Abstract
Mycoplasmas are minimal bacteria whose genomes barely exceed the smallest amount of information required to sustain autonomous life. Despite this apparent simplicity, several mycoplasmas are successful pathogens of humans and animals, in which they establish intimate interactions with epithelial cells at mucosal surfaces. To identify biological functions mediating mycoplasma interactions with mammalian cells, we produced a library of transposon knockout mutants in the ruminant pathogen Mycoplasma agalactiae and used this library to identify mutants displaying a growth-deficient pheonotype in cell culture. M. agalactiae mutants displaying a 3-fold reduction in CFU titers to nearly complete extinction in coculture with HeLa cells were identified. Mapping of transposon insertion sites revealed 18 genomic regions putatively involved in the interaction of M. agalactiae with HeLa cells. Several of these regions encode proteins with features of membrane lipoproteins and/or were involved in horizontal gene transfer with phylogenetically distant pathogenic mycoplasmas of ruminants. Two mutants with the most extreme phenotype carry a transposon in a genomic region designated the NIF locus which encodes homologues of SufS and SufU, two proteins presumably involved in [Fe-S] cluster biosynthesis in Gram-positive bacteria. Complementation studies confirmed the conditional essentiality of the NIF locus, which was found to be critical for proliferation in the presence of HeLa cells and several other mammalian cell lines but dispensable for axenic growth. While our results raised questions regarding essential functions in mycoplasmas, they also provide a means for studying the role of mycoplasmas as minimal pathogens.
Often portrayed as minimal bacteria, mycoplasmas have evolved from low-G+C content Gram-positive ancestors by massive losses of genetic material and extensive genome downsizing (37, 44). As a consequence of this reductive evolution, mycoplasmas are lacking a significant number of biological functions found in more complex bacteria and have increased their dependence on the host for many nutrients. The absence of a cell wall, small size, fastidious growth in cell-free environments, and complex requirements for nutrients are among the most emblematic features of these particular organisms (37). Their minute genomes, which for some species are close to the minimal requirements for sustaining autonomous life, are used as experimental platforms to explore the concept of a minimal cell and as a model system for the design of synthetic bacterial genomes (17, 18, 25, 35). While significant progress has been made in understanding the biology of these minimal organisms under laboratory conditions, little is known regarding mycoplasma factors involved in virulence and host interaction. Recent genomic studies indicated that several mycoplasma species have retained sexual competence, a trait that may provide some pathogenic species with a high potential for adaptation (44, 45).
Mycoplasmas are widely distributed in nature, and several species are successful pathogens, capable of establishing persistent infections and causing debilitating diseases in humans and a wide range of animal species (37). Mycoplasmas are also recurrently found associated with cultures of mammalian cells, where they can survive for long periods, often without apparent signs of contamination but with potential consequences for the reliability of experimental results and the safety of biological products. Classified by the World Organization for Animal Health (OIE) as notifiable diseases, a number of mycoplasma infections in domestic animals can have a significant impact on livestock production (16). Among those, the ruminant pathogen Mycoplasma agalactiae is the main etiological agent of contagious agalactia in sheep and goats, a syndrome that is characterized by mastitis, arthritis, keratoconjunctivitis, and pneumonia (3). Although phylogenetically distant from M. agalactiae, several members of the so-called mycoides cluster, such as Mycoplasma mycoides subsp. mycoides large colony (MmmLC) and Mycoplasma capricolum subsp. capricolum (Mcc), are also able to induce similar symptoms in the same ruminant species. Remarkably, whole-genome sequence analysis has revealed that extensive horizontal gene transfer (HGT) events, affecting up to 18% of the M. agalactiae genome, occurred between M. agalactiae and members of the mycoides cluster, illustrating the unexpected plasticity and adaptability of the mycoplasma genome (44, 45).
Recent advances in whole-genome sequencing have greatly facilitated the study of mycoplasmas (2, 33, 34, 44, 45). Unfortunately, these data alone have been of little help in identifying the mechanism underlying diseases caused by mycoplasmas. The main reason is that the predicted mycoplasma gene products, other than those involved in housekeeping functions, display little or no homology to those known for classical bacteria (45). Among the few exceptions is the ADP-ribosylating cytotoxin found in the human respiratory pathogen Mycoplasma pneumoniae that displays some similarity with the pertussis toxin (27).
Factors that may contribute to the pathogenic process in mycoplasmal infections include the capacity to adhere and invade host cells, the production of immunomodulatory molecules, and a highly variable antigenic structure, as well as the formation of biofilm and the release of metabolic hydrogen peroxide (5, 10, 22, 29, 41). Gene products presumably involved in M. agalactiae-host interaction include the P40 adhesion protein (13), a family of phase-variable surface proteins, designated Vpma, which are encoded by a locus subjected to high-frequency DNA rearrangements (19, 20), and the immunomodulatory P48 protein (40).
