Abstract
The tremendous success of Staphylococcus aureus as a pathogen is due to the controlled expression of a diverse array of virulence factors. The effects of host environments on the expression of virulence factors and the mechanisms by which S. aureus adapts to colonize distinct host tissues are largely unknown. Vertebrates have evolved to sequester nutrient iron from invading bacteria, and iron availability is a signal that alerts pathogenic microorganisms when they enter the hostile host environment. Consistent with this, we report here that S. aureus senses alterations in the iron status via the ferric uptake regulator (Fur) and alters the abundance of a large number of virulence factors. These Fur-mediated changes protect S. aureus against killing by neutrophils, and Fur is required for full staphylococcal virulence in a murine model of infection. A potential mechanistic explanation for the impact of Fur on virulence is provided by the observation that Fur coordinates the reciprocal expression of cytolysins and a subset of immunomodulatory proteins. More specifically, S. aureus lacking fur exhibits decreased expression of immunomodulatory proteins and increased expression of cytolysins. These findings reveal that Fur is involved in initiating a regulatory program that organizes the expression of virulence factors during the pathogenesis of S. aureus pneumonia.
Staphylococcus aureus is one of the most significant infectious threats to human health. This fact is reinforced by the increasing incidence of nosocomial as well as community-acquired infections worldwide (23). S. aureus is responsible for an array of diseases ranging from minor skin and soft tissue infections to more invasive and serious infections, such as pneumonia, osteomelytis, and endocarditis. The ability of S. aureus to cause such diverse diseases is due primarily to an arsenal of virulence factors encoded in the staphylococcal genome (21, 47).
During infection of mammalian hosts, pathogens are exposed to a variety of environmental signals that have the potential to influence the expression of virulence factors. These signals include, but are not limited to, changes in nutrient availability, temperature, pH, osmolarity, and oxygen tension. S. aureus senses these and other cues to alter the expression of virulence factors (2, 11, 12, 15, 26, 37, 50, 56, 69); however, the molecular mechanisms employed by staphylococci to sense signals present in thejr hosts are not well understood.
One key environmental signal that pathogens encounter during infection of vertebrates is alterations in the iron status (10). In vertebrates bioavailable iron is scarce, and most of the iron is in erythrocytes bound by hemoglobin. S. aureus, like most pathogens, requires iron to multiply and cause disease (62). To acquire iron from its host, S. aureus encodes two heme acquisitions systems (Isd and Hts) (62) and produces siderophores (17, 19). The importance of iron acquisition to S. aureus pathogenesis is highlighted by the observation that disruption of iron acquisition results in reduced virulence in systemic animal models of infection (19, 61, 66). Notably, staphylococcal strains defective in heme uptake are not attenuated for virulence in murine models of pneumonia, suggesting that heme iron is not a critical nutrient for S. aureus in the murine lung (39).
Bacteria are known to sense iron-limited environments via the ferric uptake regulator (Fur) (31). Fur-mediated sensing of iron availability is conserved across Gram-positive and Gram-negative bacteria (31, 35). Generally, Fur is a repressor that binds to DNA, inhibiting the expression of target genes when iron is abundant in the bacterium. Conversely, in iron-limited environments, Fur-mediated repression is lifted and target genes are expressed. Staphylococcal Fur regulates the expression of genes encoding iron acquisition systems (5, 19, 41, 66, 71), influences the expression of a large number of cytoplasmic proteins (25), is involved in biofilm formation (33), and affects the expression of antioxidative stress proteins (25, 32, 56).
In the present study we determined the contribution of Fur to the expression of a subset of secreted staphylococcal virulence factors and evaluated the impact of Fur on S. aureus-host interactions. Our data demonstrate that S. aureus senses iron limitation via Fur to coordinate increased production of hemolysins and cytotoxins. Notably, our study also revealed that staphylococci lacking Fur are more susceptible to host-mediated clearance, a phenotype associated with decreased production of immunomodulatory proteins involved in avoidance of neutrophil-mediated killing. Taken together, our results indicate that Fur is an important regulator produced by S. aureus during infection to modulate a potent cytotoxic and immunity-modulating response.
MATERIALS AND METHODS
Ethics statement.
Mouse infections were approved by Vanderbilt University's Institutional Animal Care and Use Committee (IACUC). All experiments conformed to regulatory guidelines for animal infections.
Bacterial strains and growth conditions.
S. aureus clinical isolate Newman was used in all experiments unless stated otherwise. The other S. aureus strains were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus and Frank DeLeo (Rocky Mountain Laboratories, NIAID, NIH). The Newman Δfur::tet and Δfur hla::ermC strains were created by transduction of previously created mutant loci (42, 49) to strain Newman with the transducing phage φ-85 as previously described (60).
S. aureus strains were grown in tryptic soy broth (TSB) (Difco) or Roswell Park Memorial Institute culture medium 1640 (RPMI) (Invitrogen) supplemented with 1% Casamino Acids (RPMI+CAS) and 300 to 400 μM dipyridyl (iron-restricted medium). When required, the culture medium was supplemented with 10 μg/ml of chloramphenicol and/or 10 μg/ml of erythromycin. Overnight cultures were grown in 5 ml medium in 15-ml tubes kept at a 45° angle and incubated at 37οC with shaking at 180 rpm. Following overnight growth, bacteria were subcultured using a 1:100 dilution for 4 to 6 h as described above. Escherichia coli DH5α was used to propagate plasmids and was grown on Luria-Bertani (LB) agar and in LB broth supplemented with 100 μg/ml of ampicillin.
Construction of complementation vectors.
For construction of the fur (S. aureus Newman NWMN_1406) complementation plasmid, we generated a plasmid that contains S. aureus fur under control of the lipoprotein diacylglycerol transferase (lgt) promoter (9). To this end, a primer annealing to the 5′ end of the S. aureus fur open reading frame and containing an NdeI site (GGGCATATGGGACATCGTTGGAAGAACG) and a 3′ primer containing a BamHI site (CCCGGATCCGCAATTTACTATCCTTTACC) were used to amplify fur from S. aureus strain Newman genomic DNA. The amplicon was inserted into pCR2.1 (Promega), generating plasmid pCR2.1-fur. pCR2.1-fur was digested with NdeI and BamHI, and fur was inserted into pOS1-plgt that had been digested with the same enzymes, generating pOS1plgt-fur. Ligation products were transformed into E. coli, and transformants were selected on LB agar supplemented with ampicillin. Colonies were examined via restriction mapping and PCR to isolate a correct fur complementation plasmid. Complementation plasmids were electroporated into the restriction-deficient S. aureus RN4220 strain (48), after which they were electroporated into the appropriate S. aureus Newman strain. The hla complementation vector has been described previously (49).
SDS-PAGE of secreted proteins.
For proteomic studies S. aureus strains were grown as described above. Exoproteins in S. aureus culture supernatants were precipitated with 10% (vol/vol) trichloroacetic acid (TCA) and incubated for ∼15 h at 4°C. The precipitated proteins were washed once with 100% ethanol, air dried, resuspended with 25 μl of SDS-Laemmli buffer, and boiled at 95°C for 10 min. Proteins were separated using 12 to 15% SDS-PAGE gels and stained with colloidal blue (Invitrogen) or Coomassie blue.
2D-DIGE analysis.
For two-dimensional difference in-gel electrophoresis (2D-DIGE) analysis three independent cultures of the wild-type and Δfur S. aureus strains were inoculated into RPMI+CAS and grown to late exponential phase at 37°C. Culture supernatants were then collected, filtered, and concentrated using a Millipore centrifugal device with a 5-kDa cutoff. Concentrated proteins were then washed two times with Tris-buffered saline (TBS) (50 mM Tris [pH 7.5], 150 mM NaCl, 100 μM phenylmethylsulfonyl fluoride [PMSF]), and the protein concentration was adjusted to 1 mg/ml. For each sample, 0.25 mg of protein was precipitated and labeled as described previously (25). The 2D-DIGE gels were prepared and 2D-DIGE and principal component analysis (PCA) were performed as described previously (24, 25). Proteins were identified as described below.
Liquid chromatography-MS/MS analysis and protein identification.
