Abstract
The gastrointestinal tract is colonized by an enormous array of microbes that are known to have many beneficial effects on the host. Previous studies have indicated that stressor exposure can disrupt the stability of the intestinal microbiota, but the extent of these changes, as well as the effects on enteric infection, has not been well characterized. In order to examine the ability of stressors to induce changes in the gut microbiota, we exposed mice to a prolonged restraint stressor and then characterized microbial populations in the intestines using both traditional culture techniques and bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). Exposure to the stressor led to an overgrowth of facultatively anaerobic microbiota while at the same time significantly reducing microbial richness and diversity in the ceca of stressed mice. Some of these effects could be explained by a stressor-induced reduction in the relative abundance of bacteria in the family Porphyromonadaceae. To determine whether these alterations would lead to increased pathogen colonization, stressed mice, as well as nonstressed controls, were challenged orally with the enteric murine pathogen Citrobacter rodentium. Exposure to the restraint stressor led to a significant increase in C. rodentium colonization over that in nonstressed control mice. The increased colonization was associated with increased tumor necrosis factor alpha (TNF-α) gene expression in colonic tissue. Together, these data demonstrate that a prolonged stressor can significantly change the composition of the intestinal microbiota and suggest that this disruption of the microbiota increases susceptibility to an enteric pathogen.
The gastrointestinal tract is colonized by a vast array of microbes that reside as part of a complex, and relatively stable, ecosystem. The number of commensal bacteria in the body is estimated to be about 100 trillion, which is 10-fold greater than the number of human cells in the body. Moreover, the use of molecular techniques to study the microbiota has led to the finding that the microbial genome (or microbiome) in the body contains approximately 100 times more genes than does the human genome (23). The use of germfree and conventional mice clearly demonstrates that the microbiota are essential for a variety of gastrointestinal functions, such as the synthesis of vitamins and the metabolism of carcinogens, and many structural/host defense functions such as the regulation of intestinal epithelial cell development and stimulation and regulation of the mucosal immune response (reviewed in reference 44). In fact, the ability of the commensal microbiota to inhibit enteric pathogen colonization (a phenomenon generally referred to as competitive exclusion) has been recognized for many years (53). And it is now realized that enteric infections, either natural or experimental, can be exacerbated in humans, or animals, with a disrupted intestinal microbiota.
Exposure to stressors has been shown to significantly change microbial populations in the gastrointestinal tract of both humans and laboratory animals (4, 5, 20, 28, 34, 36, 48). For example, the stress response to maternal separation causes a significant reduction in lactobacillus levels in young monkeys (4). Moreover, stressor exposure during gestation reduced developmental colonization by lactobacilli and bifidobacteria in monkeys during the first 6 months of life (5). These previous studies all utilized traditional culture techniques to quantify the intestinal microbiota. While there are many advantages to using culture-based methods, approximately 90% of the microbiota are strict anaerobes that have not been characterized by traditional culture-based methods due to undefined culture requirements (43). As a result, traditional culture techniques can provide only a limited description of the microbiota. Thus, one purpose of this study was to use bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) to more fully characterize stressor-induced changes in microbial diversity.
Because the intestinal microbiota can inhibit the ability of enteric pathogens to colonize the intestines and to cause gastrointestinal disease, the impact of the stress response on enteric infection was assessed. Rodents are naturally susceptible to Citrobacter rodentium, which can cause an attaching and effacing (A/E) lesion to the brush border surface of intestinal epithelial cells that is virtually indistinguishable from lesions caused by enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC, respectively) in humans. In resistant mice, including the Swiss-derived CD-1 mice used in the current study, C. rodentium infection is self-limiting and the colonization results in little morbidity and mortality (9). However, pathogen colonization still leads to cytokine production in the colon and can induce a mild inflammatory response and mild colonic hyperplasia characterized by epithelial cell hyperproliferation in the descending colon (7, 9). Importantly, EPEC, EHEC, and C. rodentium must colonize the intestinal epithelium in order to induce the inflammatory response and colonic hyperplasia.
The use of germfree mice challenged with C. rodentium has indicated that the intestinal microbiota can inhibit C. rodentium colonization and the resultant disease progression (6, 29-31). Citrobacter rodentium colonization and pathology can also be significantly decreased by administering probiotic microbes; administering probiotic Lactobacillus acidophilus to suckling mice reduced colonic hyperplasia and inflammation following C. rodentium infection (13). In addition to exogenous probiotic microbes, indigenous microbiota levels have been associated with C. rodentium levels; a low relative abundance of bacteria in the family Porphyromonadaceae was found to be associated with high C. rodentium levels (37). Thus, we hypothesized that stressors that change the microbiota prior to infection would also result in increased C. rodentium levels upon oral challenge. Our data demonstrate that a prolonged restraint stressor significantly reduces microbial richness in the intestines and decreases the relative abundance of bacteria in the family Porphyromonadaceae, specifically within the genus Tannerella, prior to infection. Oral challenge with C. rodentium after exposure to the restraint stressor resulted in significantly elevated pathogen levels in the stool, which were associated with altered cytokine gene expression in the colon. Together, the data suggest that stressor-induced changes in the microbiota significantly enhance the ability of enteric pathogens to colonize in the intestines.
MATERIALS AND METHODS
Mice.
Male CD-1 mice (aged 6 to 8 weeks) were purchased from Charles River Laboratories (Kingston, NY) and allowed to acclimate to the animal facility for 1 week prior to experimentation. Mice were kept in an AAALAC-approved vivarium with food and water available ad libitum except during experimental procedures. The lights were maintained on a 12-h:12-h light-dark schedule with lights on at 0600 h. All procedures were approved by The Ohio State University's Animal Care and Use Committee.
Stressor paradigm.
Mice were randomly assigned to the nonstressed home cage control (HCC), food and water deprivation (FWD) control, and the prolonged restraint stress (RST) conditions. The restraint stressor is a widely used stressor that is largely psychological in nature, resulting from the perception of burrow collapse and confinement (8, 24). It induces a physiological stress response characterized by activation of the autonomic nervous system and the hypothalamic-pituitary-adrenal axis (35, 45). The restraint stressor entails placing the mice in well-ventilated 50-ml conical centrifuge tubes at the beginning of their active cycle (i.e., at 1800 h) and removing them the following morning at 0800 h. During the inactive cycle, the mice were undisturbed in their home cage with food and water available ad libitum. The restraint stressor was repeated every night for 7 consecutive nights. Because the RST mice do not have access to food and water while in the restraining tube, FWD control mice were also deprived of food and water during those periods in which the RST mice were in the restraining tubes. The nonstressed HCC mice always had food and water available ad libitum and were not exposed to any stressors during the experiment. In experiments involving the characterization of the intestinal microbiota, HCC mice were used to set baseline or prestudy values.
Microbiota characterization.
Mice were humanely euthanized with CO2 asphyxiation, and the intestines were excised using a sterile technique. The contents of the lowest 3 cm of the small intestine, the cecum, and the entire colon were weighed by being emptied into preweighed sterile phosphate-buffered saline (PBS). The fecal suspension was homogenized and serially diluted so that the number of bacteria in these intestinal segments that could grow aerobically could be determined via standard pour plate analysis. The bacteria were grown in brain heart infusion agar (BBL, Cockeysville, MD) to quantify both Gram-positive and Gram-negative microbes and eosin methylene blue agar (BBL) to quantify Gram-negative microbes only. The agar plates were incubated aerobically at 37°C for 24 h.
bTEFAP.