Transposon mutagenesis has been used extensively as a tool for the identification of virulence genes in pathogenic bacteria. Similar approaches have been developed with a few mycoplasma species, mainly with the aim of defining the minimal set of essential genes required to sustain autonomous life under axenic conditions (12, 15, 18, 25). However, the genetic information necessary to develop interactions of mycoplasma with its animal host is likely to differ from the minimal set of essential genes required for laboratory growth. New opportunities to investigate factors involved in mycoplasma-host interaction have emerged through the development of genomic tools that facilitate the manipulation of animal mycoplasmas including M. agalactiae (8, 9). However, in vivo screening of mutant libraries of ruminant mycoplasmas involves difficulties inherent in experiments with infections in large animals.
The present study combines large-scale transposon mutagenesis of the pathogen M. agalactiae and an appropriate model of bacteria-HeLa cell interactions in coculture. Genomic regions of M. agalactiae specifically required for survival under cell culture conditions, while dispensable for axenic growth, were thus identified, indicating that mycoplasma cocultivation with mammalian cells represents an original and efficient system for high-throughput screening of large mutant libraries.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. agalactiae reference strain PG2 (45) was cultured at 37°C in Aluotto or SP4 medium (46) supplemented with 500 μg/ml cefalexin (Virbac). Gentamicin (50 μg/ml) was added to the medium for the propagation of M. agalactiae mutants generated by transposon mutagenesis. Mycoplasma cultures were stored at −80°C as 10-μl aliquots. CFU titers were determined by serial dilutions in Dulbecco's phosphate-buffered saline (Invitrogen) supplemented with 1% heat-inactivated horse serum (Invitrogen). Dilutions were spotted (10 μl) onto solid Aluotto or SP4 medium, and mycoplasma colonies were counted after 2 to 5 days incubation at 37°C. E. coli DH10B [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara leu)7697 galU galK rpsL endA1 nupG] (Invitrogen) was used for DNA cloning and plasmid propagation. E. coli bacteria were grown in Luria broth supplemented with ampicillin (50 μg/ml) and/or tetracycline (5 μg/ml) when required. Sheep and horse erythrocytes were derived from defibrinated blood (bioMérieux).
Plasmids and DNA constructions.
Plasmid pMT85, which contains a modified version of transposon Tn4001 (mini-Tn), was kindly provided by Richard Herrmann (48). The gentamicin resistance marker encoded by the aacA-aphD gene is located between the two inverted repeats (IRs) that define the extremities of the transposed fragment, while the transposase gene (tnpA) is located outside the transposable elements to prevent reexcision events. Plasmid p20-1miniO/T was used as a shuttle vector for complementation studies of M. agalactiae. Plasmid p20-1miniO/T was derived from pMM20-1 (8) by partial deletion of both the tetM region and the 6.9-kb DNA fragment carrying the M. agalactiae origin of replication. For complementation of M. agalactiae mutants T07.082 (clone 82 isolated from transformation T07) and T07.134, the genomic region designated the NIF locus (MAG0720 and MAG0730) was cloned downstream of the P40 gene promoter region. These two regions were assembled by PCR amplification using an overlapping primer. The promoter region was amplified first by using oligonucleotide primers p40RF-CC (5′-ACGGGGCTAAAGAAGCTGAT-3′) and P40-nifS-R (5′-GATCTAATCGATTTAGGCATAATTATTTATATCCTTTTC-3′) to generate a 200-bp DNA fragment overlapping coding sequence (CDS) MAG0720 at the ATG codon. The NIF locus was then amplified by using the overlapping DNA fragment and the primer 88595_R (5′-CTGTGCGCGCTTACAAAGTA-3′). The resulting PCR product was cloned into pGEM-T Easy (Promega) before subcloning at the NotI site of the p20-1miniO/T plasmid to generate pO/T-NIF. Cloned sequences were verified by DNA sequencing. PCRs were performed using an Expand High Fidelity PCR System (Roche).
Cell lines.
Human epithelial HeLa cells (ATCC CCL2; cervical carcinoma) were kindly provided by P. Mason (University of Texas Medical Branch, Galveston, TX). The bovine turbinate cells (BT; ATCC CRL-1390) were purchased at the ATCC. The bovine cell line KOP (esophageal tissue of a calf) was obtained from the Friedrich Loeffler Institute (Greifswald-Insel Riems, Germany). The caprine cell lines including simian virus 40 (SV40) large T-antigen immortalized goat embryo fibroblasts (TIGEF) (11) and similarly immortalized milk epithelial cells (TIGMEC) (31) were kindly provided by C. Leroux (INRA, Lyon, France). Cells were grown in Dulbecco's modified Eagle's medium (DMEM)-based medium. This medium is composed of DMEM (high glucose, sodium pyruvate, and GlutaMAX-I; Invitrogen) supplemented with nonessential amino acids (NEAA; Invitrogen) and 10% heat-inactivated fetal calf serum (FCS; Eurobio). Cells were incubated at 37°C in an atmosphere with 5% CO2 and subcultured every 2 to 3 days by seeding one-third to one-sixth the number of cells reached at confluence.