Exoproteins from wild-type and Δfur cultures were prepared as described above. Proteins were electrophoresed 2 cm into a 15% SDS-PAGE gel and stained with colloidal blue (Invitrogen). The 2-cm gel regions were excised and subjected to in-gel trypsin digestion and peptide extraction as previously described (30). The resulting peptides were analyzed using a Thermo Finnigan LTQ ion trap instrument equipped with a Thermo MicroAS autosampler and a Thermo Surveyor high-performance liquid chromatography (HPLC) pump, a nanospray source, and an Xcalibur 2.0 SR2 instrument control. Peptides were separated using a packed capillary tip (100 mm by 11 cm; Polymicro Technologies) with Jupiter C18 resin (5 mm; 300 Å; Phenomenex) and an in-line trapping column (100 μm by 6 cm) packed with the same C18 resin (using a frit generated with liquid silicate Kasil) similar to the column described previously (65). The flow from the HPLC pump was split prior to the injection valve to obtain flow rates of 700 nl min−1 to 1,000 μl min−1 at the column tip. Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 0.1% formic acid in acetonitrile. A 95-min gradient was used with a 15-min washing period (100% mobile phase A for the first 10 min, followed by a gradient to 98% mobile phase A at 15 min) to allow loading and flushing of any residual salts. Following the washing period, the gradient was changed to 25% mobile phase B at 50 min and then to 90% mobile phase B by 65 min, which was used for 9 min before the conditions were returned to the initial conditions. Tandem spectra were acquired using a data-dependent scanning mode in which one full mass spectrometry (MS) scan (m/z 400 to 2,000) was followed by nine MS/MS scans. Tandem spectra were compared with data for the Newman strain of the S. aureus subset in the UniRef100 database using the SEQUEST algorithm. The database was concatenated with the reverse sequences of all proteins in the database to allow determination of false-positive rates. The Sequest outputs were filtered through the ID Picker suite, which allows the user to set a false discovery rate threshold (e.g., 0.05 or 5%) based on reverse sequence hits in the database, and proteins were required to be identified by two or more unique peptides. Reassembly of a protein from identified peptide sequences was done with the aid of a parsimony method recently described by Zhang et al. (73), which identifies and clusters together indiscernible proteins (protein groups) that can account for the identified peptides.
Hemolysis and cytotoxicity assay.
The hemolytic activity of staphylococcal exoproteins was determined as described previously (6). For cytotoxicity assays, HL-60 cells (ATCC CCL-240) were grown as recommended by ATCC in a 5% CO2 atmosphere in an incubator at 37°C. Cells were seeded at a concentration of ∼1.2 × 105 cells per well on 96-well plates. HL-60 cells were routinely intoxicated with 2 to 10 μl of staphylococcal culture supernatant for 3 h. Cell viability was examined by inverted light microscopy and was quantified using the CellTiter 96 reagent (Promega). The HL-60 viability data are expressed below as percentages of viable cells (optical densities at 490 nm [OD490]); the value for cells treated with medium was defined as 100%.
Western blotting.
Precipitated exoproteins obtained as described above were loaded on 12% SDS-PAGE gels and then transferred to nitrocellulose membranes. The membranes were blotted with antibody against Hla as the primary antibody (44) and Alexa Fluor 680-conjugated anti-rabbit secondary antibodies (Invitrogen). Membranes were dried and scanned using an Odyssey infrared imaging system (LI-COR Biosciences). The entire experiment was carried out three times independently.
Mouse model of infection.
The wild-type and fur mutant strains were grown overnight in 5 ml of RPMI+CAS. Overnight cultures were then subcultured 1:100 in RPMI+CAS and grown to late log phase until the optical densities of the cultures were similar. To create the individual inocula, the cultures were centrifuged, the supernatants were removed, and the bacterial pellets were washed in 5 ml of endotoxin-free phosphate-buffered saline (PBS) twice. The pellets were then resuspended in 1.5 ml of endotoxin-free PBS, and the OD600 was determined.
Seven- to 8-week-old female C57BL/6J mice (Jackson Laboratories) were infected intranasally with the wild-type and fur mutant strains (3 × 108 to 5 × 108 CFU resuspended in 30 μl of PBS) as described previously (8). Briefly, mice were anesthetized, and 30 μl of a bacterial culture was inoculated into the right nare of each mouse; the mice were held upright for 30 s following inoculation. The mice were monitored closely over the course of infection. At 6 and 18 h after infection, mice were euthanized with CO2, the lungs were removed and homogenized in PBS, and bacterial loads were determined based on colony formation on tryptic soy agar (TSA). Statistical analyses were performed using the Student t test.
Neutrophils were depleted by intraperitoneal injection of 250 μg rat IgG2b anti-Gr-1 monoclonal antibody (MAb) RB6-8C5 (anti-neutrophil antibody) in 100 μl PBS 24 h prior to infection and again at the time of infection. The appropriate inoculum was determined in preliminary experiments (data not shown) by infecting groups of three neutrophil-depleted C57BL/6J mice with S. aureus wild-type strain Newman using the following inocula: 1.2 × 108 CFU, 2.2 × 107 CFU, 6 × 106 CFU, 1.2 × 106 CFU, and 1.5 × 105 CFU. Mice were euthanized at 18 h postinfection, and lungs were harvested for enumeration of CFU. Based on the results of this preliminary experiment, an inoculum of 2 × 107 CFU was selected. To compare the virulence of the Δfur strain in neutropenic mice with that of the wild type, groups of 10 mice were infected by intranasal inoculation of 2.9 × 107 CFU (wild type) or 1.5 × 107 CFU (Δfur strain) bacteria in 30 μl PBS. Mice were euthanized at 18 h postinfection, and lungs were harvested for enumeration of CFU.
Flow cytometry.
Erythrocyte-free total lung homogenates from C57BL/6J animals infected with S. aureus or uninfected animals were stained for four-color flow cytometric analysis as described previously using a FACSCalibur instrument (Becton Dickinson) (67). The data were analyzed using FlowJo software (Treestar Inc.).
Opsonophagocytic killing assay.
The opsonophagocytic killing assay was performed as described previously (15). To test the role of exoproteins in protecting S. aureus against peritoneal exudate cell-mediated killing, we used a protocol similar to that described above, but the cultures were supplemented with 10% (vol/vol) exoproteins harvested from stationary-phase cultures of the S. aureus wild-type strain and the isogenic strain lacking fur grown in RPMI+CAS. Exoproteins were collected from normalized cultures (based on OD600) grown to stationary phase. Equal amounts (volume/volume) of culture supernatants were used in each experiment. The percentage of viable bacteria was calculated by normalizing the values for the samples to the number of input bacteria added to the cultures; the values for wild-type cultures and S. aureus cultures supplemented with exoproteins produced by the wild-type bacterium were defined as 100%.
RESULTS
Fur alters the production of staphylococcal exoproteins.
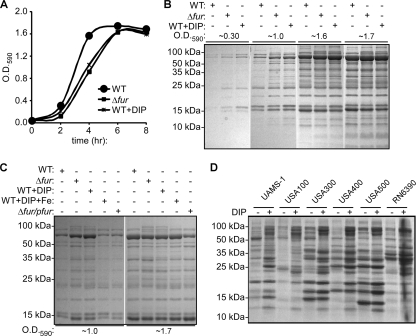
Iron availability is an environmental signal sensed by microorganisms that is used to regulate the expression of virulence factors (36, 51, 59). Recently, it was demonstrated that S. aureus senses iron limitation to alter the expression of iron uptake systems, as well as the expression of a series of virulence factors (2). S. aureus is known to monitor iron availability via Fur (25, 32, 41, 66, 71); however, the global contribution of iron and Fur to staphylococcal exoprotein production has not been evaluated. Therefore, we decided to investigate the effects of Fur-mediated iron sensing on the production of secreted staphylococcal virulence factors. To this end, we analyzed the exoprotein profiles of normalized samples collected at exponential, late-exponential, and stationary phases from S. aureus wild-type strain Newman and an isogenic strain lacking fur (Δfur strain) grown in iron-sufficient medium and iron-limited medium (medium supplemented with 300 to 400 μM 2,2′-dipyridyl [DIP], an iron chelator). As observed previously, iron-starved S. aureus and S. aureus lacking fur exhibited delayed growth in liquid culture (Fig. 1A) (32). After controlling for the growth differences by normalization using the optical density, we found that iron limitation (i.e., medium with DIP) and disruption of fur altered the abundance of exoproteins, particularly at early time points in the growth curve (Fig. 1B). Expression of fur in trans complemented the exoprotein phenotype exhibited by the fur mutant (Fig. 1C). Furthermore, the exoprotein phenotype exhibited by staphylococci grown in iron-limited medium (i.e., medium with DIP) was chemically complemented by supplementing the medium with excess iron (Fig. 1C). An altered exoprotein phenotype in response to iron limitation was also observed when we used both rich and defined media (i.e., tryptic soy broth [TSB] and Tris-minimal succinate [TMS] medium) and the iron chelator ethylene diamine-di(omega-hydroxyphenol acetic acid (EDDHA) (data not shown). These results demonstrated that S. aureus senses iron availability via Fur to regulate the production and/or secretion of exoproteins.