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) was used to determine the relative percentages of the primary populations of microbes in the cecal contents. This technique is a semiquantitative universal detection and identification method for bacteria based upon the Roche Titanium 454 FLX pyrosequencing platform and was performed as described previously (17) at the Research and Testing Laboratory (Lubbock, TX).
DNA extraction.
Cecal contents were emptied into sterile 1.8-ml microcentrifuge tubes and snap-frozen in liquid nitrogen. The samples were stored at −80°C until being shipped overnight on dry ice for bTEFAP analysis. After thawing, the samples were centrifuged at 14,000 rpm for 30 s and resuspended in 500 μl RLT buffer (Qiagen, Valencia, CA) (with β-mercaptoethanol). Sterile 5-mm steel beads (Qiagen, Valencia, CA) and 0.1-mm glass beads (Scientific Industries, Inc.) were added for complete bacterial lysis in a Qiagen TissueLyser (Qiagen, Valencia, CA), run at 30 Hz for 5 min. Samples were centrifuged, and 100 μl of 100% ethanol was added to a 100-μl aliquot of the sample supernatant. This mixture was added to a DNA spin column, and DNA recovery protocols were followed as instructed in the QIAamp DNA minikit (Qiagen, Valencia, CA). DNA was eluted from the column with 30 μl water, and sample concentration was adjusted to 20 ng/μl. DNA samples were quantified using a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France).
PCR to create tag-encoded amplicons.
A 100-ng aliquot of each sample's DNA was used for a step 1 PCR. The 16S universal eubacterial primers 530F (5′ GTG CCA GCM GCN GCG G) and 1100R (5′ GGG TTN CGN TCG TTG) were used for amplifying the 600-bp region of 16S rRNA genes and sequencing through the V4 region. HotStarTaq Plus Master Mix kit (Qiagen, Valencia, CA) was used for PCR under the following conditions: 94°C for 3 min followed by 30 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 5 min. A step 2 PCR was performed for 454 amplicon sequencing under the same conditions by using designed special fusion primers with different tag sequences as described previously (16).
Massively parallel bTEFAP.
In preparation for FLX sequencing, the DNA fragments' sizes and concentrations were measured using DNA chips under a Bio-Rad Experion automated electrophoresis station (Bio-Rad Laboratories) and a TBS-38 fluorometer (Turner Biosystems). A sample of 9.6 × 106 double-stranded DNA molecules/μl with an average size of 625 bp was combined with 9.6 × 106 DNA capture beads and then amplified by emulsion PCR. After bead recovery and bead enrichment, the bead-attached DNAs were denatured with NaOH, and sequencing primers were annealed. A two-region 454 sequencing run was performed on a 70 × 75 GS PicoTiterPlate using the Genome Sequencer FLX system (Roche, Nutley, NJ). Twenty tags were used per region on the PicoTiterPlate. Based upon the number of reads per sample, the depth of sequencing was sufficient to evaluate primary gastrointestinal populations at a dissimilarity of >0.1%.
Bacterial diversity data analysis.
Following sequencing, all failed sequence reads, low-quality sequence ends, and tags were removed and sequences were depleted of any nonbacterial ribosome sequences and chimeras using custom software (16) and the Black Box Chimera Check software B2C2 (described and freely available at http://www.researchandtesting.com/B2C2.html). Sequences of less than 150 bp were removed. This resulted in approximately 1,500 to 4,000 sequences per sample. To determine the identities of bacteria in the remaining sequences, sequences were first queried using a distributed BLASTn .NET algorithm (18) against a database of high-quality 16S bacterial sequences derived from NCBI. Database sequences were characterized as high quality based upon the criteria originally described for RDP version 9 (14). Using a .NET and C# analysis pipeline, the resulting BLASTn outputs were compiled and validated using taxonomic distance methods, and data reduction analysis was performed as described previously (17). Rarefaction, abundance of coverage estimate (ACE), and Chao1 analyses to estimate mathematically predicted diversity and richness in the treatments using 200-bp trimmed, nonribosomal-sequence-depleted, chimera-depleted, high-quality reads were performed as described previously (1).
Bacterial ID.
Based upon the above BLASTn-derived sequence identity (percentage of total length query sequence which aligns with a given database sequence) and validation using taxonomic distance methods, the bacteria were classified at the appropriate taxonomic levels based upon the following criteria. Sequences with identity scores, to known or well-characterized 16S sequences, greater than 96.5% identity (<3% divergence) were resolved at the species level when possible, those with scores between 94.5% and 96.4% were resolved at the genus level, those with scores between 89.5% and 94.4% were resolved at the family level, and those with scores between 80% and 89.4% were resolved at the order level. After resolution based upon these parameters, the percentage of each bacterial identification (ID) was individually analyzed for each cecal sample, providing relative abundance information within and among the samples based upon relative numbers of reads within a given sample. When multiple identifications were found (e.g., Bacteroides caccae and Bacteroides stercoris), such identifications were resolved arbitrarily to the top hit. Evaluations presented at a given taxonomic level, except species level, represent all sequences resolved to their primary genus identification or their closest relative (where indicated).
Infection.
Citrobacter rodentium strain DBS120(pCRP1::Tn5) (49) was grown in Trypticase soy broth (TSB) overnight at 37°C. Cultures were centrifuged, and pellets were resuspended in sterile PBS and adjusted to a concentration of 3 × 109 to 5 × 109 CFU/ml. Mice were challenged via oral gavage with 100 μl of the bacteria (i.e., 3 × 108 to 5 × 108 CFU) in PBS and were deprived of food and water for 2 h after inoculation. Fecal shedding of C. rodentium was determined by plating stool on MacConkey lactose agar supplemented with kanamycin (40 μg/ml).
Histopathology.
On days 1, 6, 12, and 24 postchallenge, the entire colons of the mice were excised, cleaned of stool, and transected longitudinally. One section was used for semiquantitative real-time PCR, with the remaining section being formalin fixed in 10% neutral buffered saline for 24 to 48 h. These fixed tissues were processed routinely, paraffin embedded, sectioned at 5 μm, and stained with hematoxylin and eosin. The sections were scored by a board-certified veterinary pathologist (Nicola Parry), who was blinded to experimental groups. Inflammation, hyperplasia, dysplasia, edema, and epithelial defects within intestinal tissue sections were graded on a scale of 0 to 4 at 0.5 intervals as described previously (10). These scores were then summed to provide a colonic histological index that was used for statistical analyses.
Semiquantitative real-time PCR.
Interleukin-1β (IL-1β), IL-10, tumor necrosis factor alpha (TNF-α), inducible nitric oxide synthase (iNOS), and mouse β-defensin 1 gene expression were quantified using real-time PCR. Total RNA was isolated from colonic tissue on days 1, 6, 12, and 24 postchallenge using Trizol reagent per the manufacturer's instructions (Invitrogen, Carlsbad, CA). A total of 2 μg of RNA was reverse transcribed to make complementary cDNA using a commercially available kit (Promega, Madison, WI) according to the manufacturer's instructions. Real-time PCR primers and probes were previously published and were synthesized by Applied Biosystems. In all cases, 18S rRNA was used as a housekeeping gene. The 5′-3′ sequences are listed in Table 1.