Cocultivation of M. agalactiae with mammalian cells.
Cocultivations were carried out in DMEM-based medium, supplemented with gentamicin (50 μg/ml), for M. agalactiae mutants. Since Aluotto broth (up to 0.1%) has no apparent toxic effect on mammalian cells (data not shown), mycoplasma inocula were prepared by direct dilution of frozen mycoplasma cultures in DMEM-based medium. Mammalian cells were prepared by trypsin-EDTA treatment of nearly confluent monolayers. After low-speed centrifugation, cells were resuspended in DMEM-based medium, seeded in 24-well plates (Falcon) at a density of 2 × 104 cells/cm2, and inoculated with M. agalactiae at different multiplicities of infection (MOI). Mycoplasma and mammalian cells were then allowed to grow at 37°C under 5% CO2. At different times postinoculation, mycoplasma titers were determined by CFU titrations following one freeze-thaw (−80°C/+37°C) cycle to disrupt mammalian cells.
Transformation of M. agalactiae with plasmid DNA.
Transformation of mycoplasma cells (108 to 109 CFU) was performed by electroporation using 1 to 3 μg of plasmid DNA, as described previously (9). Following electroporation, mycoplasma cells were incubated in nonselective SP4 medium for 3 h at 37°C. Cells were then allowed to grow in the presence of the appropriate antibiotic for an additional period of 3 to 12 h before they were plated on selective SP4 agar. Gentamicin and tetracycline were used at a concentration of 50 μg/ml and 2 μg/ml, respectively. Isolated colonies were picked after 4 to 7 days, and transformants were subcultured in 1 ml of selective SP4 medium.
Transposon mutagenesis in M. agalactiae.
Transposon mutagenesis in M. agalactiae was carried out using plasmid pMT85. Colonies were collected from independent transformations and subcultured in 1 ml of selective SP4 medium. Cultures of individual mutants were distributed in 96-well plates, and the pMT85-based library was stored at −80°C. Transposon insertion sites in the M. agalactiae chromosome were mapped by sequencing the junction between M. agalactiae genomic DNA and the 3′ end of the transposon, using the orientation of the gentamicin resistance gene as a reference. Genomic DNA (5 μg) was sequenced using BigDye terminator chemistry and oligonucleotide SG8 (5′-GAGTCAGTGAGCGAGGAAGC-3′) as a primer. Direct sequencing of genomic DNA was performed at the sequencing facility of the Bio-Medical Research Federative Institute of Toulouse (Toulouse, France).
Identification of growth-deficient mutants under coculture conditions.
A 96-pin replicator (Boekel Scientific) was used for high-throughput screening of the library and the identification of mutants displaying reduced growth capacity under cell culture conditions. Cocultivation of individual mutants with HeLa cells was performed in 96-well plates. Cells were seeded in DMEM-based medium at a density of 2 × 104 cells/cm2 and inoculated with individual mutants using the 96-pin replicator. The volume of the sample transferred by one pin of the replicator was estimated at about 1 μl. After 3 days of cocultivation, plates were submitted to one freeze-thaw (−80°C/+37°C) cycle and spotted onto solid medium using the 96-pin replicator. The development of mycoplasma colonies was observed after 5 to 10 days of incubation at 37°C. Growth-deficient mutants failed to produce detectable numbers of CFU upon cocultivation with HeLa cells. The presence of viable CFU in culture stocks of M. agalactiae mutants was controlled by direct spotting onto solid medium.
PCR-based screening of the mutant library.
The detection of mutants with transposon insertion events at specific genomic regions in the whole library was performed by PCR amplification using genomic DNA prepared from individual mutants or mutant pools containing up to 96 individual mutants. Mutant pools were constituted by the addition of a 15- to 30-μl aliquot of each mutant culture. Mutants with a transposon inserted at genomic position 180349 (MAG1540) were identified using the M. agalactiae-specific primer 181025_TIG_R (5′-TCTCCACAGGAACAGTTGCTTA-3′), which spans genomic positions 181025 to 181004, and the transposon-specific oligonucleotide SG8 (5′-GAGTCAGTGAGCGAGGAAGC-3′) priming at the 3′ end of the integrated transposon sequence. PCR amplifications (25 μl) were performed according to the recommendations of the Taq DNA polymerase supplier (New England Bioloabs).
Southern blot hybridization.
Genomic DNA (1 μg) was digested by HindIII, and hybridization was performed in the presence of digoxigenin (DIG)-labeled DNA probes as described previously (28).
RESULTS
Proliferation of M. agalactiae in cell culture depends on HeLa cells for nutrients.