FIG. 1.
S. aureus senses iron availability via Fur to modulate the production of exoproteins. (A) Representative growth curves for the S. aureus wild-type (WT) and Δfur (Δfur) strains grown in iron-sufficient medium and for the S. aureus wild-type strain grown in iron-depleted medium (medium containing 400 μM 2,2′ dipyridyl) (WT+DIP). (B) Exoprotein profiles of the S. aureus wild-type (WT) and Δfur (Δfur) strains grown to early exponential phase (OD590, ∼0.3), mid-exponential phase (OD590, ∼1.0), early stationary phase (OD590, ∼1.6), and mid-stationary phase (OD590, ∼1.7) in iron-sufficient medium and exoprotein profiles of the S. aureus wild-type strain in iron-depleted medium (medium containing 400 μM 2,2′-dipyridyl) (WT+DIP). The results for culture supernatants were normalized based on optical density and CFU data, and exoproteins were precipitated with TCA, separated using SDS-PAGE, and stained with Coomassie blue. (C) Exoprotein profiles of the S. aureus wild-type strain (WT), the Δfur strain (Δfur), and the Δfur strain transformed with a fur complementation plasmid (Δfur/pfur) grown to mid-exponential phase (OD590, ∼1.0) and stationary phase (OD590, ∼1.7) in iron-sufficient medium, exoprotein profiles of the S. aureus wild-type strain grown in iron-depleted medium (medium containing 400 μM DIP) (WT+DIP), and exoprotein profiles of the S. aureus wild-type strain grown in medium supplemented with DIP and excess iron chloride (medium containing 400 μM DIP and 45 μM FeCl3) (WT+DIP+Fe). The results for supernatants were normalized and analyzed as described above for panel B. (D) Exoprotein profiles for different S. aureus strains grown to stationary phase in iron-sufficient medium and iron-limited medium (medium containing 300 μM 2,2′-dipyridyl) analyzed as described above for panel B.
To confirm that the effect of iron limitation on the production and/or secretion of staphylococcal exoproteins is not strain specific, we analyzed the effect of iron deprivation on the production of exoproteins using a panel of S. aureus strains. Iron limitation altered the exoprotein profiles of all strains tested, including S. aureus strains associated with osteomyelitis infections (UAMS-1) (28), lineages associated with hospital-acquired infections (USA100 and USA500), and strains associated with community-acquired infections (USA300 and USA400), as well as a commonly used laboratory strain (RN6390) (Fig. 1D). These results support the notion that the observed response to iron limitation is conserved across staphylococcal strains.
Fur senses iron limitation to enhance staphylococcal hemolytic and cytotoxic activities.
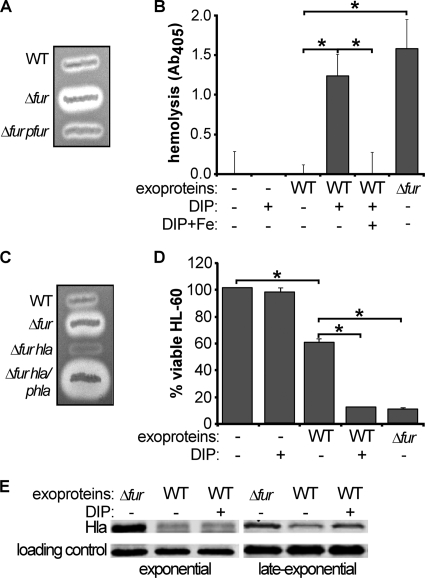
S. aureus secretes a large number of proteins into the extracellular milieu, including hemolysins and cytotoxins, which are pivotal for staphylococcal pathogenesis (7, 8, 21, 44, 46, 64). We found that S. aureus lacking fur exhibits increased hemolysis on blood agar plates compared to the wild type, a phenotype that was complemented by providing fur in trans (Fig. 2A). Liquid hemolysis assays revealed that iron limitation induces levels of hemolysis similar to those observed with S. aureus lacking fur (Fig. 2B). Supplementation of dipyridyl-treated S. aureus cultures with iron eliminated the increased hemolytic activity, confirming that the increase in hemolysis was due to dipyridyl-mediated iron chelation (Fig. 2B). Similar results were observed when the experiments were repeated with both rich and defined media (i.e., TMS medium and TSB) or when cultures were exposed to the iron chelator EDDHA (data not shown).
FIG. 2.
S. aureus senses iron availability via Fur to modulate hemolytic and cytotoxic activities. (A) Hemolytic activity exhibited by the S. aureus wild-type strain (WT), the S. aureus Δfur strain (Δfur), and the S. aureus Δfur strain harboring a fur complementation plasmid (Δfur/pfur) after 24 h of growth on sheep blood-TSA plates. (B) Hemolysis of erythrocytes intoxicated with exoproteins harvested from the wild-type and S. aureus Δfur strains grown in iron-sufficient medium, from the wild-type strain grown in iron-limited medium (medium containing 300 μM 2,2′-dipyridyl) (DIP), and from the wild-type strain grown in medium containing DIP and 10 μM iron chloride (DIP+Fe). The data are the means and standard deviations for triplicate determinations. Asterisks indicate statistically significant differences between the samples indicated, as determined by Student's t test (P < 0.05). (C) Hemolytic activity exhibited by the S. aureus wild-type strain (WT), an isogenic strain lacking fur (Δfur), a double-mutant isogenic strain lacking fur and hla (Δfur hla), and the fur hla double-mutant strain harboring an hla complementation plasmid (Δfur hla/phla) after 24 h of growth on sheep blood-TSA plates. (D) Viability of HL-60 mammalian cells intoxicated with staphylococcal exoproteins harvested from the wild-type strain (WT) grown in iron-sufficient and iron-limited medium (DIP) or with exoproteins harvested from S. aureus lacking fur (Δfur). Cell viability was measured using CellTiter (Promega). The data are expressed as percentages determined by comparing the number of viable cells with the number of cells grown with medium alone. The data are the means and standard deviations for triplicate determinations. Asterisks indicate statistically significant differences between the samples indicated as determined by Student's t test (P < 0.05). (E) Exoproteins secreted by the S. aureus wild-type strain (WT) and an isogenic strain lacking fur (Δfur) were collected at exponential phase and late exponential phase, and the data were normalized based on optical density. Exoproteins were precipitated with TCA, separated using SDS-PAGE, and transferred onto nitrocellulose membranes. The membranes were immunoblotted with anti-Hla antibodies (44). A nonspecific band that cross-reacted with the anti-Hla antisera was used as a loading control.
Disruption of hla, the gene that codes for alpha-toxin, eliminated the increased hemolytic activity exhibited by S. aureus lacking fur (Fig. 2C), suggesting that alpha-toxin was responsible for the observed increase in hemolysis. Consistent with this, expression of hla in trans in a fur- and hla-deficient background rescued the enhanced hemolytic activity exhibited by the fur mutant (Fig. 2C). These data suggest that S. aureus senses iron limitation via Fur to regulate the expression of alpha-toxin. To confirm this, we performed immunoblot analyses to monitor the abundance of alpha-toxin in culture supernatants of the wild-type strain and the isogenic fur mutant strain. This experiment revealed that alpha-toxin was more abundant in the culture supernatants of the fur mutant than in the culture supernatants of the wild-type strain at both the exponential and late-exponential phases of growth (Fig. 2E). Notably, the level of alpha-toxin production in iron-starved S. aureus was not as high as the level of production in the fur mutant during exponential growth, suggesting either that alpha-toxin is subjected to both iron-dependent and iron-independent regulation by Fur or that DIP-mediated iron starvation is less absolute than mutation of fur.