TABLE 1.
Primer and probe 5′-3′ sequences
| Gene | Sequence |
||
|---|---|---|---|
| Forward | Reverse | Probe | |
| 18S | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT | TGCTGGCACCAGACTTGCCCTC |
| IL-1β | GGCCTCAAAGGAAAGAATCTATACC | GTATTGCTTGGGATCCACACTCT | ATGAAAGACGGCACACCCACCCTG |
| iNOS | CAGCTGGGCTGTACAAACCTT | TGAATGTGATGTTTGCTTCGG | CGGGCAGCCTGTGAGACCTTTGA |
| TNF-α | CTGTCTACTGAACTTCGGGGTGAT | GGTCTGGGCCATAGAACTGATG | TGAGAAGTTCCCAAATGGCCTCCCTC |
| IL-10 | TTTGAATTCCCTGGGTGAGAA | ACAGGGGAGAAATCGATGACA | NAa |
| Mouse β-defensin 1 | GGCTGCCACCACTATGAAAACTC | GAGACAGAATCCTCCATGTTGAA | NA |
| Mouse β-defensin 3 | GCATTTGAGGAAAGGAACTCCACAAC | GTCTCCACCTGCAGCTTTTAGCAA | NA |
NA, not applicable because SYBR green was used in place of a fluorescent probe.
The PCR mixture consisted of 2.5 μl of probe (or SYBR green for IL-10 and mouse β-defensin 1), 5 μl of sterile distilled H2O, and 12.5 μl of TaqMan Universal Master Mix (PE Applied Biosystems, Foster City, CA) for a final volume of 25 μl. After an initial 2-min cycle at 50°C followed by 10 min at 95°C, the reaction ran for 40 total cycles, which consisted of a 15-s denaturing phase (90°C) and a 1-min annealing/extension phase at 60°C. The change in fluorescence was measured using an Applied Biosystems 7000 Prism sequence detector (PE Applied Biosystems) and analyzed using Sequence Detector version 1.0. The relative amount of transcript was determined using the comparative cycle threshold (CT) method as described by the manufacturer.
MLN cultures.
On days 1, 6, 12, and 24 postchallenge, mesenteric lymph nodes (MLN) were sterilely excised and homogenized by grinding the organs through a 70-μm nylon cell strainer. The strainer was rinsed with RPMI plus 10% heat-inactivated fetal bovine serum (FBS), and the cells were washed via centrifugation (600 × g for 10 min at 4°C). Red blood cells were lysed using lysis buffer (0.16 M NH4Cl, 10 mM KHCO3, and 0.13 mM EDTA) followed by a second centrifugation wash in RPMI plus 10% FBS. The cells were suspended in RPMI plus 10% FBS, and 5 × 105 cells were plated in flat-bottomed 96-well plates. The cells were cultured with 1 × 108 CFU of heat-killed C. rodentium at 37°C in 5% CO2 for 20 h. TNF-α levels were assessed in the supernatants using enzyme-linked immunosorbent assay (ELISA; BD Pharmingen, San Diego, CA).
Quantification of IgA levels.
Colonic contents were sterilely removed from mice and suspended in sterile PBS at a concentration of 1 g/4 ml. The suspension was diluted 1:200, and IgA levels were assessed via immunosorbent assay (Bethyl Laboratories, Montgomery, TX).
Statistical analyses.
Stressor-induced changes in bacterial numbers and gene expression were analyzed with a two-factor analysis of variance (ANOVA). The dependent variable for the real-time PCR was the ΔCT, which is calculated by determining the amplification cycle number at which the gene of interest begins exponential amplification minus the amplification cycle number at which the internal housekeeping gene (i.e., 18S) begins exponential amplification. For descriptive purposes, ΔCT was standardized and transformed to depict the n-fold change in gene expression compared with gene expression in the FWD control animals on day 1 postchallenge.
For all analyses, the level of statistical significance was set at P < 0.05 and was determined using SPSS for Windows version 17.0 (SPSS, Chicago, IL). Multivariate hierarchical clustering methods were performed using NCSS 2007 (NCSS, Kaysville, UT). Self-organizing maps were performed within Acuity 4.0 (Molecular Devices Corp., Sunnyvale, CA). Maximum operational taxonomic units (OTU) were predicted using Richard's equation to model the rarefaction curve as described previously (1) predicting Y up to 30,000 reads, based upon the changes in X (observed OTU). The maximum bootstrap iterations were 300 to provide the best fit for the observed curves. Richard's equations were computed within NCSS 2007 (NCSS, Kaysville, UT).
Data accession number.
Curated (size-trimmed, chimera-screened, etc.) data are available for download from http://www.researchandtesting.com/baileytrim.zip and will be available through NCBI SRA via accession number SRA010593.1.
RESULTS
The prolonged restraint stressor significantly changes the intestinal microbiota.
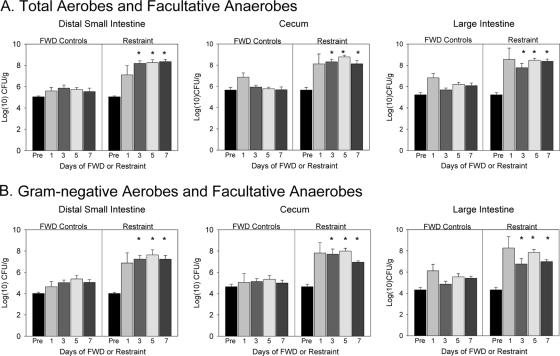
Exposure to the restraint stressor, as well as prolonged food and water deprivation, significantly changed microbial populations in the intestines. When bacteria in the distal small intestine, cecum, and colon were aerobically cultured, the RST mice had significantly higher bacterial levels than did the FWD controls (Fig. 1). This was evident when the contents of the small intestine [F(1, 37) = 63.14, P < 0.001], cecum [F(1, 37) = 54.10, P < 0.001], and large intestine [F(1, 37) = 62.03, P < 0.001] were grown in a nonselective nutrient-rich agar (Fig. 1A) or on a differential agar for Gram-negative bacteria {small intestine [F(1, 37) = 38.45, P < 0.001], cecum [F(1, 37) = 59.89, P < 0.001], and large intestine [F(1, 37) = 41.24, P < 0.001]} (Fig. 1B). When the microbiota were cultured, bacterial levels in the FWD controls were never significantly higher than baseline levels taken during quiescent nonstress conditions (Fig. 1); however, culturing did not allow for a more thorough description of changes in the microbiota. As a result, we used molecular sequencing to identify microbial populations (primarily anaerobic microbial populations) that were affected by exposure to the stressor.
FIG. 1.
Exposure to prolonged restraint significantly changed the levels of bacteria that could be cultured from the distal small intestine, cecum, and large intestine. The data are the means + standard errors, with panel A showing levels of total (i.e., both Gram-positive and Gram-negative) aerobic and facultatively anaerobic bacteria and panel B showing Gram-negative aerobes and facultative anaerobes. n = 3 preexperiment animals; n = 9 per group at days 1, 3, 5, and 7. *, P < 0.05 versus FWD levels on the same culture day. In this experiment, nonstressed home cage controls were used to set preexperiment values.