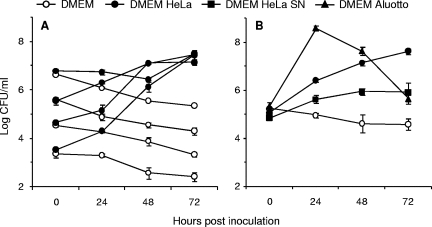
To assess the capacity of M. agalactiae to proliferate under cell culture conditions, HeLa cells were infected with serial dilutions of mycoplasma cultures, and the growth of mycoplasmas over a period of 72 h was determined by enumerating CFU. Unless incubated with HeLa cells, M. agalactiae was unable to grow in DMEM-based medium alone, despite supplementation with fetal bovine serum (Fig. 1A). HeLa cells dramatically enhanced M. agalactiae proliferation under cell culture conditions, yet CFU titers were about 100-fold lower than those produced in axenic media, which are classically used to propagate M. agalactiae under laboratory conditions (data not shown). Conversely, M. agalactiae had no visible effect on cell monolayer development, at least during the first 72 h of infection (data not shown). At the end of the incubation period, M. agalactiae reached an average titer of ca. 107 CFU/ml, with a mycoplasma/cell ratio estimated at about 50 to 100 bacteria per cell, regardless of the starting inoculum. This offered the possibility to compare simultaneously a large collection of individual mutants of unknown CFU titers, as performed below.
FIG. 1.
Growth and survival of M. agalactiae under cell culture conditions. Serial dilutions from mycoplasma stocks (A) or defined dilutions (B) were inoculated to HeLa cells seeded at a density of 2 × 104 cells per cm2 in DMEM-based medium (DMEM HeLa), DMEM-based medium alone (DMEM), DMEM-based medium preincubated with HeLa cells (DMEM HeLa SN), or DMEM-based medium supplemented with 10% Aluotto broth (DMEM Aluotto). Cultures were incubated at 37°C under 5% CO2, and mycoplasma titers (log CFU/ml) were determined by CFU titrations following one freeze-thaw cycle of to disrupt HeLa cells. The data are the means of at least three independent assays. Standard deviations are indicated by error bars.
The growth-promoting effect of HeLa cells on M. agalactiae was further examined by using a cell culture medium preincubated with HeLa cells or supplemented with Aluotto broth. As shown in Fig. 1B, M. agalactiae growth was observed under both conditions, indicating that a deficiency in essential nutrients, rather than the presence of growth inhibitors, was probably responsible for the absence of proliferation in DMEM-based medium. This suggests that HeLa cells may provide a number of nutrients or growth factors that are required for M. agalactiae proliferation under cell culture conditions.
Although human epithelial surfaces are probably not a natural environment for M. agalactiae, these results indicate that HeLa cells may provide a useful model system to study basic interactions of mycoplasmas with mammalian cells.
Isolation of M. agalactiae growth-deficient mutants in cell culture.
An M. agalactiae library of 1,813 gene-disrupted mutants was generated by transposition mutagenesis using plasmid pMT85 (48), which does not replicate in mycoplasma and contains a modified version of transposon Tn4001 (mini-Tn). Because in this plasmid the transposase gene has been placed outside the transposon, the random insertion of the mini-Tn in the mycoplasma genome is stable in addition to conferring gentamicin resistance. Given the low transformation efficiency of M. agalactiae, mutants were collected from multiple individual transformations to produce a representative library (Table 1).
TABLE 1.
Transposon mutagenesis in M. agalactiae
| Transformation no.a | No. of clones isolated | No. (%) of mutants in: |
|
|---|---|---|---|
| Group Ab | Group Bc | ||
| T01 | 52 | 0 (0.0) | 5 (9.6) |
| T02 | 65 | 0 (0.0) | 8 (12.3) |
| T05 | 149 | 8 (5.4) | 30 (20.1) |
| T06 | 169 | 3 (1.8) | 35 (20.7) |
| T07 | 175 | 2 (1.1) | 1 (0.6) |
| T08 | 270 | 2 (0.7) | 23 (8.5) |
| T09 | 247 | 1 (0.4) | 47 (19.0) |
| T10 | 686 | 7 (1.0) | 27 (3.9) |
| Total | 1,813 | 23 (1.3) | 176 (9.7) |
Independent transformation assays with plasmid pMT85.
Growth-deficient mutants in cell culture.
Mutants found to be moderately inhibited upon high-throughput screening of the mutant library in cell culture.
A high-throughput screening strategy was developed to identify mutants displaying a growth-deficient pheontype in cell culture. Using this approach, a first set of 23 mutants was selected (group A) whose multiplication in cell culture failed to produce detectable titers (with a detection limit of about 104 to 105 CFU/ml); a second set of 176 mutants (group B) displaying apparently reduced, but still detectable, CFU production was also selected. The distribution of these mutants obtained from independent transformation events ranged from 0 to 5% for group A and from 1 to 21% for group B (Table 1) of the total number of transformants.
