centrifuged at 500 × g for 10 min, and trypan blue-excluding cells were quantified using a hemacytometer. Cells were incubated with CCF2-AM and anti-CD45 as described above and then analyzed by flow cytometry. CD45+ cells were excluded from the FACS analysis of type II pneumocyte preparations.
Statistical methods.
Analyses of bacterial load differences were performed by using Student's t test. Prior to analysis, all colonization data were natural log transformed so that they fit a normal distribution. The use of parametric tests on transformed colonization data was justified by analysis of a large set of control data that confirmed that colonization data were log normally distributed. The proportions of the cell types injected were compared by analysis of variance (ANOVA). For ANOVA comparisons with a P value of <0.05, adjustment for multiple unplanned comparisons was performed using the Tukey-Kramer honestly significant difference (HSD) test with an α value of 0.05.
RESULTS
ExoU-Bla fusions are secreted in vitro.
To analyze translocation of ExoU, we generated C-terminal translational fusions of full-length catalytically active ExoU or a catalytically inactive noncytotoxic ExoU variant, designated ExoU(S142A), with the mature fragment of the TEM-1 β-lactamase (Bla). The constructs encoding these fusion proteins were introduced into a neutral site in the chromosome of PA99null, a strain with an intact type III secretion system but with disruptions in each of its effector-encoding genes (37). We assessed the abilities of the resulting strains, designated PA99null+ExoU-Bla and PA99null+ExoU(S142A)-Bla, to secrete the corresponding fusion proteins by immunoblot analysis of supernatants from cultures grown under type III secretion-inducing conditions. Both fusion proteins were detected in culture supernatants (Fig. 1A), indicating that addition of Bla does not prevent movement of ExoU through the type III secretion needle.
FIG. 1.
ExoU-Bla fusion proteins are injected into host cells in vitro. (A) Immunoblot analysis of culture supernatants from P. aeruginosa strains secreting ExoU-Bla or ExoU(S142A)-Bla grown under type III secretion-inducing conditions using polyclonal ExoU antiserum. The same volume of supernatant was used for each sample. α-ExoU, anti ExoU antibody. (B to I) J774 cells were incubated with P. aeruginosa bacteria at an MOI of 10, which was followed by treatment with CCF2-AM for 1 h. Cells were examined using a filter set that allows visualization of green fluorescence and blue fluorescence simultaneously. Green cells contain intact CCF2-AM, and blue cells contain cleaved CCF2-AM, indicating translocation of the ExoU-Bla fusion. Representative fields are shown for the following conditions: uninfected J774 cells (B) without or (C) with CCF2-AM substrate; cells incubated with CCF2-AM after infection with (D) PA99null+ExoU, (E) PA99null+ExoU(S142A), (F) PA99null+ExoU-Bla, (G) PA99null+ExoU(S142A)-Bla, (H) PA99secr−+ExoU(S142A)-Bla, or (I) PA99null+GST-Bla. Scale bars, 100 μm.
We next examined translocation of the ExoU-Bla fusion proteins into J774 macrophage-like cells in vitro using fluorescence microscopy (Fig. 1B to I). Following coincubation with PA99null+ExoU-Bla, PA99null+ExoU(S142A)-Bla, or control strains, J774 cells were treated with the substrate CCF2-AM, which fluoresces upon diffusion into host cells. Intact CCF2-AM exhibits FRET resulting in green fluorescence, and cleavage of this substrate by a β-lactamase molecule disrupts FRET, resulting in a shift to blue fluorescence. Cells containing translocated ExoU-Bla (Fig. 1F) or ExoU(S142A)-Bla (Fig. 1G) were identified by blue fluorescence in contrast to uninfected cells (Fig. 1C) and cells infected with P. aeruginosa secreting ExoU or ExoU(S142A) without the Bla tag (Fig. 1D and 1E, respectively). Notably, few cells remained after 3 h of incubation with either PA99null+ExoU (Fig. 1D) or PA99null+ExoU-Bla (Fig. 1F) as a result of rapid cell lysis induced by the effector, and the remaining cells appeared to be severely damaged. Importantly, neither ExoU-Bla nor ExoU(S142A)-Bla was associated with blue fluorescence when it was expressed from PA99secr−, a PA99 mutant with a deletion in the pscJ gene, which encodes an essential component of the type III secretion apparatus (Fig. 1H and data not shown). Likewise, P. aeruginosa expressing a glutathione S-transferase-Bla fusion protein (GST-Bla) did not cause blue fluorescence (Fig. 1I), confirming that cleavage of the CCF2-AM substrate requires type III translocation of ExoU and is not due to engulfment of bacteria by J774 cells or nonspecific movement of the β-lactamase moiety into the cells. These observations confirm that ExoU-Bla fusion proteins are translocated into host cells in vitro and that cells into which ExoU is injected can be detected in vitro using this reporter assay.