Next we evaluated the impact of iron availability and Fur on the cytolytic activity of S. aureus with nonerythroid cells. We found that iron limitation or fur inactivation increased the cytotoxic potential of S. aureus with HL-60 cells, which are human promyelocytic cells that differentiate into leukocyte-like cells upon stimulation with a variety of agents (14) (Fig. 2D). Similar results were obtained when human epithelial cells from lungs, kidneys, and livers were examined (data not shown). Taken together, the results suggest that expression of staphylococcal hemolysins and cytotoxins is affected by Fur.
Fur coordinates the expression of staphylococcal virulence factors.
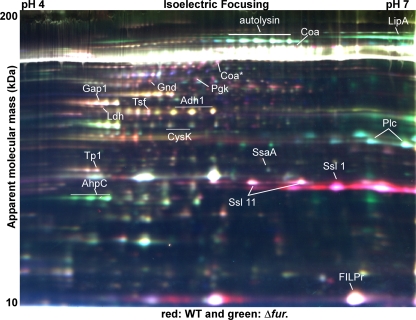
To determine the identities of secreted proteins whose expression is under Fur-mediated control, we employed two-dimensional difference in-gel electrophoresis (2D-DIGE) to compare the exoprotein profiles of the S. aureus wild-type strain and the isogenic strain lacking fur. To this end, three independent exoprotein samples were prepared for each strain and coresolved in pairs in three 2D-DIGE gels using previously described techniques (Fig. 3) (25). Because samples were coresolved on each 2D-DIGE gel with an internal standard comprised of equal aliquots of all six samples, we were able to normalize the expression values for each resolved protein feature across all samples with statistical confidence. This enabled us to visualize the variation between the six samples on a global level using principal component analysis (PCA), which reduces the variation in a data set to the two largest components (independent of the sample classification). When performed for all 1,065 features that were matched across the six samples, PCA clearly segregated the six samples based on genotype, indicating that there was a high level of reproducibility for the replicate samples (data not shown). The large number of proteins identified in this experiment was due to numerous staphylococcal exoproteins that exhibited multiple isoelectric focusing properties that resulted in three to six isoforms per protein (Fig. 3) (11, 25). 2D-DIGE analyses identified 386 distinct features that exhibited altered expression patterns in the wild-type and fur mutant strains with P values of ≤0.05 (data not shown). PCA performed for these 386 features produced results similar to the results for the unfiltered data set (data not shown), and a high level of reproducibility was maintained for the replicate samples in each group; no significant sample outliers were detected. The combination of 2D-DIGE and PCA allowed grouping of exoproteins into groups containing (i) proteins with expression patterns were not changed in the fur mutant, (ii) proteins that were less abundant in the fur mutant (Fur acted positively and thus was required for expression), and (iii) proteins that were more abundant in the fur mutant (Fur acted negatively and thus the absence of Fur led to increased production). Using these results, we identified 32 proteins by mass spectrometry-based protein identification (25) (Fig. 3 and Tables 1 and 2). These experiments revealed that disruption of fur results in decreased production of several immunomodulatory proteins, including the formyl peptide receptor-like 1 inhibitory protein (FLIPr), coagulase (Coa), and six superantigen-like exotoxins (Ssl 1, Ssl 2, Ssl 6, Ssl 7, Ssl 9, and Ssl 11) (Fig. 3) (21, 22, 47). On the other hand, disruption of fur resulted in increased production of several proteins that are thought to be involved in virulence (lipase [LipA], phospholipase C [Plc], alkyl hydroperoxide reductase [AhpC], iron surface determinants [IsdAB], and leukotoxin [LukED]) (13, 16, 29, 45, 50, 53, 66) (Tables 1 and 2).
FIG. 3.
2D-DIGE analysis of exoproteins secreted by the S. aureus wild-type strain and an isogenic strain lacking fur: false-color representative gel from a three-gel set containing differentially labeled samples. Secreted proteins from three independent cultures of the S. aureus wild-type strain (WT) and an isogenic fur mutant (Δfur strain) were collected and concentrated, and proteins were labeled with fluorescent dye as described in Materials and Methods. Samples were separated using a pI 4 to 7 gel. For the proteins indicated there were statistically significant differences in expression between the wild-type and Δfur strains, and the proteins were identified by a mass spectrometry-based protein identification method.
TABLE 1.
2D-DIGE data for pI 4 to 7
| Protein | Locusa | Mol wt (103)b | pIb | Combined MS and MS/MS search scoresc | No. of matched peptides (no. of unmatched peptides) | No. of peptides with MS/MS data | % of amino acids accounted for by matching peptides (coverage) | Avg wild-type/fur mutant volume ratio | Pd |
|---|---|---|---|---|---|---|---|---|---|
| Lipase (LipA) | NWMN_2569 | 76.7 | 77.0 | 7103 | 15 | 73 | 24 | −4.24 | 0.00038 |
| N-Acetylmuramoyl-l-alanine amidase | NWMN_2543 | 69 | 6.0 | 114 | 16 | 3 | 32 | −4.41 | 9.60E−04 |
| Coagulase (Coa) | NWMN_0166 | 71.6 | 8.4 | 131 | 13 | 5 | 34 | 1.88 | 3.10E−03 |
| Dihydrolipoamide dehydrogenase (PdhD) | NWMN_0962 | 49.5 | 4.95 | 68 | 10 | 1 | 22 | 2.47 | 0.0038 |
| 6-Phosphogluconate dehydrogenase | NWMN_1417 | 52 | 5.0 | 77 | 5 | 0 | 11 | 1.97 | 3.50E−03 |
| Phosphoglycerate kinase | NWMN_0742 | 42.6 | 5.2 | 79 | 7 | 3 | 23 | 3.54 | 2.10E−05 |
| Glyceraldehyde 3-phosphate dehydrogenase 1 (GapA) | NWMN_0741 | 36.4 | 4.9 | 245 | 14 | 8 | 39 | 4.08 | 1.30E−05 |
| l-Lactate dehydrogenase (Ldh) | NWMN_2499 | 34.5 | 4.8 | 62 | 4 | 3 | 13 | 5.60 | 1.90E−04 |
| Elongation factor TS | NWMN_1167 | 32.5 | 5.2 | 177 | 12 | 5 | 46 | 2.74 | 0.001 |
| Alcohol dehydrogenase I | NWMN_0577 | 36.4 | 5.5 | 52 | 5 | 3 | 18 | 4.53 | 7.30E−05 |
| Cysteine synthase | NWMN_0475 | 33 | 5.2 | 43 | 3 | 2 | 11 | 2.73 | 6.10E−04 |
| 1-Phosphatidylinositol phosphodiesterase (PI-PLCc) | NWMN_0041 | 35 | 6.5 | 78 | 10 (1) | 1 | 26 | −3.00 | 1.40E−03 |
| Triose phosphate isomerase | NWMN_0743 | 27.4 | 4.8 | 71 | 5 (1) | 3 | 23 | 2.25 | 9.70E−05 |
| Secretory antigen SsaA homolog | NWMN_0634 | 28.2 | 6.1 | 46 | 3 (2) | 1 | 14 | −1.71 | 0.00071 |
| Ssl 11 | NWMN_0400 | 25.3 | 8.5 | 135 | 13 | 6 | 66 | 4.45 | 2.20E−05 |
| Ssl 1e | NWMN_0388 | 25.6 | 8.5 | 146 | 15 (3) | 5 | 54 | 11.61 | 6.00E−05 |
| Ssl 2e | NWMN_0389 | 25 | 9.0 | 67 | 7 | 1 | 30 | 11.61 | 6.00E−05 |
| Alkyl hydroperoxide reductase subunit C (AphC) | NWMN_0372 | 21 | 4.9 | 90 | 5 | 3 | 40 | −3.06 | 4.40E−05 |
| Hypothetical protein | NWMN_0272 | 21.4 | 5.3 | 82 | 5 | 1 | 25 | 1.48 | 0.026 |
| FLIPr | NWMN_1067 | 15.2 | 9.1 | 256 | 10 (1) | 5 | 48 | 4.27 | 5.90E−06 |
Gene annotation in the NCBI database for S. aureus strain Newman.
The theoretical molecular weights and isoelectric points were calculated using the database entries, which often contain precursor sequences not present in the mature form migrating on the gel.