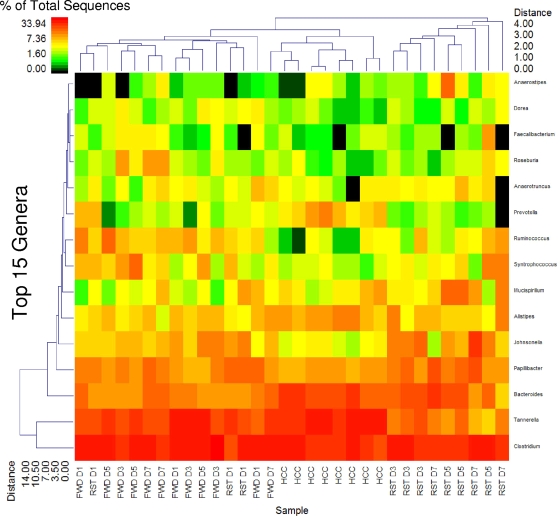
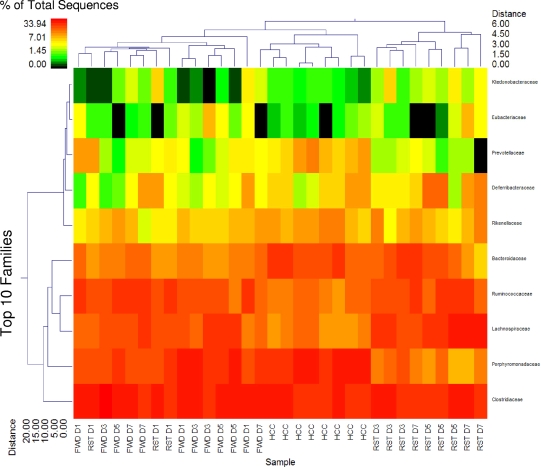
Figure 2 provides a summary of the top 15 genera among the RST, FWD, and HCC treatments that represent approximately 95% of identified sequences. This double clustering indicates that the RST D1 samples clustered with the FWD samples while all RST D3 to D7 samples clustered separately. The HCC samples also clustered separately but were more closely associated with the FWD and RST D1 treatments than with the RST D3 to D7 samples. And RST D5 and D7 samples resolved at a greater distance from RST D3. The primary populations leading to the marked clustering of RST D3 to D7 were Tannerella spp. At the family taxonomic level, the primary variable involved in distinguishing RST treatments from all others was the reduction in the Porphyromonadaceae, with changes in the Lachnospiraceae and Ruminococcaceae being primarily responsible for the separate clustering of the HCC samples (Fig. 3).
FIG. 2.
Exposure to prolonged restraint significantly changes community structure at the genus level. The double dendrogram describes the top 15 genera detected among the FWD, RST, and HCC samples. The heat map indicates the relative percentage of the given genera within each sample ID with a color legend and scale provided. The distance of the samples based upon weighted pair linkage and Manhattan distance methods with no scaling is provided at the top of the figure along with a distance score. The bacterial genera and the associated clustering are provided along the y axis, and their associated distance scores are indicated. n = 3 per group on days 1, 3, 5, and 7; n = 8 HCC controls.
FIG. 3.
Exposure to prolonged restraint significantly changes community structure at the family level. The double dendrogram describes the top 10 classes detected among the FWD, RST, and HCC samples. The heat map indicates the relative percentage of the given classes within each sample ID with a color legend and scale provided. The distance of the samples based upon weighted pair linkage and Manhattan distance methods with no scaling is provided at the top of the figure along with a distance score. The bacterial classes and the associated clustering are provided along the y axis, and their associated distance scores are indicated. n = 3 per group on days 1, 3, 5, and 7; n = 8 HCCs.
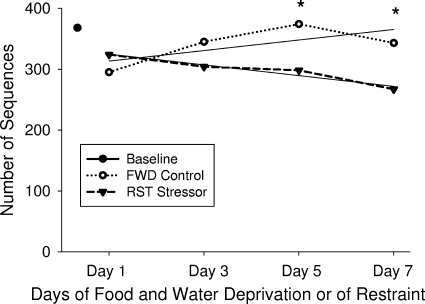
Mathematical evaluation of diversity and richness estimates based upon rarefaction, Chao1, and ACE methods as well as the use of curve fitting equations to predict maximum potential operational taxonomic units indicates that there are significant differences between the FWD and RST samples as a whole. There is little significance in D1 and D3 FWD compared to RST D1 and D3, and yet when FWD D5 and D7 are compared to RST D5 and D7, the data show a significantly lower diversity and species richness in the RST samples (P < 0.05) (Table 2 and Fig. 4). In general, diversity tended to increase in the FWD mice across the sampling days, whereas diversity decreased in the RST mice during this same time period (Fig. 4). There were no significant differences among the FWD and HCC samples over the course of the study when D1 and D3 were compared to D5 and D7, providing additional evidence that this is indeed a treatment effect.
TABLE 2.
Diversity analysis based upon rarefaction, OTU, ACE, Chao1, and modeling of rarefaction to predict maximum OTUa
| Group | Diversity |
Maximum modeled OTU species | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Rarefaction |
OTU |
ACE |
Chao1 |
||||||
| Species | Genus | Species | Genus | Species | Genus | Species | Genus | ||
| Baseline | 588 | 387 | 601 | 393 | 1,102 | 572 | 1,011 | 545 | 621 |
| FWD D1 | 451 | 295 | 456 | 297 | 741 | 376 | 738 | 373 | 478 |
| FWD D3 | 529 | 345 | 531 | 346 | 843 | 435 | 836 | 438 | 571 |
| FWD D5b | 598 | 374 | 602 | 376 | 992 | 488 | 968 | 493 | 667 |
| FWD D7b | 534 | 343 | 537 | 344 | 874 | 435 | 854 | 419 | 577 |
| RST D1 | 534 | 324 | 539 | 326 | 876 | 402 | 807 | 395 | 588 |
| RST D3 | 458 | 304 | 461 | 305 | 756 | 402 | 746 | 413 | 485 |
| RST D5b | 468 | 298 | 470 | 299 | 776 | 369 | 763 | 368 | 501 |
| RST D7b | 413 | 267 | 415 | 268 | 633 | 320 | 646 | 312 | 438 |
Species designations are based upon 3% sequence divergence, and genus designations are based upon 5% sequence divergence. The data are the means from 3 animals per treatment, i.e., food and water deprivation control (FWD) and restraint stressor (RST), at each time point (i.e., after D1, D3, D5, and D7 under each condition).
Based upon Student's t test, there is a significant difference in the predictions between FWD D5 and D7 and RST D5 and D7 (P < 0.05) for all estimations.
FIG. 4.
Rarefaction analysis of microbial communities on different sampling days. The number of phylotypes, identified with >95% similarity, is plotted for FWD controls (open circles) and the RST mice (closed inverted triangles) after 1, 3, 5, and 7 exposures. The data indicate that, over time, the number of phylotypes in the FWD control mice slightly increased, with the number of phylotypes in the RST mice decreasing with repeated exposure to RST. n = 3 per group at each time point; n = 4 to calculate baseline values. RST mice had a decreased number of phylotypes across the sampling days [F(1, 15) = 4.40, P < 0.05], with the largest differences occurring on days 5 and 7 (*, P < 0.05). In this experiment, nonstressed HCC mice were used to set baseline values.