Secretion of ExoU(S142A)-Bla, but not secretion of ExoU-Bla, is detectable in vivo.
ExoU is injected into many cell types in vitro; however, the relevant cellular targets in vivo during infection have not been determined. We infected mice intranasally with P. aeruginosa secreting either wild-type or catalytically inactive ExoU-Bla fusions or appropriate control strains. After 18 h, cells were recovered by bronchoalveolar lavage (BAL), incubated with CCF2-AM, and analyzed by flow cytometry. Infection with PA99null+ExoU(S142A) without the Bla moiety resulted in only green fluorescence (Fig. 2A). Similar to what we observed in vitro, blue cells were not detected in lavage fluid from mice infected with a nonsecreting P. aeruginosa strain expressing either ExoU(S142A)-Bla or ExoU-Bla (Fig. 2B and data not shown) or with P. aeruginosa expressing GST-Bla (Fig. 2C). During infection with PA99null+ExoU-Bla, we were unable to detect cells containing translocated ExoU-Bla in BAL fluid (Fig. 2D) or whole lung tissue (data not shown). In contrast, a significant proportion (40%) of cells recovered from the lungs of mice infected with PA99null+ExoU(S142A)-Bla were injected with fusion protein (Fig. 2E). The inability to detect cells containing catalytically active ExoU-Bla was likely due to the rapid cell death induced by this molecule since in vitro observations have shown that ExoU induces fast and complete cell lysis (6). Consistent with this explanation is the fact that our in vitro CCF2-AM experiments using PA99null+ExoU-Bla also resulted in relatively few cells with blue fluorescence (Fig. 1F). Because ExoU-induced cell lysis precluded in vivo detection of injected cells, we utilized the noncytotoxic ExoU(S142A)-Bla fusion for quantification and identification of injected cells in vivo.
FIG. 2.
Detection of cells injected with ExoU-Bla fusion proteins in vivo. FACS analysis was performed with cells recovered by BAL at 18 h postinfection. Green fluorescence (y axis) indicates the presence of CCF2-AM in the cell, and blue fluorescence (x axis) indicates translocation of the ExoU(S142A)-Bla fusion protein. The percentage of blue (injected) cells is indicated in the upper right corner of each dot plot. (A) Infection with PA99null+ExoU(S142A), (B) PA99secr−+ExoU(S142A)-Bla, and (C) PA99null+GST-Bla resulted in only green cells. (D) Infection with PA99null+ExoU-Bla did not yield a significant proportion of blue cells compared to control strains. (E) ExoU(S142A)-Bla translocation was detectable in vivo. Approximately 40% of cells recovered from BAL fluid were intoxicated with ExoU(S142A)-Bla. The data are from a representative experiment; similar results were obtained in at least three independent experiments. The results varied from day to day, and the mean percentage of injected cells ranged from 15 to 40%.
Since secretion of catalytically active ExoU facilitates bacterial persistence in the lung, higher numbers of bacteria are present over the course of infection with an ExoU+ strain than over the course of infection with an ExoU− strain or with P. aeruginosa expressing the noncytotoxic ExoU variant [ExoU(S142A)] (6). To more accurately model infection with P. aeruginosa secreting catalytically active ExoU, we infected mice with a higher number of PA99null+ExoU(S142A)-Bla cells (1.2 × 107 CFU/mouse), which resulted in numbers of bacteria in the lungs after 18 h of infection similar to the numbers observed with a standard inoculum (1.2 × 106 CFU) of PA99null+ExoU (data not shown). Infection with this inoculum resulted in recruitment of high numbers of inflammatory cells to the lung over time (data not shown). These conditions approximate the bacterial burden and bacterium-to-host cell ratio observed during infection with P. aeruginosa secreting catalytically active ExoU (6), indicating that secretion of ExoU(S142A) under these conditions is a suitable model to study ExoU secretion. Therefore, we used the larger inoculum for all experiments in this study.
A substantial portion of phagocytic cells are injected with ExoU(S142A) in the lung.