Combined MS and MS/MS search (MOWSE) scores greater than 79 are within the 95% confidence interval. Scores were calculated using the MASCOT v1.9 database search algorithms.
P values determined by Student's t test. Analysis of variance (ANOVA) P values were calculated using DeCyder software version 6.5 and the mixed-sample internal standard methodology.
The sample used was a mixed sample containing peptides for Ssl 1 and Ssl 2.
TABLE 2.
2D-DIGE data for pI 7 to 11
| Protein | Locusa | Mol wt (103)b | pIb | Combined MS and MS/MS search scoresc | No. of matched peptides (no. of unmatched peptides) | No. of peptides with MS/MS data | % of amino acids accounted for by matching peptides (coverage) | Avg wild-type/fur mutant volume ratiod |
|---|---|---|---|---|---|---|---|---|
| IsdB | NWMN_1040 | 72 | 9.06 | 60 | 4 | 1 | 8 | −10.62 |
| IsdAe | NWMN_1041 | 38.6 | 9.6 | 67 | 4 (1) | 2 | 15 | −3.50 |
| LukD | NWMN_1718 | 36.8 | 9.2 | 106 | 10 | 2 | 40 | −4.55 |
| LukE | NWMN_1719 | 34.8 | 9.5 | 205 | 13 | 4 | 41 | Downf |
| GlpQ | NWMN_0830 | 35.2 | 8.67 | 103 | 9 | 3 | 31 | 5.26 |
| Ssl 6 | NWMN_0393 | 26.6 | 9.2 | 88 | 6 | 2 | 25 | 6.40 |
| Ssl 9 | NWMN_0396 | 26.7 | 9.3 | 56 | 3 | 3 | 15 | 4.12 |
| Ssl 2 | NWMN_0389 | 26.4 | 9.1 | 204 | 14 | 6 | 61 | 6.62 |
| Ssl 7 | NWMN_0394 | 26.1 | 8.92 | 407 | 16 | 8 | 67 | 8.65 |
| Efb | NWMN_1069 | 18.8 | 9.85 | 154 | 9 | 3 | 36 | 8.80 |
Gene annotation in the NCBI database for S. aureus strain Newman.
The theoretical molecular weights and isoelectric points were calculated using the database entries, which often contain precursor sequences not present in the mature form migrating on the gel.
Combined MS and MS/MS search (MOWSE) scores greater than 79 are within the 95% confidence interval. Scores were calculated using the MASCOT v1.9 database search algorithms.
Average volume ratios for only one gel based on the high reproducibility of triplicate samples for the pI 4 to 7 gels.
Coverage for the middle of the protein and running at a lower molecular weight.
The protein was found to be upregulated in culture supernatants of S. aureus lacking fur.
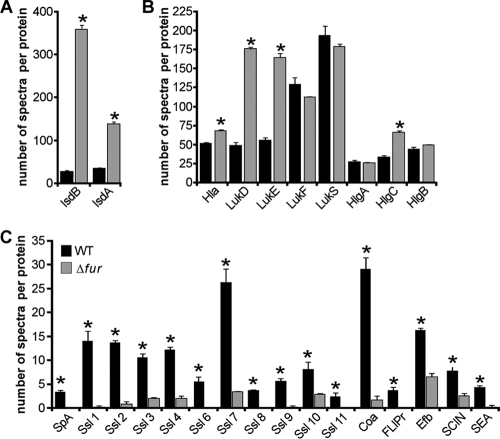
The number of proteins that can be identified using gel-based proteomic techniques is limited by the molecular weight and pH ranges of the gel and by the abundance of each protein in the proteome. In an effort to identify a larger subset of iron-regulated exoproteins, we employed shotgun proteomic analysis to compare the exoproteome profiles of the S. aureus wild-type and fur mutant strains. Differences in exoprotein abundance between samples were determined by label-free quantitation utilizing spectral counting of tandem spectra acquired for each protein from three independent samples (73). We identified 108 proteins in the culture supernatant of the wild-type S. aureus strain and 92 proteins in the culture supernatant of the S. aureus strain lacking fur (see Table S1 in the supplemental material). Consistent with previous reports (11, 74), we identified a series of cytoplasmic, membrane, and cell wall-associated proteins thought to be released into the extracellular milieu during autolysis. We also identified a group of proteins that have been predicted to be secreted but have no ascribed function (see Table S1 in the supplemental material). In addition, we identified a large number of staphylococcal exoproteins with known or predicted functions. These exoproteins could be divided into cytotoxins (e.g., Hla, HlgABC, LukED, and LukSF), hydrolases (e.g., nuclease, lipase, and Spl proteases), and a large number of known immunomodulatory proteins (e.g., staphylococcal enterotoxin A [SEA], chemotaxis-inhibiting protein [CHIP], staphylococcal complement inhibitor [SCIN], and Ssl 1 to Ssl 11). We identified 58 proteins that exhibited altered expression patterns in the wild-type and fur mutant strains with P values of ≤0.05 (see Table S1 in the supplemental material). As expected, S. aureus lacking fur overproduced IsdA, IsdB, and IsdH (Fig. 4A; see Table S1 in the supplemental material), which are proteins involved in iron acquisition and in protection against host defenses (13, 41, 50, 66, 68). We found that, among other proteins affected by Fur, α-hemolysin (Hla), γ-hemolysin (HlgC), and leukocidin ED (LukED) were upregulated in the isogenic fur strain (Fig. 4B). Furthermore, we found that Fur influences the expression of several immunomodulatory exoproteins and exoproteins known to protect staphylococci from host-mediated clearance, as demonstrated by the results for S. aureus lacking fur, which produced lower levels of protein A (SpA), staphylococcal immunoglobulin G-binding protein (Sbi), Ssl proteins (Ssl 1, Ssl 2, Ssl 3, Ssl 4, Ssl 6, Ssl 7, Ssl 8, Ssl 9, Ssl 10, and Ssl 11), Coa, FLIPr, SCIN, extracellular fibrinogen-binding protein (Efb), SEA, and CHIP (Fig. 4C; see Table S1 in the supplemental material) (21, 22, 34, 55, 67a). Taken together, these results suggest that S. aureus coordinates the production of iron acquisition systems together with the production of cytolysins and hemolysins in a Fur-dependent manner.
FIG. 4.
Global analysis of exoproteins secreted by the S. aureus wild-type strain (WT) and an isogenic strain lacking fur (Δfur). The differences in abundance of exoproteins produced by the S. aureus wild-type and isogenic strains lacking fur were determined by liquid chromatography-MS/MS and spectral counting. Exoprotein profiles were subdivided into profiles for iron acquisition (A), cytotoxins and hemolysins (B), and immunomodulatory proteins (C). The data are the means and standard errors of the means for three independent samples. The statistical significance of differences between the samples was analyzed by Student's t test (*, P < 0.05).
Fur alters the ability of S. aureus to avoid killing by peritoneal murine neutrophils.
Our proteomics data suggest that S. aureus lacking fur exhibits decreased expression of a number of immunomodulatory proteins thought to be involved in protection against killing by neutrophils (Fig. 4). Therefore, we sought to determine the role of iron sensing in the ability of S. aureus to avoid neutrophil-mediated clearance. We compared the abilities of wild-type and fur mutant strains to survive in a previously described opsonophagocytic killing assay using peritoneal murine neutrophils (15). These experiments revealed that disruption of fur decreases the ability of S. aureus to avoid neutrophil-dependent killing (Fig. 5A).
FIG. 5.