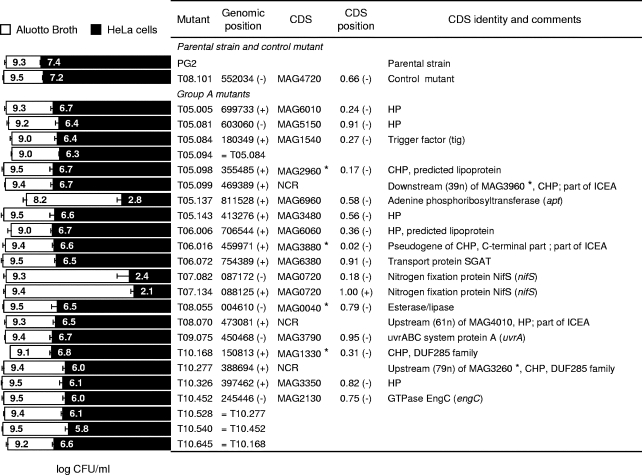
In this study, only mutants of group A were further examined because of their marked phenotypes. After subcloning, the growth phenotype of each mutant was determined under both axenic and cell culture conditions (Fig. 2). Compared to the parental strain PG2 or to the control mutant, namely, T08.101, these mutants displayed a 3-fold reduction in CFU titers to nearly complete extinction in the presence of HeLa cells while producing wild-type CFU titers in Aluotto broth. One exception was mutant T05.137, whose multiplication was affected under both cell culture and axenic conditions.
FIG. 2.
Characterization of M. agalactiae mutants displaying altered growth in cell culture. Mutants were designated according to transformation and clone numbers (e.g., T05.081 designates clone 81 isolated from transformation T05). PG2 and T08.101 refer to the parental strain and the control mutant, respectively. Group A mutants were selected from the mutant library by high-throughput screening on HeLa cells, as described in the Results section. Mycoplasma titers (log CFU/ml) at 48 h in Aluotto broth (open bars) and at 72 h in cell culture (solid bars) are compared. The data are the means of at least three independent assays. Standard deviations are indicated by error bars. Transposon insertion sites were determined by direct sequencing of genomic DNA, and their positions were defined based on the published sequence (NC_009497). The orientation of the inserted sequences is indicated in parenthesis. Mutants sharing an identical insertion are indicated. CDSs found disrupted in M. agalactiae mutants are indicated by the mnemonic codification (45). Noncoding regions are indicated. CDSs involved in horizontal gene transfer between M. agalactiae and mycoplasmas of the mycoides cluster (45) are designated by asterisks (*). For each CDS, the relative position and orientation of the inserted transposon are indicated (CDS position). Hypothetical proteins (HP) have no homologs outside the M. agalactiae species. Conserved hypothetical proteins (CHP) share sequence similarity with proteins of unknown function identified in mollicutes or other bacteria. Several insertion sites mapped within a 20-kb locus that contains a vestige of an integrative conjugative element (ICEA).
Mapping of transposon insertion sites in M. agalactiae growth-deficient mutants.
Direct sequencing of genomic DNA from group A mutants revealed single transposon insertion events in each mutant and identified 19 insertion sites mapping within 15 different coding sequences (CDSs), and three noncoding regions (NCR) (Fig. 2). CDSs found to be disrupted in group A mutants corresponded to proteins belonging to a broad spectrum of COG (cluster of orthologous groups) functional categories including chaperones (trigger factor; O-COG0544), nucleotide metabolism (adenine phosphoribosyltransferase; F-COG0503), amino acid metabolism (nitrogen fixation protein NifS; E-COG0520), and DNA repair (UvrABC system protein A; L-COG0178), as well as proteins with poorly characterized functions (transport protein SGAT [S-COG3037], esterase/lipase [R-COG0596], and GTPase EngC [R-COG1162]) and hypothetical proteins (HP) of unknown functions, some of which have features of membrane-bound lipoproteins.
Localization of transposon insertion sites in the M. agalactiae chromosome of group A mutants failed to reveal any hot spot for the transposition of mini-Tn. However, three mutants (13% of the total number of selected mutants) were found to have a transposon inserted within a 20-kb locus containing a vestige of an integrative conjugative element, ICEA, encompassing CDS MAG4060 to MAG3860 in M. agalactiae strain PG2 (45). Insertions were found in pseudogene MAG3880 (mutant T06.016) and two NCR located upstream of MAG4010 (mutant T08.070) and downstream of MAG3960 (mutant T05.099). Reexamination of the M. agalactiae genome using AMIGene software (6) predicted the presence of a short CDS (spanning genomic position 469474 to 469319) which was disrupted in mutant T05.099 and was not previously annotated.