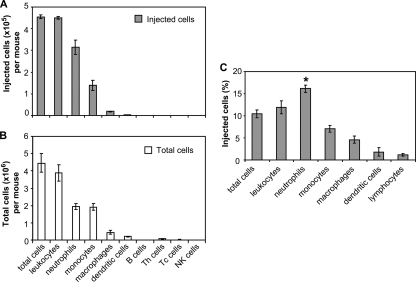
Previous studies indicated that ExoU contributes to virulence by compromising phagocytic cells intended to eradicate bacteria from the lung (6). Based on these observations, we aimed to directly identify the types of cells that were injected with ExoU during acute pneumonia. For these experiments, cells were recovered from whole lung tissue of infected mice, incubated with CCF2-AM, stained with cell discriminatory antibodies, and analyzed by flow cytometry. We initially investigated the types of cells injected with ExoU(S142A) at 18 h postinfection, when large numbers of inflammatory cells are present in the lung (6). In this experiment, approximately 0.5 × 106 cells recovered from the lung of an infected mouse were injected with ExoU(S142A) (Fig. 3A). In this population of injected cells, 99% (452,000/458,000) of the cells were inflammatory cells and over 80% (383,000/458,000) were phagocytic cells (Fig. 3A). In the injected phagocytic cell population, neutrophils were the predominant cell type (60%), followed by monocytes (27%) and macrophages (5%), while dendritic cells comprised less than 1% of the injected cell population (Fig. 3A). In addition to these phagocytic cell types, we also examined CD4+ and CD8+ T cells, B cells, and NK cells. Together, lymphocytes comprised <2% of the injected cells recovered from the lung (Fig. 3A). The majority of cells (>85%) incorporated CCF2-AM efficiently, and no obvious difference in CCF2-AM uptake by different cell types was observed (data not shown). Therefore, a single cell type was not specifically targeted for injection of ExoU, but phagocytic cells, particularly neutrophils, comprised the majority of cells injected with ExoU(S142A) in vivo.
FIG. 3.
Analysis of injected and total cells recovered from the lungs of mice infected with P. aeruginosa. FACS analysis was performed with cells recovered from the whole lungs of mice infected with PA99null+ExoU(S142A)-Bla at 18 h postinfection. (A and B) Numbers of injected cells (A) and total cells (B) for each immune cell type recovered from whole lungs per mouse. The numbers of cells per mouse were determined by equating the number of total cells with normal scatter characteristics measured by flow cytometry to the number of trypan blue-negative cells counted with the hemacytometer. (C) Proportion of each of the cell subpopulations purified from the lungs that was injected with ExoU(S142A)-Bla. A significantly higher proportion of neutrophils than of monocytes, macrophages, dendritic cells, or lymphocytes was injected. The data are means ± standard errors of the means (≥3 mice per group); similar results were obtained in at least three independent experiments. The results varied from day to day, and the mean percentage of injected cells ranged from 7 to 15%. *, P < 0.01.
To examine whether the sizes of the various cell populations in the lung dictated the number of cells of a cell type injected with ExoU(S142A), we determined the total number of cells of each cell type recovered from the lungs (Fig. 3B) and calculated the proportion of each cell type that was injected (Fig. 3C). Although the composition of the injected cells largely mirrored the composition of the overall population of cells in the lung (compare Fig. 3A to Fig. 3B), there was a modest preference for neutrophils. Of the neutrophils recovered from the lung, 16% were injected with ExoU(S142A), compared to 7% of the monocytes, 5% of the macrophages, 2% of the dendritic cells, and 1% of the lymphocytes (P < 0.01 for neutrophils compared to each other cell type) (Fig. 3C). Therefore, a higher proportion of neutrophils than of other cell types was injected with ExoU(S142A).