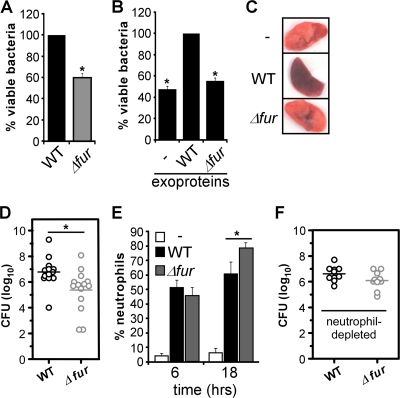
Inactivation of fur alters S. aureus virulence. (A) Peritoneal neutrophils were infected with the wild-type (WT) and Δfur strains, and the survival of S. aureus, expressed as a percentage, was determined by plating. The value for the wild-type strain was defined as 100%. The data are the means and standard errors of the means for at least three independent experiments. An asterisk indicates that the value is statistically significantly different from the value for the wild-type strain as determined by Student's t test (P < 0.05). (B) Primary murine peritoneal neutrophils were infected with washed wild-type S. aureus supplemented with medium (−) or with staphylococcal exoproteins from stationary cultures of the wild-type strain (WT) or the isogenic fur mutant strain (Δfur). The S. aureus burden was determined by plating, and the value for the sample supplemented with exoproteins produced by the wild-type strain was defined as 100%. The data are the means and standard errors of the means for at least three independent experiments. An asterisk indicates that the value is statistically significantly different from the value for the sample supplemented with exoproteins produced by the wild-type strain as determined by Student's t test (P < 0.05). (C to F) C57BL/6J mice were infected for 6 and 18 h via the intranasal (i.n.) route with the S. aureus wild-type strain (WT) and an isogenic strain lacking fur (Δfur). (C) Photographs of lungs dissected from uninfected animals (−) and from animals infected with the wild-type strain and the fur mutant for 18 h. (D) C57BL/6J mice were infected with the wild-type strain (3.78 × 108 CFU) or the isogenic strain lacking fur (3.51 × 108 CFU) as described above for panel C. Eighteen hours postinfection lungs were harvested, and the S. aureus burden was measured. Each circle represents one infected animal, and each horizontal line indicates the mean of the log values. The asterisk indicates that the data for the two strains are statistically significantly different as determined by Student's t test (P ≤ 0.05). (E) C57BL/6J animals were not infected (−) or were infected as described above, the lungs were dissected and homogenized, and the infiltration of neutrophils (B220− CD11b+ Ly6G+) was determined by multiparametric FACS analysis. The data are the means and standard errors of the means for three independent experiments in which at least two animals were used for each experiment. The asterisk indicates that the data for the two strains are statistically significantly different as determined by Student's t test (P < 0.05). (F) C57BL/6J mice were made neutropenic by treatment with IgG2b anti-Gr-1 MAb RB6-8C5 and infected with the wild-type strain (2.9 × 107 CFU) or the isogenic strain lacking fur (1.5 × 107 CFU) as described above for panel C. Eighteen hours postinfection lungs were harvested, and the S. aureus burden was measured. Each circle represents one infected animal, and each horizontal line indicates the mean of the log values. The observed differences were not statistically significant in this analysis (P ≥ 0.05).
To specifically test the role of iron-regulated exoproteins in S. aureus-neutrophil interactions, we evaluated the ability of exoproteins produced by a wild-type or fur mutant strain to protect S. aureus from neutrophil-mediated killing. We infected peritoneal murine neutrophils with wild-type S. aureus cells that had been extensively washed, resulting in removal of exoproteins that had accumulated in the supernatant. These bacteria were acutely sensitive to neutrophil-dependent killing (Fig. 5B). The effect of adding exoproteins harvested from stationary-phase cultures of the S. aureus wild-type or Δfur strain was then evaluated to determine the ability of these proteins to protect wild-type S. aureus in this assay. These experiments demonstrated that exogenous addition of exoproteins produced by the wild-type strain, but not exogenous addition of exoproteins produced by the fur mutant, protected S. aureus against killing by peritoneal murine neutrophils (Fig. 5B).
Fur is critical to the pathogenesis of staphylococcal pneumonia.
S. aureus strains lacking fur exhibit alterations in exoprotein production that correlate with increased susceptibility to neutrophils. This observation suggests that Fur may contribute to the host-pathogen interaction by modulating virulence factor expression in vivo. To investigate the contribution of Fur to S. aureus pathogenesis, we used a murine model of S. aureus pneumonia described by Bubeck Wardenburg et al. (8). We infected a cohort of C57BL/6J mice with the S. aureus wild-type and isogenic fur mutant strains via the intranasal route. Eighteen hours after infection animals were euthanized, the lungs were aseptically removed, and the bacterial burden was determined by plating preparations on solid medium. We observed that 41% more animals infected with wild-type S. aureus than with the fur mutant succumbed to infection (data not shown). Infection with wild-type S. aureus was associated with differences in the gross pathology of infected lungs (Fig. 5C), and the bacterial burden in animals infected with wild-type S. aureus was approximately 1.5-log greater than the bacterial burden in animals infected with the fur mutant (Fig. 5D).
The data described above suggest that S. aureus strains lacking fur are not able to protect themselves against the innate immune response of a host. To evaluate whether a host is able to respond equivalently to wild-type and Δfur strains, we performed multiparametric fluorescence-activated cell sorting (FACS) analyses to characterize the innate immune cell profile and the chemokine and cytokine production in lungs infected with wild-type S. aureus and in lungs infected with S. aureus lacking fur. S. aureus infection resulted in time-dependent infiltration of granulocytes composed primarily of neutrophils (Fig. 5E and data not shown). We observed no significant differences in the immune cell population or the percentage of recruited cells 6 h after infection between lungs infected with the wild-type strain and lungs infected with the fur mutant (Fig. 5E). In contrast, a comparison of the immune cell profiles for infected animals at 18 h after infection revealed an increase in the number of neutrophils in the lungs infected with the fur mutant strain despite the fact that the number of cells of this strain was less than the number of cells of the wild-type strain (Fig. 5E). We observed no significant differences in the concentrations of a subset of cytokines and chemokines involved in inflammation and neutrophil recruitment (interleukin-6 [IL-6], IL-10, IL-12p70, KC, MIP-1α, MIP-1β, monocyte chemoattractant protein 1 [MCP-1], and RANTES) in the lungs of animals infected with the wild-type and fur mutant strains (data not shown). Together, these results reveal that Fur is required for full virulence in a mouse model of pneumonia and support a model in which S. aureus expressing Fur exhibits a secreted protein response that is important for the ability of S. aureus to avoid host-mediated clearance.
The virulence defect of the Δfur strain is partially dependent on the presence of neutrophils.
Based on the requirement for neutrophils for protection of murine lungs against staphylococcal challenge (58), the observed increase in the total number of neutrophils may partially explain the reduction in the bacterial burden in animals infected with the fur mutant (Fig. 5D and E). To test this hypothesis, we compared the virulence of the wild-type strain and the virulence of the Δfur strain in a murine model of pneumonia using mice that had been depleted of neutrophils. These experiments revealed that neutrophils contribute to the virulence defect of the Δfur strain in wild-type mice. Specifically, we observed an approximately 0.5-log difference in virulence between the Δfur and wild-type strains in neutropenic mice, which was not statistically significant. This is in contrast to the statistically significant 1.5-log decrease for similar infections in wild-type animals (Fig. 5D and F). The results presented here are consistent with the demonstrated role of Fur-regulated exoproteins in protecting S. aureus from neutrophil-mediated killing (Fig. 5A and B). Taking all of the data into consideration, it is likely that the attenuation of the Δfur strain in a murine pneumonia model is the result of a combination of the decreased fitness and dysregulated exoprotein production of this strain.
DISCUSSION
The importance of S. aureus as a threat to human health is highlighted by the recent increase in infection of otherwise healthy individuals (community-associated infections) and by the emergence of antibiotic-resistant strains. The success of S. aureus as a pathogen is due in part to the expression of an arsenal of virulence factors (21, 47). At this point it is not well understood how S. aureus coordinates gene expression during infection or what host signals are sensed by S. aureus to regulate the expression of virulence factors. Here we show that S. aureus senses iron availability through Fur to modulate the expression of a variety of virulence factors. Specifically, we show that Fur affects the reciprocal expression of secreted cytolysins and a subset of immunomodulatory factors, which supports the hypothesis that iron-starved S. aureus expresses cytolysins, whereas staphylococci with sufficient iron secrete numerous immunomodulatory factors. The pathophysiological relevance of this coordinated expression program is highlighted by the observation that S. aureus lacking Fur is less virulent in a murine model of pneumonia and by the fact that the differential expression of staphylococcal virulence factors has also been observed in vivo (2). Despite extensive studies of other organisms, this is the first report of the global impact of Fur on staphylococcal exoprotein production. In addition, this is the first demonstration that Fur is required for pathogenesis in a murine model of staphylococcal pneumonia and the first time that the immune response to a staphylococcal Δfur mutant has been studied.