A detailed analysis of transposition events in group A mutants revealed an important bias in the orientation of the integrated mini-Tn sequence. Using the mini-Tn gentamicin resistance gene as a reference (48), almost all CDSs found disrupted were harboring a transposon inserted in an antisense direction relative to the transcription of the mutated CDS. One exception was mutant T07.134, which has a positively oriented mini-Tn in CDS MAG0720 (see below). The lac promoter and the promoter of the gentamicin resistance-encoding gene are the only regulating sequences provided by the mini-Tn. Both are transcribed in the same direction (48). Experiments are in progress to determine whether transcription from these two promoter regions can extend beyond mini-Tn-inserted sequences and influence the orientation of the integrated mini-Tn sequence.
Several growth-deficient mutants identified by our screening strategy shared identical insertion sites (Fig. 2). As expected, mutants with identical insertions produced similar CFU titers under cocultivation conditions, suggesting that these mutants were probably siblings derived from the same parental clone. To rule out a possible bias in our screening strategy due to the outgrowth of mutants with higher degrees of fitness, we used a PCR-based screening method to determine the frequency of mutants sharing identical insertion sites. Analysis of the 149 mutants derived from transformation T05 identified two mutants with a transposon inserted at genomic position 180349 (MAG1540). These two mutants were those selected upon cocultivation with HeLa cells, which indicates that the screening procedure was efficient.
The NIF locus is essential for M. agalactiae growth under cell culture conditions but is dispensable for axenic growth.
Two mutants (T07.082 and T07.134) with the most extreme phenotypes under coculture conditions have a transposon inserted in a locus composed of two genes with homology to nitrogen fixation proteins NifS and NifU (MAG0720 and MAG0730). The insertion site identified in mutant T07.082 was found to disrupt the nifS gene at a region corresponding to the N-terminal part of the protein (Fig. 2). Mutant T07.134 had a transposon inserted within the same gene, right into the stop codon (TAG). Interestingly, in this mutant the mini-Tn insertion is such that it restored the sequence of the termination codon, suggesting that the phenotype of mutant T07.134 cannot be explained simply by disruption of the gene encoding the NifS protein.
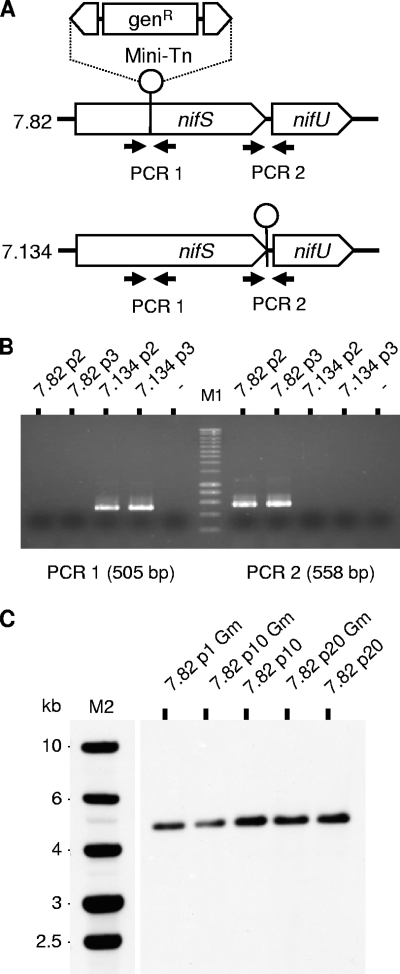
In contrast to the situation described in Mycoplasma genitalium (18, 25), the ability to obtain such mutants suggests that the NIF locus (MAG0720 and MAG0730) is dispensable for M. agalactiae growth under axenic conditions. The dispensability of the NIF locus for axenic growth of mutants T07.82 and T07.134 was further confirmed in Aluotto and SP4 media, both of which are classically used to propagate M. agalactiae under laboratory conditions (data not shown). BLAST analysis of the M. agalactiae genome sequence failed to reveal an additional copy of CDS MAG0720 or the presence of a paralog, ruling out the possibility of an insertion event having inactivated one copy of a duplicated nifS gene in mutants T07.82 and T07.134. However, essential genes can also be found disrupted if gene products are supplied by other mutants in mixed populations. To rule out this hypothesis, the presence of wild-type sequences in cloned nifS mutants propagated in medium without gentamicin was tested by PCR assay using oligonucleotide primers flanking the transposon insertion site in mutants T07.82 and T07.134 (Fig. 3). The absence of wild-type sequences was confirmed in all populations tested. Finally, Southern blotting performed on nifS mutant populations at passages 1 to 20 in selective or nonselective medium confirmed the stability of the inserted sequences (Fig. 3).
FIG. 3.