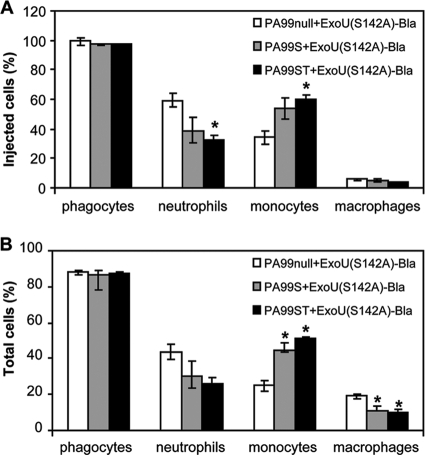
Most P. aeruginosa isolates secrete multiple effector molecules. To assess the effect of cosecretion of ExoS and/or ExoT on translocation of ExoU in vivo, we examined the types of cells injected with ExoU(S142A)-Bla during infection with P. aeruginosa secreting ExoU(S142A)-Bla along with ExoS alone [PA99S+ExoU(S142A)-Bla] or along with both ExoT and ExoS [PA99ST+ExoU(S142A)-Bla] (Fig. 4). For these experiments, we used a smaller bacterial inoculum (6 × 106 CFU/mouse) since secretion of ExoS results in more severe illness (37). Similar to the results for infection with PA99null+ExoU(S142A)-Bla, phagocytic cell types comprised over 95% of the injected cells in the lung (Fig. 4A). We assessed the proportions of neutrophils, monocytes, and macrophages in the injected cells at 18 h postinfection. Interestingly, the repertoire of recruited inflammatory cells was somewhat different when ExoS alone or ExoS and ExoT were secreted with ExoU. We observed slightly higher numbers of injected monocytes than of neutrophils when ExoS was secreted compared to the results for infection with PA99null+ExoU(S142A)-Bla, a result which was also reflected in all cells recovered from the lungs of these animals (Fig. 4B). The difference may be due to previously undescribed effects of ExoS. Still, the relative sizes of the various cell populations dictated the numbers of cells of the different cell types injected with ExoU(S142A) even in the presence of other effectors (compare Fig. 4A and 4B). Thus, secretion of additional effectors does not dramatically alter the specificity of injection in vivo.
FIG. 4.
Secretion of ExoU(S142A)-Bla in the presence of other effectors in vivo. FACS analysis was performed with cells recovered from the lungs of mice infected with the PA99null strain engineered to secrete ExoU(S142A)-Bla alone [PA99null+ExoU(S142A)-Bla], ExoU(S142A)-Bla along with ExoS [PA99S+ExoU(S142A)-Bla], or ExoU(S142A)-Bla along with ExoS and ExoT [PA99ST+ExoU(S142A)-Bla] at 18 h postinfection. (A) Percentages of phagocytic cell types in the injected cell population recovered from infected lungs. (B) Percentages of phagocytic cell types in the total cells recovered from infected lungs. The data are means ± standard errors of the means for a representative experiment (3 mice per group). Similar results were obtained in two independent experiments. *, P < 0.05.
ExoU intoxication preferentially occurs in the airways of infected animals.
We next addressed the question of where in the lungs ExoU(S142A) injection occurred. Since P. aeruginosa causes a bronchopneumonia in which the majority of bacteria reside in the airways and alveoli (7, 22) and since type III secretion requires direct contact between the bacterium and a host cell, we anticipated that injection would occur primarily in these lung compartments. The proportions of injected cells in the airways (recovered by BAL) were compared to the proportions in the entire lung (recovered from whole lung tissue) (Fig. 5). A significantly higher proportion of ExoU(S142A)-intoxicated cells was recovered from the airways and alveoli (29%) (Fig. 5A) than from whole lung tissue (11%) (Fig. 5B) (P < 0.05). The relative abundance of cells into which ExoU was injected in the airway indicates that type III secretion occurs predominantly in the airspace of infected lungs.
FIG. 5.
ExoU injection occurs in the airspace of infected lungs. FACS analysis was performed with cells recovered from BAL fluid (A) or whole lungs (B). Green fluorescence (y axis) indicates the presence of CCF2-AM in the cells, and blue fluorescence (x axis) indicates translocation of the ExoU(S142)-Bla fusion protein. The percentage of blue (injected) cells is indicated in the upper right corner of each density plot. The insets show histograms of blue fluorescence. The proportion of injected cells in the BAL fluid (29%) was approximately three times that in the whole lung sample (11%) (P < 0.05). Representative plots are shown; similar results were obtained in at least three experiments with a total of 8 mice.
Relatively few type II alveolar epithelial cells are injected with ExoU(S142A).