The prolonged restraint stressor significantly increases C. rodentium levels.
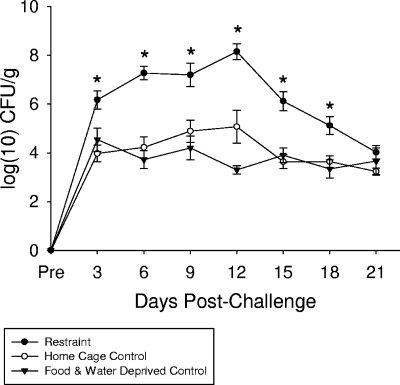
Exposure to the restraint stressor significantly changed fecal shedding of C. rodentium during the first 21 days of infection [F(2, 234) = 78.33, P < 0.001; Fig. 5]. This stressor-induced difference was first evident on day 3 postinfection and lasted until day 18 postinfection (P < 0.05). At the peak of colonization, i.e., day 12 postinfection, C. rodentium levels were increased by nearly 10,000-fold. Importantly, the levels of C. rodentium shed from nonstressed HCCs and from FWD controls were equivalent during the 21-day infection [F(1, 157) = 1.83, not significant], indicating that the increased colonization by C. rodentium was due to the effects of the restraint stressor and not the associated food and water deprivation.
FIG. 5.
Exposure to prolonged restraint prior to oral challenge with C. rodentium significantly increased the number of C. rodentium bacteria shed in the stool. Mice were exposed to prolonged restraint on 7 consecutive nights prior to oral challenge with 3 × 108 to 5 × 108 CFU of C. rodentium. The C. rodentium bacteria were then cultured from the stool during the first 21 days of infection. The data are the means ± standard errors of 10 to 16 mice per group. *, P < 0.05 versus both FWD and HCC at the specified time point.
The restraint stressor significantly alters gene expression in the colonic tissue after challenge with C. rodentium.
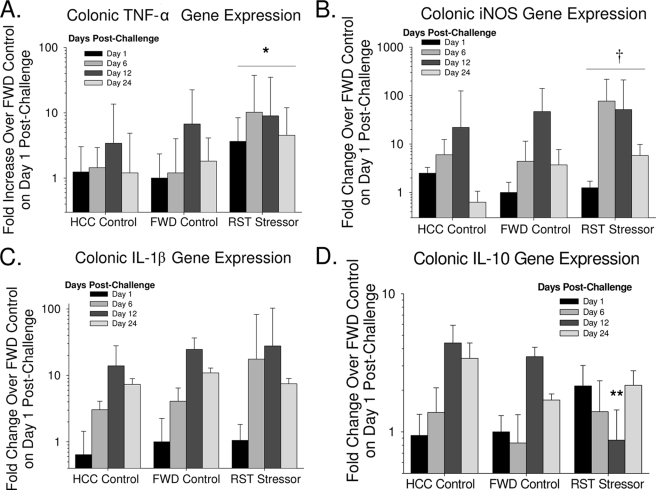
During the first 3 weeks postchallenge, mice exposed to the restraint stressor prior to oral challenge with C. rodentium had significantly higher levels of TNF-α than did mice in the HCC and FWD control groups [F(2, 33) = 5.64, P < 0.01; Fig. 6A]. There was a nearly 10-fold increase in TNF-α levels in the RST mice compared to the HCC and FWD mice on day 6 postchallenge (Fig. 6A). There was also a trend for iNOS gene expression to be higher in the mice exposed to RST prior to oral challenge [F(2, 33) = 2.64, P = 0.08; Fig. 6B]). Like TNF-α, iNOS gene expression was primarily different on day 6 postchallenge, with iNOS being nearly 100-fold higher in the RST mice than in the HCC and FWD controls (Fig. 6B). Although IL-1β levels increased during the 3 weeks postchallenge [F(3, 33) = 10.34, P < 0.001], this increase occurred regardless of whether the mice were exposed to RST or were in the HCC or FWD control groups prior to challenge as indicated by the lack of a statistically significant difference between the groups [F(2, 33) = 0.81, P = 0.45; Fig. 6C]. In contrast to gene expression for these inflammatory mediators, gene expression for the anti-inflammatory cytokine IL-10 tended to be suppressed in mice exposed to RST prior to oral challenge with C. rodentium (Fig. 6D). This effect, however, did not occur on all days postchallenge. Post hoc testing indicated that IL-10 gene expression was significantly lower in the RST mice than in the HCC and FWD control mice specifically on day 12 postchallenge (P < 0.05) (Fig. 6D).
FIG. 6.
Exposure to prolonged restraint prior to oral challenge with C. rodentium significantly altered colonic gene expression. Mice were exposed to prolonged restraint on 7 consecutive nights prior to oral challenge with 3 × 108 to 5 × 108 CFU of C. rodentium. Mice were euthanized on the indicated days postinfection, and gene expression for TNF-α (A), iNOS (B), IL-1β (C), and IL-10 (D) in colonic tissue was determined. The data are the means ± standard errors of the fold increase in gene expression over the FWD controls on day 1 postinfection. n = 6 mice per group on day 1 postchallenge; n = 3 mice per group on days 6, 12, and 24 postchallenge. The asterisk indicates a main effect of RST having higher gene expression than HCC and FWD across all days of infection (P < 0.05). **, P < 0.05 versus HCC and FWD controls at the same time point; †, marginally significant main effect of RST having higher gene expression than HCC and FWD across all days of infection (P = 0.07).
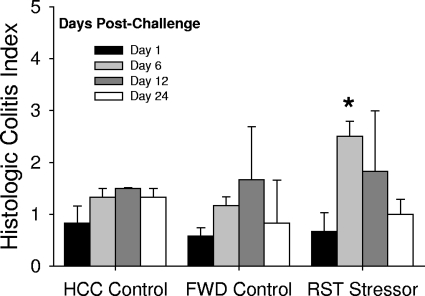
Colonic histopathology was significantly increased during the 3 weeks postinfection [F(3, 32) = 3.38, P < 0.05], but overall colonic histopathology was relatively mild, with the histologic colitis index remaining below 4 for all animals (Fig. 7). Even with this low level of inflammation, there was a statistically significant difference in the colitis index [F(2, 9) = 11.40, P < 0.01]. Mice exposed to the restraint stressor prior to oral challenge with C. rodentium had significantly higher colitis scores than did nonstressed HCC or FWD control mice on day 6 postchallenge (P < 0.05).
FIG. 7.
Colonic histopathology is increased in mice exposed to prolonged restraint prior to C. rodentium challenge. The data are the means ± standard errors of the cumulative histologic colitis score. n = 6 per group on day 1 postchallenge; n = 3 per group on days 6, 12, and 24 postchallenge. *, P < 0.05 versus HCC and FWD controls on day 6 postchallenge.
Secretory immunity and mesenteric lymph node reactivity to C. rodentium is unaffected by prolonged restraint.