Clonality and stability of M. agalactiae NIF mutant populations. (A) The genomic regions surrounding the transposon insertion site in mutants T07.82 and T07.134 were analyzed by PCR amplification to detect the presence of contaminating transposon-free sequences. PCR1 and PCR2 were performed using the primer pair 86768F (5′-TCAGCCGACATTATTCATGG-3′) and 87272R (5′-CACCGGCTTTTAATTTTTGC-3′) and the pair 88037F (5′-AGGGTTTCGCTAGGGGTTTA-3′) and 88595R (5′-CTGTGCGCGCTTACAAAGTA-3′), respectively. (B) The PCR1 product (505 bp) amplified from mutant T07.134 populations at passages 2 (7.134p2) and 3 (7.134p3) was not detected upon amplification of the corresponding populations of mutant T07.82 (7.82p2 and 7.82p3). The opposite result was observed for the PCR2 product (558 bp). These negative results suggest the absence of detectable contaminating sequences in all the populations tested. M1, molecular weight markers (SmartLadder; Eurogentec). (C) Southern blot analysis of genomic DNA derived from NIF mutant T07.82 (7.82) at passages 1, 10, and 20 in selective (Gm) or nonselective medium. Genomic DNA was digested by HindIII and Southern blot hybridized with DIG-labeled amplicons derived from plasmid pMT85. M2, molecular weight markers (1 Kb DNA ladder; Promega).
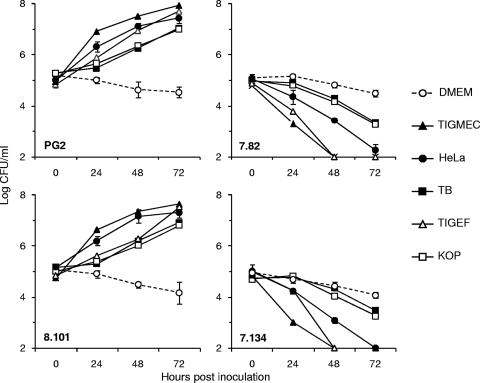
The growth phenotype of nifS mutants T07.82 and T07.134 under cell culture conditions was further characterized. Growth experiments were carried out using HeLa cells and a number of animal cell lines derived from ruminant species (see Materials and Methods). Incubation with HeLa cells, while producing a growth-promoting effect on the M. agalactiae parental strain and control mutant T08.101, had the opposite effect on nifS mutants T07.82 and T07.134 (Fig. 4). This dual effect, growth-promoting and death-inducing, was not restricted to HeLa cells and was also observed with all mammalian cell lines tested in this study, although to different degrees (Fig. 4). These results suggest that components, most likely nutrients released by mammalian cells, were required for M. agalactiae proliferation in DMEM-based medium but were toxic for the nifS mutants. This was further confirmed by reproducing this dual effect in the absence of mammalian cells, using a DMEM-based medium preincubated with HeLa cells (data not shown).
FIG. 4.
Comparative growth of M. agalactiae NIF mutants under cell culture conditions. M. agalactiae parental strain PG2, the control mutant T08.101 (8.101), and NIF mutants T07.082 (7.82) and T07.134 (7.134) were assessed for survival over a 72-h incubation in DMEM-based medium (DMEM) or in coculture with a number of mammalian cell lines including HeLa, goat fibroblast (TIGEF), goat epithelial (TIGMEC), bovine turbinate (TB), and bovine esophageal (KOP) cells. The data are the means of two or three independent assays. Standard deviations are indicated by error bars.
Finally, disruption of the NIF locus was also found to affect M. agalactiae growth in the presence of erythrocytes. Whereas M. agalactiae development on blood agar plates produced hemolytic zones surrounding colonies, the development of the nifS mutants T07.82 and T07.134 was inhibited in a dose-dependent manner by erythrocytes or erythrocyte lysates (Fig. 5) but not by erythrocyte ghosts (data not shown). This inhibition was not species specific since similar results were obtained using horse or sheep erythrocytes. Attempts to further characterize the nifS mutant growth-inhibiting factors present in erythrocyte lysates failed. These results further illustrate the vulnerability of the nifS mutants when they are exposed to mammalian cells and the critical role played by the NIF locus for M. agalactiae survival under these conditions.
FIG. 5.
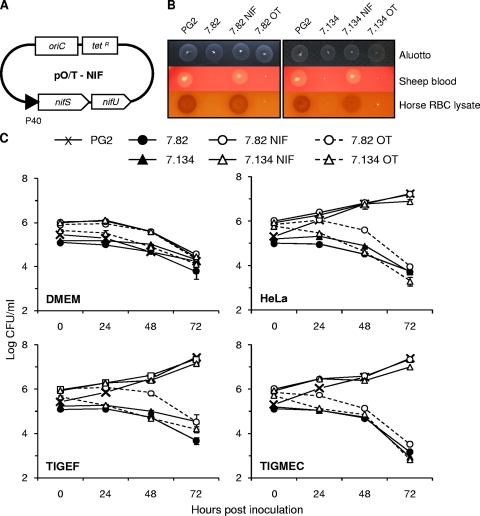
Complementation of M. agalactiae NIF mutants. Schematic representation of the plasmid pO/T-NIF used for complementation studies (A). The NIF locus was cloned under the control of the P40 protein promoter region. M. agalactiae parental strain PG2, mutants T07.082 (7.82) and T07.134 (7.134), and mutants transformed with plasmid pO/T-NIF (7.82 NIF and 7.134 NIF) or the control plasmid p21-1miniO/T (7.82 OT and 7.134 OT) were assessed for colony development on Aluotto, 10% horse red blood cell (RBC) lysates, or 5% sheep blood agar plates (B) and for survival over a 72-h incubation in DMEM-based medium (DMEM) or in coculture with HeLa, goat fibroblast (TIGEF), or goat epithelial (TIGMEC) cells (C). The data are the means of at least three independent assays. Standard deviations are indicated by error bars. Serial dilutions of mycoplasma stocks were spotted on blood agar plates, and colony development was observed after 4 to 6 days of incubation at 37°C.