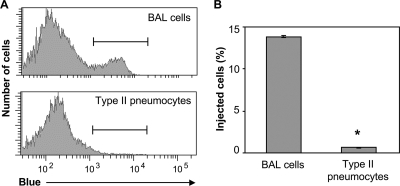
The protocols used in the experiments described above are not conducive for purification of cell types other than immune cells. Thus, approximately 99% of cells recovered from the lungs of infected mice were resident or recruited leukocytes (Fig. 3B and data not shown). Since ExoU is injected into and lyses numerous types of epithelial cells in vitro and since secretion appears to occur primarily in the airways and alveoli of infected mice, we wished to determine whether alveolar epithelial cells (pneumocytes) were injected with ExoU during infection. Although type I pneumocytes comprise the majority of the alveolar surface area, these cells are not amenable to isolation from mouse lungs. Type II pneumocytes comprise much less of the epithelial surface but nonetheless are extremely important in lung physiology and the response to infection. Type II pneumocytes differentiate into type I cells to maintain the epithelium and also provide protection from pathogens by secreting surfactant into the alveolar space. To assess secretion of ExoU into cell types other than leukocytes, we isolated type II pneumocytes from the lungs of mice infected with PA99null+ExoU(S142A)-Bla by digestion with elastase, followed by removal of leukocytes by magnetic separation. Cells were incubated with CCF2-AM, and injection was measured by flow cytometry (Fig. 6). Any contaminating leukocytes were removed from the analysis of type II pneumocytes by exclusion of CD45+ cells. BAL samples, which contained mostly leukocytes, were analyzed in parallel. Interestingly, while 15% of the cells in the BAL fluid were intoxicated with ExoU(S142A), minimal injection of ExoU(S142A)-Bla into type II pneumocytes (1%) was observed (Fig. 6B). The two cell populations incorporated CCF2-AM with similar efficiencies (>85%). This result indicates that not all cell types present in the lungs are substantially injected with ExoU during acute pneumonia.
FIG. 6.
Few type II alveolar epithelial cells (pneumocytes) are injected with ExoU(S142A). FACS analysis was performed with type II pneumocytes isolated from lungs of mice infected with PA99null+ExoU(S142A)-Bla at 18 h postinfection. Cells recovered by BAL were analyzed in parallel. (A) Histogram showing blue fluorescence of cells from BAL fluid (top panel) or type II pneumocyte preparations (bottom panel). Blue cells indicate injection with ExoU(S142A)-Bla. Brackets indicate cells exhibiting blue fluorescence. Representative plots from a total of three experiments are shown. (B) Percentages of cells in BAL fluid or type II pneumocyte preparations that were injected with ExoU(S142A)-Bla. The data are means ± standard errors of the means for triplicate samples of cells pooled from three mice; similar results were obtained in three independent experiments. *, P < 0.00001.
ExoU injection specificity changes over time.
Finally, we examined whether the types of cells injected with ExoU(S142A) changed over time (Fig. 7). For these experiments, we infected mice with PA99null+ExoU(S142A)-Bla or appropriate control strains and sacrificed animals after 3, 6, 12, or 18 h. Cells were recovered from the whole lungs, incubated with CCF2-AM, stained with cell discriminatory antibodies, and analyzed by flow cytometry. The number of cells recovered from the infected mouse lungs steadily increased over the first 18 h following inoculation (Fig. 7A), reflecting the recruitment of inflammatory cells to the site of infection. At all time points examined, the majority of cells recovered from the lungs were leukocytes (data not shown). At 3 h postinfection, a substantial number of recovered cells were resident (alveolar) macrophages (Fig. 7B). A robust neutrophilic infiltrate was apparent at 6 h, and the maximum level was reached at 12 h after infection. Monocyte recruitment was somewhat more delayed and continued to increase through 18 h postinfection. The increase in the monocyte level may reflect the recruitment of macrophage precursors intended to replenish the resident macrophage population. The timing of mononuclear cell recruitment observed in our model is similar to that described by other investigators (15). Relatively few dendritic cells and lymphocytes were recovered at all time points.
FIG. 7.
Cells injected with ExoU(S142A) in the lung over time. FACS analysis was performed with cells recovered from whole lungs of mice infected with PA99null+ExoU(S142A)-Bla at 3, 6, 12, and 18 h postinfection. (A) Total leukocytes recovered from the lungs of mice infected with the ExoU(S142A)-Bla strain. The number of total lung leukocytes increased over the course of infection. (B) Proportions of cell types in total cells recovered from the lung. (C) Percentages of total cells that were injected with ExoU(S142A). *, P < 0.05 compared to the data for the preceding time point. (D) Percentages of immune cell types in the injected cell population. Data for lymphocyte subpopulations (T cells, B cells, and NK cells) are combined. The data are means ± standard errors of the means (≥3 mice per group); similar results were obtained in at least two independent experiments.