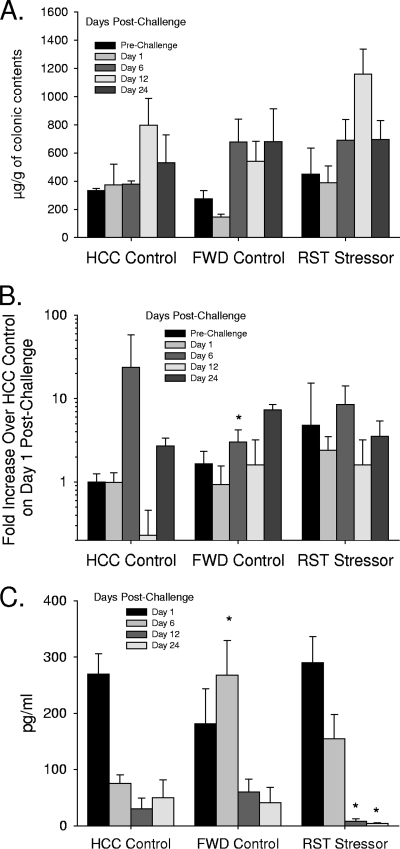
To determine whether the stressor-induced increase in C. rodentium colonization could be due to stressor-induced immunosuppression, two measures of secretory immunity (i.e., colonic IgA levels and colonic gene expression for mouse β-defensin 1) were assessed by ELISA and real-time PCR (respectively). Although both IgA and mouse β-defensin 1 changed during the course of infection (Fig. 8A and B), there were no statistically significant differences between levels found in RST mice and those found in either the FWD controls or the HCCs [F(2, 55) = 0.65, not significant, for IgA and F(2, 34) = 2.21, not significant, for mouse β-defensin 1]. Only the FWD control mice on day 6 postchallenge were found to have lower mouse β-defensin 1 gene expression than HCC mice. Gene expression for mouse β-defensin 3 was too low to be reliably quantified in all of the samples. Reactivity of MLN cultures to C. rodentium was unaffected by exposure to the stressor early during the infectious challenge (i.e., days 1 and 6 postchallenge), but by days 12 and 24 postchallenge MLN cultures from mice exposed to RST prior to C. rodentium challenge produced lower levels of TNF-α (P < 0.05). In the absence of infectious challenge, C. rodentium-induced TNF-α production by MLN cultures was below our level of detection in HCCs, FWD controls, and RST mice.
FIG. 8.
Prolonged restraint had little effect on measures of mucosal immunity. (A) Colonic IgA levels were unaffected by restraint exposure prior to challenge. n = 3 per group prechallenge; n = 5 per group on days 1, 6, 12, and 24 postchallenge. (B) Gene expression for mouse β-defensin 1 was similar in RST mice and in HCCl mice, with FWD control mice having lower gene expression on day 6 postchallenge (*, P < 0.05). n = 5 per group prechallenge; n = 3 per group on days 1, 6, 12, and 24 postchallenge. (C) TNF-α levels were suppressed in RST mice compared to HCC mice on days 12 and 24 postchallenge (*, P < 0.05 versus HCC mice). n = 3 per group prechallenge; n = 5 per group on days 1, 6, 12, and 24 postchallenge. In all cases the data are the means ± standard errors.
DISCUSSION
The gastrointestinal microbiota have been reported to comprise a relatively stable community of microbes residing at their climax levels within specific segments of the intestines. Our results indicated that exposure to prolonged restraint, as well as to food and water deprivation, results in both quantitative and qualitative differences in microbial populations in the intestines. This was partially manifest as an overgrowth of aerobically cultured bacteria in the distal small intestine, cecum, and large intestine. The types of bacteria that were responsible for the overgrowth were not determined in this experiment, but it was evident that both Gram-positive and Gram-negative microbes overgrew in the intestines due to growth patterns on nonselective and differential agars. Critically, the number of bacteria cultured from the intestines was similar in HCC and FWD mice, leading to the conclusion that the restraint stressor, and not the associated food and water deprivation, was responsible for the overgrowth.
We also used a high-throughput 16S tag pyrosequencing approach, termed bTEFAP, to characterize the intestinal microbiota of stressed and nonstressed mice. This method helped to identify several important stressor-induced changes in community structure of the gut microbiota. Using the relative percentage of bacteria classified at the family or genus level and dual hierarchical clustering methods, individual animals tended to fall into 3 groups based upon the microbial diversity, with the FWD condition primarily being found in one group, the HCC condition primarily being found in the second group, and the RST condition primarily being found in the third group. Importantly, the phylogenetic analyses also indicated that the FWD and HCC groups were more similar than the RST group. This was not evident immediately, however, and it was only when the animals were exposed to at least 3 overnight cycles of restraint stress (i.e., RST D3) that the microbiota of the RST mice were noted to be dissimilar from those of the FWD mice. Microbial diversity, as determined using rarefaction, OTU, Chao1, ACE, and maximum predicted OTU analysis, also indicated that changes in the gut microbial community were more pronounced after repeated cycles of RST (i.e., after days 5 and 7). The reason that repeated cycles of stress were necessary for this shift to occur is not immediately clear, but studies in humans, nonhuman primates, and rodents have found that changes in the microbiota were more pronounced several days after a stressor than on the day of or the day after stressor exposure (4, 20, 34). Thus, although stressor-induced changes to the microbiota may be transient, prolonged or repeated stressors can have lasting effects on the microbiota.
Nonstressed control mice harbored a stable baseline community of bacteria at the family and genus taxonomic level of analysis, which is consistent with previous reports (2). However, reliable stressor-induced changes in the microbiota did emerge. The relative abundance of bacteria in the family Porphyromonadaceae was lower in the RST mice sampled on days 3, 5, and 7 than in the HCC and FWD control groups. This was primarily due to a decrease in the relative abundance of bacteria in the genus Tannerella in the RST mice. Importantly, the levels of the Porphyromonadaceae, and specifically the Tannerella bacteria, between the FWD and the HCC mice were similar. Thus, only the stressed mice had a microbiota profile that consisted of an overgrowth of aerobically cultured Gram-positive and Gram-negative bacteria and a reduction in microbial richness that could in part be explained by a reduction in bacteria in the genus Tannerella. The mechanisms through which the stressor changed the microbiota are not immediately clear, but other studies have shown that intestinal inflammation, such as inflammation induced by 3% dextran sodium sulfate, significantly reduces the relative abundance of Tannerella spp., resulting in an overgrowth of the Enterobacteriaceae (37). To determine whether the stressor could be inducing intestinal inflammation prior to infection, we measured gene expression for IL-1β, TNF-α, IL-6, IFN-γ, and IL-10 in small intestinal, cecal, and colonic tissue using semiquantitative real-time PCR. However, in the absence of an infectious challenge, the restraint stressor did not significantly affect cytokine gene expression (data not shown). Thus, it is likely that factors other than inflammation, such as stressor-induced changes to gastrointestinal physiology or direct neuroendocrine-bacterial interactions (12, 25, 26, 38-41), led to microbiota changes. These are topics for future study.
The intestinal microbiota are well known for their ability to inhibit the colonization of enteric pathogens, like E. coli and Salmonella enterica serovar Typhimurium, through competitive exclusion (11, 27). The members of the microbiota that can inhibit C. rodentium colonization are not known, but a previous study found an association between low relative abundance of bacteria in the genus Tannerella, along with a high relative abundance of Enterobacteriaceae, and increased C. rodentium levels (37). The authors rationalized that C. rodentium-induced disruption of the microbiota would further promote C. rodentium colonization and proliferation due to the reduced ecological competition by commensals (37). Because we observed stressor-induced reductions in the abundance of bacteria in the genus Tannerella and an associated aerobic overgrowth of Gram-negative bacteria prior to infectious challenge, it is likely that the stressor-induced increase in C. rodentium colonization was the result of reduced competition with commensal microbes.