We have performed extensive bioinformatic analyses in a search for putative staphylococcal Fur boxes in the S. aureus genomes available (32, 72). Similar to the results of previous studies (1, 32, 71), we have identified Fur boxes in known Fur-regulated genes (e.g., isd, fhu, hts, and sbn promoters), but we were unable to identify Fur boxes in the intergenic regions of genes encoding transcription factors or other Fur-regulated exoproteins (hemolysins, cytotoxins, and immunomodulatory proteins). These results suggest that staphylococcal Fur regulates protein abundance using both direct and indirect mechanisms. In keeping with this, we recently reported that Fur influences the levels of a large number of cytoplasmic proteins, including several proteins that are involved in the regulation of staphylococcal virulence factors (e.g., RsbU and CodY) (25, 37). In addition, several groups have shown that in other bacterial genera, Fur regulates the expression of small regulatory RNAs to alter gene expression (27, 40, 43, 70). S. aureus is known to express several small and stable RNAs (3, 52, 57), and whether these regulatory RNAs are controlled by Fur and/or play a role in the phenotypes described in this study is a question now being investigated in our laboratories. Based on the data presented in this study, we propose that Fur is an important regulatory protein that potentially interfaces with other regulatory systems to coordinate the expression of staphylococcal virulence factors during infection.
We demonstrated in this study that S. aureus Fur coordinates the production of iron acquisition systems and hemolysins involved in the release of intracellular iron from the host. Increased production of hemolysins in response to iron limitation has been reported for several Gram-negative pathogens, including Yersinia ruckeri (20), Vibrio cholerae (63), Serratia marcescens (54), Vibrio parahaemolyticus (18), Helicobacter pylori (38), and Aggregatibacter actinomycetemcomitans (4), among others. Taken together, these studies support a model in which the release of Fur-mediated repression increases the secretion of hemolysins that lyse erythrocytes, resulting in liberation of hemoglobin at the site of infection. In the case of S. aureus, this likely increases the efficiency of hemoglobin capture by the Isd system, facilitating nutrient iron acquisition during pathogenesis (41, 53, 66). Hence, Fur affects the expression of toxins that are exquisitely suited to increase the amount of available nutrient iron in the host.
Our data also demonstrate that Fur influences the expression of proteins that modulate S. aureus-host interactions, tilting the balance in favor of the host's efforts to combat infection. It seems that under iron starvation conditions S. aureus devotes considerable energy to producing the machinery required for the acquisition of iron. This comes at the expense of decreased production of factors involved in escaping from cells of the immune system, which in turn results in increased susceptibility to immune cell clearance. We envision that during infection of vertebrates S. aureus transitions from iron-sufficient environments to iron-limited environments and thus undergoes an adaptive response. It is conceivable that S. aureus is able to efficiently acquire iron in specific anatomic sites and therefore, when colonizing these sites, focuses its energy on the production of factors that prevent immune cell chemotaxis to the site of infection. However, when S. aureus becomes iron starved, it must acquire iron in order to survive, and thus Fur coordinates a gene expression profile that leads to significant cell lysis and iron uptake. The fact that strains lacking fur are more sensitive to neutrophil-mediated killing in the murine lung suggests that wild-type S. aureus may not be iron starved in this pneumonia model. This hypothesis is supported by the lack of a role for iron uptake systems in staphylococcal pathogenesis using this animal model (39). Further understanding of how S. aureus senses host environments to coordinate the expression of virulence factors and the mechanism employed by S. aureus to alter host immune responses may uncover novel targets for the development of new treatments for use against this important pathogen.
Supplementary Material
Acknowledgments
We thank members of the Skaar lab for critical reading of the manuscript, Corbin W. Whitwell for help with the 2D-DIGE experiments, and Amanda McCoy for assistance during experiments. We thank Douglas Kernodle for providing an hla-deficient staphylococcal strain, the hla complementation vector, and the alpha-toxin antisera. We also thank Svetlana Gerdes of Argonne National Laboratory for performing the genomic searches for Fur boxes.
This work was supported by Vanderbilt University Medical Center development funds and by United States Public Health Service grants AI69233 and AI073843 from the National Institute of Allergy and Infectious Diseases (to E.P.S.). E.P.S. was also supported by NIH grant U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense and received an Investigator in Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. V.J.T. and B.D.C. were supported by Ruth L. Kirschstein NRSA postdoctoral fellowships from the National Institute of Allergy and Infectious Diseases (grants AI071487 and AI074278, respectively). W.J.M. was supported by grant T32 AI-07474-13. D.L.S. was supported by grant T32 HL069765 from the National Institute of Allergy and Infectious Diseases. M.I.H. was supported by Public Health Service award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program. D.E.H. was supported by a grant from the Canadian Institutes of Health Research. P.M.D. was supported by University of Nebraska Medical Center development funds, by American Heart Association award 0535037N, and by National Institute of Allergy and Infectious Diseases award AI073780.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 25 January 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alkema, W. B., B. Lenhard, and W. W. Wasserman. 2004. Regulog analysis: detection of conserved regulatory networks across bacteria: application to Staphylococcus aureus. Genome Res. 14:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard, M., H. Moisan, E. Brouillette, A. L. Gervais, M. Jacques, P. Lacasse, M. S. Diarra, and F. Malouin. 2006. Transcriptional modulation of some Staphylococcus aureus iron-regulated genes during growth in vitro and in a tissue cage model in vivo. Microbes Infect. 8:1679-1690. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, K. L., C. Roberts, T. Disz, V. Vonstein, K. Hwang, R. Overbeek, P. D. Olson, S. J. Projan, and P. M. Dunman. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balashova, N. V., R. Diaz, S. V. Balashov, J. A. Crosby, and S. C. Kachlany. 2006. Regulation of Aggregatibacter (Actinobacillus) actinomycetemcomitans leukotoxin secretion by iron. J. Bacteriol. 188:8658-8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beasley, F. C., E. D. Vines, J. C. Grigg, Q. Zheng, S. Liu, G. A. Lajoie, M. E. Murphy, and D. E. Heinrichs. 2009. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Mol. Microbiol. 72:947-963. [DOI] [PubMed] [Google Scholar]
- 6.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bubeck Wardenburg, J., T. Bae, M. Otto, F. R. Deleo, and O. Schneewind. 2007. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13:1405-1406. [DOI] [PubMed] [Google Scholar]
- 8.Bubeck Wardenburg, J., R. J. Patel, and O. Schneewind. 2007. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect. Immun. 75:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bubeck Wardenburg, J., W. A. Williams, and D. Missiakas. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 103:13831-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullen, J. J., and E. Griffiths. 1999. Iron and infection: molecular, physiological and clinical aspects. John Wiley and Sons, New York, NY.