The total number of cells injected with ExoU(S142A) increased over the first 18 h of infection. After only 3 h, approximately 2% of the total recovered cells were already injected, but the proportion rose to approximately 10% by 18 h (Fig. 7C). The increase was statistically significant (P < 0.05). Thus, even as more inflammatory cells entered the lungs, a larger proportion of these cells were injected with ExoU(S142A).
At all time points tested, neutrophils, monocytes, and macrophages comprised the majority of injected cells recovered from the lungs, whereas dendritic cells and lymphocytes each comprised less than 5% of injected cells (Fig. 7D). Therefore, the proportion of each cell type in the injected cell population reflected to a large extent the composition of the total leukocytes recovered from the lungs at each time point (compare Fig. 7B and 7D). At 3 h postinfection, alveolar macrophages were the predominant cell type among the injected cells (58%), while neutrophils and monocytes accounted for only 17% and 18% of the cells, respectively (Fig. 7D). Likewise, alveolar macrophages represented approximately 40% of the total cells recovered from the lungs at this early time point, before substantial numbers of neutrophils and monocytes had been recruited to the lungs. At 6 h postinfection, 70% of the injected cells recovered from the lungs were neutrophils and 18% were monocytes, while only 5% were macrophages. Again, these proportions closely reflected the relative abundance of each cell type in the total cell population at this time point (Fig. 7B). At 12 and 18 h, neutrophils remained the predominant cell type injected with ExoU(S142A) even though the proportion of monocytes present in the lung had increased substantially (Fig. 7B). In summary, alveolar macrophages, which are resident cells of the lung, were injected early during infection prior to the recruitment of neutrophils and monocytes, but as neutrophils and mononuclear phagocytes were recruited to the lungs, they became the most commonly injected cell types.
DISCUSSION
Using a FRET-based reporter assay, we directly showed that phagocytic cells are extensively injected with ExoU during acute pneumonia caused by P. aeruginosa. Resident alveolar macrophages were injected early during infection, while recruited neutrophils and monocytes were injected slightly later as these cells entered the lungs in response to the invading microbes. To our knowledge, this is the first identification of cell types targeted by a P. aeruginosa type III effector in vivo and the first examination of injection into cells during pneumonia for any type III secretion system. Thus, our results provide important insights into the mechanisms by which bacteria cause pneumonia.
Using a noncytotoxic ExoU variant, we found that the types of cells injected with ExoU reflect the overall repertoire of immune cells in the lungs over the first 18 h of infection. Injection was observed as early as 3 h postinfection, at a time when few inflammatory cells were present in the lungs. Thus, resident alveolar macrophages comprised the majority of the cells initially injected in the lung. The subsequent influx of neutrophils and monocytes into the lung correlated with an increase in injection of these cell types, suggesting that cell type targeting of ExoU is to a large extent determined by the availability of cells rather than a specific preference of the type III secretion apparatus or P. aeruginosa adhesins for particular cell types. However, a somewhat higher proportion of neutrophils than of other cell types in the lungs were injected with ExoU. This may have simply reflected increased numbers of P. aeruginosa bacteria binding to neutrophils because of the large surface area and phagocytic nature of these cells or may have been because a larger proportion of neutrophils migrate into the airways and come into contact with bacteria. In any case, our results indicate that phagocytic cells are a primary target of ExoU injection in vivo. Since most ExoU+ clinical isolates also secrete ExoT and many ExoU+ clinical isolates secrete ExoY, it may be that these effector proteins are similarly targeted. ExoS, however, is rarely secreted by ExoU+ isolates, and ExoS-secreting strains constitute a clonal group that is distinct from ExoU+ strains (8, 40). For these reasons, it remains possible that ExoS-secreting strains differ in pathogenesis and that this effector protein is injected into distinct populations of host cells. Examination of the cell types injected with other P. aeruginosa effectors during acute pneumonia is currently under way.
Targeting of phagocytic cells, including neutrophils, has previously been implicated in the pathogenesis of other Gram-negative bacterial pathogens. Geddes and colleagues showed that Salmonella enterica targets splenic neutrophils for injection of type III effectors to promote intracellular survival in vivo (11). Similarly, Marketon et al. (24) and Koberle et al. (17) observed injection into neutrophils, macrophages, and dendritic cells in the spleen during infection with Yersinia spp. In these cases, targeting of splenic innate immune cells was shown to be a mechanism for evading or disrupting host immune responses. Our results demonstrate that neutrophils are also targets for type III secretion in the lung. Previous experimental observations suggested that impairment of recruited phagocytic cells by ExoU was important for P. aeruginosa survival in the lung and progression to severe disease. In agreement with this functional evidence, we found that P. aeruginosa injected ExoU into neutrophils and macrophages during pneumonia. Impairment or killing of these cells by direct injection of ExoU may prevent effective clearance from the lung and contribute to the enhanced disease observed with ExoU+ strains.