Systemic immune activity has been shown to be suppressed by prolonged restraint (3), but the impact of this stressor on mucosal immune responses, which are the most relevant for affecting C. rodentium colonization in the colon, is less clear. For example, some studies have reported that restraint suppresses levels of IgA in the intestinal lumen (32), while others report an enhancement (46). In our study, restraint prior to oral challenge tended to enhance IgA, as well as mouse β-defensin 1 gene expression (albeit not statistically significantly), and upon oral challenge few differences between the control and RST mice emerged. In fact, the only immune measure found to be suppressed in RST mice was the ex vivo production of TNF-α by MLN cells. Because this difference did not emerge until day 12 postchallenge, it is not likely that suppressed MLN cytokine production was a major contributing factor in the stress-induced increase in C. rodentium levels that emerged as early as day 3 postchallenge. The importance of changes in cytokine production, however, will be followed up in the future.
Colonic cytokines are important in controlling the progression of C. rodentium infection upon oral challenge (22). In our study, C. rodentium levels and cytokine levels were both elevated, but very little colonic pathology was observed during the 3 weeks post-C. rodentium challenge. This may be due to the mouse strain used in this study (i.e., outbred Swiss-derived CD-1 mice), which can be colonized by C. rodentium and can develop colonic hyperplasia but with the hyperplasia typically being asymptomatic and occurring with little inflammation (7, 10, 52). Even though the histologic colitis scores were low in this study, it is interesting that the scores were statistically significantly higher in the stressed animals on day 6 postchallenge, although the classic Citrobacter-induced lesion of hyperplasia was not a feature. This is the same time point at which cytokine levels were higher in the colonic tissue of the stressed mice, with the nonstressed control mice reaching similar pathological levels a week later (i.e., day 12 postchallenge versus day 6 postchallenge). Because previous studies have found that germfree mice develop acute intestinal inflammation by 1 week post-C. rodentium challenge, compared to 2 weeks in conventional mice (6), the earlier occurrence of increased colitis scores is consistent with the hypothesis that stressor-induced changes to the microbiota affected the progression of the C. rodentium infection.
Exposure to psychological stressors has myriad effects on the body and has long been associated with gastrointestinal functioning and diseases (19). The present study provides a molecular characterization of stressor-induced changes in the microbiota and demonstrates that stressors can influence the microbiota at many different levels of taxonomic analysis. Moreover, the finding that the stressor altered both the intestinal microbiota and susceptibility to enteric infection, while leaving IgA levels, mouse β-defensin 1 gene expression, and MLN cytokine production unaffected, suggests that the changes in the microbiota are biologically meaningful by enhancing susceptibility to infection. Because other diseases that have been linked to the microbiota, such as obesity (50), diabetes (51), irritable bowel syndrome (33), and inflammatory bowel diseases (15, 42), can be worsened during periods of psychological stress (19, 21, 47), future studies should focus on stressor-induced changes to the microbiota in these complex diseases.
Acknowledgments
David Schauer unexpectedly passed away before the completion of these studies. The manuscript was written posthumously by his colleagues, who mourn his loss.
This work was funded by Public Health Service grant 1R03AI069097 (M.T.B.) from the National Institute of Allergy and Infectious Diseases, by OSU Colleges of Dentistry and Medicine start-up funds (M.T.B.), and by Texas Tech University Health Sciences internal grants (M.L.).
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 9 February 2010.
REFERENCES
- 1.Acosta-Martinez, V., S. Dowd, Y. Sun, and V. Allen. 2008. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 40:2762-2770. [Google Scholar]
- 2.Antonopoulos, D. A., S. M. Huse, H. G. Morrison, T. M. Schmidt, M. L. Sogin, and V. B. Young. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, M., H. Engler, J. Hunzeker, and J. F. Sheridan. 2003. The hypothalamic-pituitary-adrenal axis and viral infection. Viral Immunol. 16:141-157. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, M. T., and C. L. Coe. 1999. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev. Psychobiol. 35:146-155. [PubMed] [Google Scholar]
- 5.Bailey, M. T., G. R. Lubach, and C. L. Coe. 2004. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J. Pediatr. Gastroenterol. Nutr. 38:414-421. [DOI] [PubMed] [Google Scholar]
- 6.Barthold, S. W., G. L. Coleman, P. N. Bhatt, G. W. Osbaldiston, and A. M. Jonas. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889-894. [PubMed] [Google Scholar]
- 7.Barthold, S. W., G. W. Osbaldiston, and A. M. Jonas. 1977. Dietary, bacterial, and host genetic interactions in the pathogenesis of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 27:938-945. [PubMed] [Google Scholar]
- 8.Berkenbosch, F., D. A. Wolvers, and R. Derijk. 1991. Neuroendocrine and immunological mechanisms in stress-induced immunomodulation. J. Steroid Biochem. Mol. Biol. 40:639-647. [DOI] [PubMed] [Google Scholar]
- 9.Borenshtein, D., P. R. Nambiar, E. B. Groff, J. G. Fox, and D. B. Schauer. 2007. Development of fatal colitis in FVB mice infected with Citrobacter rodentium. Infect. Immun. 75:3271-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borenshtein, D., K. A. Schlieper, B. H. Rickman, J. M. Chapman, C. W. Schweinfest, J. G. Fox, and D. B. Schauer. 2009. Decreased expression of colonic Slc26a3 (Dra) and carbonic anhydrase IV as a cause of fatal infectious diarrhea in mice. Infect. Immun. 77:3639-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U. S. A. 101:7427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C., D. R. Brown, Y. Xie, B. T. Green, and M. Lyte. 2003. Catecholamines modulate Escherichia coli O157:H7 adherence to murine cecal mucosa. Shock 20:183-188. [DOI] [PubMed] [Google Scholar]
- 13.Chen, C. C., S. Louie, H. N. Shi, and W. A. Walker. 2005. Preinoculation with the probiotic Lactobacillus acidophilus early in life effectively inhibits murine Citrobacter rodentium colitis. Pediatr. Res. 58:1185-1191. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dicksved, J., J. Halfvarson, M. Rosenquist, G. Jarnerot, C. Tysk, J. Apajalahti, L. Engstrand, and J. K. Jansson. 2008. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2:716-727. [DOI] [PubMed] [Google Scholar]