- 11.Burlak, C., C. H. Hammer, M. A. Robinson, A. R. Whitney, M. J. McGavin, B. N. Kreiswirth, and F. R. Deleo. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell. Microbiol. 9:1172-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, S. R., R. Mohamed, L. Bian, A. F. Routh, J. F. Kokai-Kun, J. J. Mond, A. Tarkowski, and S. J. Foster. 2007. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1:199-212. [DOI] [PubMed] [Google Scholar]
- 14.Collins, S. J., R. C. Gallo, and R. E. Gallagher. 1977. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature 270:347-349. [DOI] [PubMed] [Google Scholar]
- 15.Corbin, B. D., E. H. Seeley, A. Raab, J. Feldmann, M. R. Miller, V. J. Torres, K. L. Anderson, B. M. Dattilo, P. M. Dunman, R. Gerads, R. M. Caprioli, W. Nacken, W. J. Chazin, and E. P. Skaar. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962-965. [DOI] [PubMed] [Google Scholar]
- 16.Cosgrove, K., G. Coutts, I. M. Jonsson, A. Tarkowski, J. F. Kokai-Kun, J. J. Mond, and S. J. Foster. 2007. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courcol, R. J., D. Trivier, M. C. Bissinger, G. R. Martin, and M. R. Brown. 1997. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immun. 65:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai, J. H., Y. S. Lee, and H. C. Wong. 1992. Effects of iron limitation on production of a siderophore, outer membrane proteins, and hemolysin and on hydrophobicity, cell adherence, and lethality for mice of Vibrio parahaemolyticus. Infect. Immun. 60:2952-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez, L., M. Prieto, and J. A. Guijarro. 2007. The iron- and temperature-regulated haemolysin YhlA is a virulence factor of Yersinia ruckeri. Microbiology 153:483-489. [DOI] [PubMed] [Google Scholar]
- 21.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 22.Fraser, J. D., and T. Proft. 2008. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225:226-243. [DOI] [PubMed] [Google Scholar]
- 23.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 24.Friedman, D. B. 2006. Quantitative proteomics for two-dimensional gels using difference gel electrophoresis (DIGE), p. 219-239. In R. Matthiesen (ed.), Mass spectrometry data analysis in proteomics. The Humana Press, Totowa, NJ. [DOI] [PubMed]
- 25.Friedman, D. B., D. L. Stauff, G. Pishchany, C. W. Whitwell, V. J. Torres, and E. P. Skaar. 2006. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs, S., J. Pane-Farre, C. Kohler, M. Hecker, and S. Engelmann. 2007. Anaerobic gene expression in Staphylococcus aureus. J. Bacteriol. 189:4275-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaballa, A., H. Antelmann, C. Aguilar, S. K. Khakh, K. B. Song, G. T. Smaldone, and J. D. Helmann. 2008. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U. S. A. 105:11927-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravet, A., D. A. Colin, D. Keller, R. Girardot, H. Monteil, and G. Prevost. 1998. Characterization of a novel structural member, LukE-LukD, of the bi-component staphylococcal leucotoxins family. FEBS Lett. 436:202-208. [DOI] [PubMed] [Google Scholar]
- 30.Ham, A. J., R. M. Caprioli, and M. L. Gross. 2005. Proteolytic digestion protocols, p. 10-17. In The encyclopedia of mass spectrometry, vol. 2. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 31.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 32.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, M., A. Cockayne, P. H. Williams, and J. A. Morrissey. 2005. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J. Bacteriol. 187:8211-8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jongerius, I., J. Kohl, M. K. Pandey, M. Ruyken, K. P. van Kessel, J. A. van Strijp, and S. H. Rooijakkers. 2007. Staphylococcal complement evasion by various convertase-blocking molecules. J. Exp. Med. 204:2461-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, J. W., and J. D. Helmann. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485-499. [DOI] [PubMed] [Google Scholar]
- 36.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martino, M. C., R. A. Stabler, Z. W. Zhang, M. J. Farthing, B. W. Wren, and N. Dorrell. 2001. Helicobacter pylori pore-forming cytolysin orthologue TlyA possesses in vitro hemolytic activity and has a role in colonization of the gastric mucosa. Infect. Immun. 69:1697-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason, W. J., and E. P. Skaar. 2009. Assessing the contribution of heme-iron acquisition to Staphylococcus aureus pneumonia using computed tomography. PLoS One 4:e6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 42.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellin, J. R., S. Goswami, S. Grogan, B. Tjaden, and C. A. Genco. 2007. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189:3686-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menzies, B. E., and D. S. Kernodle. 1996. Passive immunization with antiserum to a nontoxic alpha-toxin mutant from Staphylococcus aureus is protective in a murine model. Infect. Immun. 64:1839-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morinaga, N., Y. Kaihou, and M. Noda. 2003. Purification, cloning and characterization of variant LukE-LukD with strong leukocidal activity of staphylococcal bi-component leukotoxin family. Microbiol. Immunol. 47:81-90. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson, I. M., O. Hartford, T. Foster, and A. Tarkowski. 1999. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect. Immun. 67:1045-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nizet, V. 2007. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 120:13-22. [DOI] [PubMed] [Google Scholar]
- 48.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 49.O'Callaghan, R. J., M. C. Callegan, J. M. Moreau, L. C. Green, T. J. Foster, O. M. Hartford, L. S. Engel, and J. M. Hill. 1997. Specific roles of alpha-toxin and beta-toxin during Staphylococcus aureus corneal infection. Infect. Immun. 65:1571-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palazzolo-Ballance, A. M., M. L. Reniere, K. R. Braughton, D. E. Sturdevant, M. Otto, B. N. Kreiswirth, E. P. Skaar, and F. R. DeLeo. 2008. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180:500-509. [DOI] [PubMed] [Google Scholar]
- 51.Pappenheimer, A. M., and S. J. Johnson. 1936. Studies in diphtheria toxin production. I. The effect of iron and copper. Br. J. Exp. Pathol. 17:335-341. [Google Scholar]
- 52.Pichon, C., and B. Felden. 2005. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U. S. A. 102:14249-14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pishchany, G., S. E. Dickey, and E. P. Skaar. 2009. Subcellular localization of the Staphylococcus aureus heme-iron transport components IsdA and IsdB. Infect. Immun. 77:2624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poole, K., and V. Braun. 1988. Iron regulation of Serratia marcescens hemolysin gene expression. Infect. Immun. 56:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prat, C., J. Bestebroer, C. J. de Haas, J. A. van Strijp, and K. P. van Kessel. 2006. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J. Immunol. 177:8017-8026. [DOI] [PubMed] [Google Scholar]
- 56.Richardson, A. R., P. M. Dunman, and F. C. Fang. 2006. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61:927-939. [DOI] [PubMed] [Google Scholar]
- 57.Roberts, C., K. L. Anderson, E. Murphy, S. J. Projan, W. Mounts, B. Hurlburt, M. Smeltzer, R. Overbeek, T. Disz, and P. M. Dunman. 2006. Characterizing the effect of the Staphylococcus aureus virulence factor regulator, SarA, on log-phase mRNA half-lives. J. Bacteriol. 188:2593-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson, C. M., E. E. Perrone, K. W. McConnell, W. M. Dunne, B. Boody, T. Brahmbhatt, M. J. Diacovo, N. Van Rooijen, L. A. Hogue, C. L. Cannon, T. G. Buchman, R. S. Hotchkiss, and C. M. Coopersmith. 2008. Neutrophil depletion causes a fatal defect in murine pulmonary Staphylococcus aureus clearance. J. Surg. Res. 150:278-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaible, U. E., and S. H. Kaufmann. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946-953. [DOI] [PubMed] [Google Scholar]
- 60.Skaar, E. P., A. H. Gaspar, and O. Schneewind. 2004. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279:436-443. [DOI] [PubMed] [Google Scholar]
- 61.Skaar, E. P., M. Humayun, T. Bae, K. L. DeBord, and O. Schneewind. 2004. Iron-source preference of Staphylococcus aureus infections. Science 305:1626-1628. [DOI] [PubMed] [Google Scholar]
- 62.Skaar, E. P., and O. Schneewind. 2004. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 6:390-397. [DOI] [PubMed] [Google Scholar]
- 63.Stoebner, J. A., and S. M. Payne. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Supersac, G., Y. Piemont, M. Kubina, G. Prevost, and T. J. Foster. 1998. Assessment of the role of gamma-toxin in experimental endophthalmitis using a hlg-deficient mutant of Staphylococcus aureus. Microb. Pathog. 24:241-251. [DOI] [PubMed] [Google Scholar]
- 65.Tabb, D. L., C. G. Fernando, and M. C. Chambers. 2007. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 6:654-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torres, V. J., G. Pishchany, M. Humayun, O. Schneewind, and E. P. Skaar. 2006. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 188:8421-8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Torres, V. J., D. L. Stauff, G. Pishchany, J. S. Bezbradica, L. E. Gordy, J. Iturregui, K. L. Anderson, P. M. Dunman, S. Joyce, and E. P. Skaar. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67a.van Wamel, W. J., S. H. Rooijakkers, M. Ruyken, K. P. van Kessel, and J. A. van Strijp. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 188:1310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Visai, L., N. Yanagisawa, E. Josefsson, A. Tarkowski, I. Pezzali, S. H. Rooijakkers, T. J. Foster, and P. Speziale. 2009. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology 155:667-679. [DOI] [PubMed] [Google Scholar]
- 69.Weinrick, B., P. M. Dunman, F. McAleese, E. Murphy, S. J. Projan, Y. Fang, and R. P. Novick. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterization of the ferric-uptake regulator, fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]
- 72.Xiong, Y. Q., J. Willard, M. R. Yeaman, A. L. Cheung, and A. S. Bayer. 2006. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 194:1267-1275. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, B., M. C. Chambers, and D. L. Tabb. 2007. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res. 6:3549-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.