Interestingly, we did not observe significant injection into type II pneumocytes during infection. There are several possible explanations for this finding. Infiltrating inflammatory cells may bind P. aeruginosa with increased affinity, thereby reducing the number of bacteria available to bind to epithelial cells. Alternatively, type II pneumocytes may be resistant to injection of ExoU. If this is the case, damage to the lung epithelium, previously observed during infection with ExoU+ P. aeruginosa (20), may be an indirect effect caused by release of damaging inflammatory mediators from ExoU-lysed phagocytic cells rather than by direct killing by injection of ExoU into epithelial cells. Additional studies are necessary to understand the interaction between pulmonary epithelial cells and ExoU.
Injection of ExoU occurred disproportionately in the airspace compared to other compartments of the lung during early pneumonia. This finding is consistent with several previous observations. First, the majority of P. aeruginosa bacteria reside in the airways and alveoli of the lungs during pneumonia (7, 22). Since type III secretion requires direct contact between the bacterium and the host cell, this implies that secretion occurs primarily in the airspace. Second, we previously observed a relative reduction in the number of viable neutrophils in the airways and alveoli compared to whole lung compartments of mice infected with ExoU+ P. aeruginosa (6), suggesting that neutrophils are injected with ExoU upon transmigration across the respiratory epithelium. Thus, in early pneumonia, type III secretion may be employed most actively against immune cells as they enter the airways and alveoli.
We were unable to detect secretion of catalytically active ExoU-Bla into host cells in vivo. This was likely due to the rapid cell lysis induced by this molecule since cells injected with ExoU(S142A)-Bla, a noncytotoxic ExoU variant, were detected at all time points examined. Previously, it has been shown that ExoU induces rapid and complete cell lysis of neutrophils in vitro, which precludes detection of cells into which ExoU has been injected in vivo (6). Similarly, other investigators have also been unable to detect cells containing catalytically active ExoU (9, 27, 33). Our inability to detect cells injected with catalytically active ExoU further supports the idea that ExoU induces rapid lysis of phagocytic cells in vivo. The use of a noncatalytic variant of ExoU rather than wild-type ExoU was one limitation of this study because the lack of cell killing by ExoU alters bacterial survival, disease progression, and the host immune response. We attempted to more accurately mimic infection with a strain producing wild-type ExoU by increasing the infecting dose to increase the overall number of bacteria in the lung and enhance recruitment of inflammatory cells to the lung. Nonetheless, it is possible that the dynamics of translocation may be somewhat different in the presence of wild-type ExoU. Another potential limitation is that throughout this study we utilized a strain which secreted ExoU(S142A)-Bla alone, without other effector proteins. Clinical isolates of P. aeruginosa usually secrete combinations of type III effector proteins. However, this does not appear to be a major limitation since the presence of ExoS and ExoT did not dramatically alter the types of cells injected with ExoU and since the injected cell populations continued to closely mirror the total phagocytic cell populations present in the lung (Fig. 4). This is consistent with reports indicating that cosecretion of ExoS or ExoT along with ExoU did not substantially alter the severity of pneumonia (36). Nonetheless, cosecretion of other effector proteins could have subtle effects on the total numbers and proportions of host cells injected with ExoU. Interestingly, the repertoire of inflammatory cells recruited to the lung did change when ExoS or ExoS and ExoT were secreted along with ExoU(S142A), indicating that the effector proteins did dictate the nature of the inflammatory response.
Based on direct analysis of injection into cells in the lung during acute infection, the results of this study suggested a model for the mechanism of ExoU activity during early pneumonia. During the initial stage of infection, ExoU is injected into alveolar macrophages, which may incapacitate these sentinel cells that are crucial in the host defense during early P. aeruginosa pneumonia (19). Later injection of ExoU into phagocytes may allow P. aeruginosa to evade phagocytosis by these immune cells, allowing large numbers of bacteria to persist in the lung. Thus, injection of ExoU creates an environment in which the bacteria can survive and cause the pathology observed during severe pneumonia.
Acknowledgments
This work was supported by the National Institutes of Health (grants AI053674, AI065615, AI053674, and AI075191 to A.R.H. and grant T32 GM008061 to M.H.D.).