- 16.Dowd, S. E., T. R. Callaway, R. D. Wolcott, Y. Sun, T. McKeehan, R. G. Hagevoort, and T. S. Edrington. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowd, S. E., R. D. Wolcott, Y. Sun, T. McKeehan, E. Smith, and D. Rhoads. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One 3:e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowd, S. E., J. Zaragoza, J. R. Rodriguez, M. J. Oliver, and P. R. Payton. 2005. Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST). BMC Bioinformatics 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drossman, D. A. 1998. Presidential address: gastrointestinal illness and the biopsychosocial model. Psychosom. Med. 60:258-267. [DOI] [PubMed] [Google Scholar]
- 20.Everson, C. A., and L. A. Toth. 2000. Systemic bacterial invasion induced by sleep deprivation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 278:R905-R916. [DOI] [PubMed] [Google Scholar]
- 21.Gaynes, B. N., and D. A. Drossman. 1999. The role of psychosocial factors in irritable bowel syndrome. Baillieres Best Pract. Res. Clin. Gastroenterol. 13:437-452. [DOI] [PubMed] [Google Scholar]
- 22.Gibson, D. L., C. Ma, K. S. Bergstrom, J. T. Huang, C. Man, and B. A. Vallance. 2008. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cell. Microbiol. 10:618-631. [DOI] [PubMed] [Google Scholar]
- 23.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glavin, G. B., W. P. Pare, T. Sandbak, H. K. Bakke, and R. Murison. 1994. Restraint stress in biomedical research: an update. Neurosci. Biobehav. Rev. 18:223-249. [DOI] [PubMed] [Google Scholar]
- 25.Green, B. T., M. Lyte, C. Chen, Y. Xie, M. A. Casey, A. Kulkarni-Narla, L. Vulchanova, and D. R. Brown. 2004. Adrenergic modulation of Escherichia coli O157:H7 adherence to the colonic mucosa. Am. J. Physiol. Gastrointest. Liver Physiol. 287:G1238-G1246. [DOI] [PubMed] [Google Scholar]
- 26.Green, B. T., M. Lyte, A. Kulkarni-Narla, and D. R. Brown. 2003. Neuromodulation of enteropathogen internalization in Peyer's patches from porcine jejunum. J. Neuroimmunol. 141:74-82. [DOI] [PubMed] [Google Scholar]
- 27.Hapfelmeier, S., and W. D. Hardt. 2005. A mouse model for S. typhimurium-induced enterocolitis. Trends Microbiol. 13:497-503. [DOI] [PubMed] [Google Scholar]
- 28.Holdeman, L. V., I. J. Good, and W. E. Moore. 1976. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl. Environ. Microbiol. 31:359-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh, K., K. Maejima, K. Ueda, and K. Fujiwara. 1979. Difference in susceptibility of mice raised under barrier-sustained (SPF) or conventional conditions to infectious megaenteron. Microbiol. Immunol. 23:909-913. [DOI] [PubMed] [Google Scholar]
- 30.Itoh, K., A. Ozaki, T. Yamamoto, and T. Mitsuoka. 1978. An autoclavable stainless steel isolator for small scale gnotobiotic experiments. Jikken Dobutsu 27:13-16. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 31.Itoh, K., K. Ueda, and K. Fujiwara. 1980. Susceptibility of germ-free mice to infectious megaenteron. Microbiol. Immunol. 24:281-290. [DOI] [PubMed] [Google Scholar]
- 32.Jarillo-Luna, A., V. Rivera-Aguilar, H. R. Garfias, E. Lara-Padilla, A. Kormanovsky, and R. Campos-Rodriguez. 2007. Effect of repeated restraint stress on the levels of intestinal IgA in mice. Psychoneuroendocrinology 32:681-692. [DOI] [PubMed] [Google Scholar]
- 33.Kassinen, A., L. Krogius-Kurikka, H. Makivuokko, T. Rinttila, L. Paulin, J. Corander, E. Malinen, J. Apajalahti, and A. Palva. 2007. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology 133:24-33. [DOI] [PubMed] [Google Scholar]
- 34.Knowles, S. R., E. A. Nelson, and E. A. Palombo. 2008. Investigating the role of perceived stress on bacterial flora activity and salivary cortisol secretion: a possible mechanism underlying susceptibility to illness. Biol. Psychol. 77:132-137. [DOI] [PubMed] [Google Scholar]
- 35.Kvetnansky, R., K. Fukuhara, K. Pacak, G. Cizza, D. S. Goldstein, and I. J. Kopin. 1993. Endogenous glucocorticoids restrain catecholamine synthesis and release at rest and during immobilization stress in rats. Endocrinology 133:1411-1419. [DOI] [PubMed] [Google Scholar]
- 36.Lizko, N. N. 1987. Stress and intestinal microflora. Nahrung 31:443-447. [DOI] [PubMed] [Google Scholar]
- 37.Lupp, C., M. L. Robertson, M. E. Wickham, I. Sekirov, O. L. Champion, E. C. Gaynor, and B. B. Finlay. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119-129. [DOI] [PubMed] [Google Scholar]
- 38.Lyte, M. 2004. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 12:14-20. [DOI] [PubMed] [Google Scholar]
- 39.Lyte, M., A. K. Erickson, B. P. Arulanandam, C. D. Frank, M. A. Crawford, and D. H. Francis. 1997. Norepinephrine-induced expression of the K99 pilus adhesin of enterotoxigenic Escherichia coli. Biochem. Biophys. Res. Commun. 232:682-686. [DOI] [PubMed] [Google Scholar]
- 40.Lyte, M., P. P. Freestone, C. P. Neal, B. A. Olson, R. D. Haigh, R. Bayston, and P. H. Williams. 2003. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet 361:130-135. [DOI] [PubMed] [Google Scholar]
- 41.Lyte, M., and K. T. Nguyen. 1997. Alteration of Escherichia coli O157:H7 growth and molecular fingerprint by the neuroendocrine hormone noradrenaline. Microbios 89:197-213. [PubMed] [Google Scholar]
- 42.Nishikawa, J., T. Kudo, S. Sakata, Y. Benno, and T. Sugiyama. 2009. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand. J. Gastroenterol. 44:180-186. [DOI] [PubMed] [Google Scholar]
- 43.Nocker, A., M. Burr, and A. K. Camper. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54:276-289. [DOI] [PubMed] [Google Scholar]
- 44.O'Hara, A. M., and F. Shanahan. 2006. The gut flora as a forgotten organ. EMBO Rep. 7:688-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotsky, P. M., and M. J. Meaney. 1993. Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Res. Mol. Brain Res. 18:195-200. [DOI] [PubMed] [Google Scholar]
- 46.Reyna-Garfias, H., A. Miliar, A. Jarillo-Luna, V. Rivera-Aguilar, J. Pacheco-Yepez, I. Baeza, and R. Campos-Rodriguez. 2010. Repeated restraint stress increases IgA concentration in rat small intestine. Brain Behav. Immun. 24:110-118. [DOI] [PubMed] [Google Scholar]
- 47.Ringel, Y., and D. A. Drossman. 2001. Psychosocial aspects of Crohn's disease. Surg. Clin. North Am. 81:231-252. [DOI] [PubMed] [Google Scholar]
- 48.Schaedler, R. W., and R. J. Dubos. 1962. The fecal flora of various strains of mice. Its bearing on their susceptibility to endotoxin. J. Exp. Med. 115:1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, R. E. Ley, M. L. Sogin, W. J. Jones, B. A. Roe, J. P. Affourtit, M. Egholm, B. Henrissat, A. C. Heath, R. Knight, and J. I. Gordon. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaarala, O., M. A. Atkinson, and J. Neu. 2008. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes 57:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallance, B. A., W. Deng, K. Jacobson, and B. B. Finlay. 2003. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect. Immun. 71:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Waaij, D., J. M. Berghuis-de Vries, and L. Lekkerkerk. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg. (Lond.) 69:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]