We thank Ciara Shaver, Cheryl Olson, Kerry Sheppard, and Karen Ridge for their advice and technical assistance with the experiments.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 25 January 2010.
REFERENCES
- 1.Allewelt, M., F. T. Coleman, M. Grout, G. P. Priebe, and G. B. Pier. 2000. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbieri, J. T., and J. Sun. 2004. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 152:79-92. [DOI] [PubMed] [Google Scholar]
- 3.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chastre, J., and J. Y. Fagon. 2002. Ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 165:867-903. [DOI] [PubMed] [Google Scholar]
- 5.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, M. H., C. M. Shaver, J. D. King, S. Musunuri, J. A. Kazzaz, and A. R. Hauser. 2008. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia in mammals. Infect. Immun. 76:4414-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMango, E., H. Zar, R. Bryan, and A. Prince. 1995. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Invest. 96:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 9.Finck-Barbançon, V., and D. W. Frank. 2001. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J. Bacteriol. 183:4330-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finck-Barbançon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 11.Geddes, K., F. Cruz, and F. Heffron. 2007. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 3:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon, S., and P. R. Taylor. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953-964. [DOI] [PubMed] [Google Scholar]
- 13.Hauser, A. R., E. Cobb, M. Bodí, D. Mariscal, J. Vallés, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 14.Hauser, A. R., P. J. Kang, and J. Engel. 1998. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 15.Henderson, R. B., J. A. Hobbs, M. Mathies, and N. Hogg. 2003. Rapid recruitment of inflammatory monocytes is independent of neutrophil migration. Blood 102:328-335. [DOI] [PubMed] [Google Scholar]
- 16.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 17.Koberle, M., A. Klein-Gunther, M. Schutz, M. Fritz, S. Berchtold, E. Tolosa, I. B. Autenrieth, and E. Bohn. 2009. Yersinia enterocolitica targets cells of the innate and adaptive immune system by injection of Yops in a mouse infection model. PLoS Pathog. 5:e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Kooguchi, K., S. Hashimoto, A. Kobayashi, Y. Kitamura, I. Kudoh, J. Wiener-Kronish, and T. Sawa. 1998. Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect. Immun. 66:3164-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, T. R. Martin, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Invest. 104:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandell, G. L., J. E. Bennett, and R. Dolin. 2005. Mandell, Douglas, and Bennett's principles and practice of infectious diseases, 6th ed. Elsevier, Philadelphia, PA.
- 24.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 309:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills, E., K. Baruch, X. Charpentier, S. Kobi, and I. Rosenshine. 2008. Real-time analysis of effector translocation by the type III secretion system of enteropathogenic Escherichia coli. Cell Host Microbe 3:104-113. [DOI] [PubMed] [Google Scholar]
- 26.Nicas, T. I., and B. H. Iglewski. 1984. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect. Immun. 45:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 28.Rabin, S. D., J. L. Veesenmeyer, K. T. Bieging, and A. R. Hauser. 2006. A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 74:2552-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabin, S. D. P., and A. R. Hauser. 2005. Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 73:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rello, J., E. Diaz, and A. Rodriguez. 2005. Etiology of ventilator-associated pneumonia. Clin. Chest Med. 26:87-95. [DOI] [PubMed] [Google Scholar]
- 31.Ridge, K. M., D. H. Rutschman, P. Factor, A. I. Katz, A. M. Bertorello, and J. L. Sznajder. 1997. Differential expression of Na-K-ATPase isoforms in rat alveolar epithelial cells. Am. J. Physiol. 273:L246-L255. [DOI] [PubMed] [Google Scholar]
- 32.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 33.Sato, H., and D. W. Frank. 2004. ExoU is a potent intracellular phospholipase. Mol. Microbiol. 53:1279-1290. [DOI] [PubMed] [Google Scholar]
- 34.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulert, G. S., H. Feltman, S. D. P. Rabin, C. G. Martin, S. E. Battle, J. Rello, and A. R. Hauser. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188:1695-1706. [DOI] [PubMed] [Google Scholar]
- 36.Shaver, C. M., and A. R. Hauser. 2006. Interactions between effector proteins of the Pseudomonas aeruginosa type III secretion system do not significantly affect several measures of disease severity in mammals. Microbiology 152:143-152. [DOI] [PubMed] [Google Scholar]
- 37.Shaver, C. M., and A. R. Hauser. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72:6969-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology (NY) 1:784-791. [Google Scholar]
- 39.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 40.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. U. S. A. 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]