Gene complementation studies.
Plasmid p20-1miniO/T was used as a shuttle vector for gene complementation studies in M. agalactiae. Mutants T07.82 and T07.134 were transformed with the same plasmid in which the NIF locus was introduced (pO/T_NIF). More specifically, the DNA region encompassing the nifS and nifU genes of PG2 was cloned in p20-1miniO/T downstream of the P40 protein promoter region (see Materials and Methods). The growth of mutants T07.82 and T07.134 on blood agar plates was restored upon transformation with construct pO/T_NIF but not with control plasmid p20-1miniO/T (Fig. 5). Complementation of these two mutants was also confirmed in coculture with mammalian cells (Fig. 5). Growth experiments failed to reveal differences between the parental strain PG2 and complemented mutants. As expected, the phenotype displayed by mutants T07.82 and T07.134 under cocultivation conditions remained unchanged following transformation with the control plasmid p20-1miniO/T. Complementation studies confirmed the conditional essentiality of the NIF locus for M. agalactiae under all of these specific culture conditions.
DISCUSSION
The development of axenic culture conditions has considerably facilitated the study of pathogenic mycoplasmas under laboratory conditions. Yet limited information is available regarding the factors that are involved in their virulence and in their interaction with the host, mainly because of the lack of cellular or small-animal models. In an attempt to fill this gap, a model system using the HeLa human epithelial cell line was developed to study the basic interactions between M. agalactiae and eukaryotic cells. We further used this model system, combined with the production of a large mutant library, in a high-throughput screening strategy for the identification of M. agalactiae growth-deficient mutants and mapped 18 regions on the M. agalactiae chromosome that are specifically required for optimal proliferation under cell culture conditions but are dispensable for propagation in axenic medium.
The number of mutants tested was 2.4 times the total number of CDSs found in the genome of the PG2 strain (877 kb; 742 CDSs). Whether the mutant library produced in M. agalactiae may be approaching saturation is unknown; however, the number of mutants collected is in agreement with the experimental estimations of the minimal size of a mutant library to reach saturation mutagenesis of all nonlethal insertion sites in other mycoplasma species (15, 18). In the human urogenital pathogen M. genitalium (580 kb; 475 CDSs), the number of unique transposon insertion sites drops dramatically after 600 mutants, corresponding to 1.3 times of the total number of CDSs found in this organism; whereas in the murine pathogen M. pulmonis (964 kb; 782 CDSs), the number of inactivating insertions in genes larger than 1 kb nearly reached a plateau at around 1,800 insertion sites (2.3 times the total number of CDSs). High-throughput screening of the M. agalactiae library using the cell system developed here identified a series of 23 mutants displaying a 3-fold reduction in CFU titers to nearly complete extinction in the presence of HeLa cells. The efficiency of this screening was confirmed by (i) the identification of several growth-deficient mutants sharing identical insertion sites, such as mutants T05.084 and T05.094 that were present at low frequency (<1.4%) in the mutant population originating from one transformation event, and (ii) the identification of growth-deficient mutants having a transposon inserted in the same CDS but at different positions.
The availability of the annotated genome sequence of M. agalactiae allowed rapid mapping of the transposon insertion sites of selected mutants. Disrupted CDSs for which a predicted function was assigned correspond to 40% of the total number of CDSs and belong to a broad number of functional categories, often with no obvious correlation between the predicted function and its potential role in M. agalactiae survival under cell culture conditions. Yet several related functions, such as protein folding, iron-sulfur cluster biosynthesis, and DNA repair, have been associated with virulence or stress tolerance in a number of pathogenic bacteria (1, 4, 24, 39, 42, 43, 47). Another 40% of CDSs encode hypothetical products, many of which were shown to display membrane lipoprotein features and/or to have undergone horizontal gene transfer (HGT) with the mycoides cluster (Fig. 2). Two of these proteins belong to a gene family, the drp, which encodes related proteins containing one or several DUF258 domains. This domain is of unknown function and is found in some bacteria but not in mollicutes, with the exception of the mycoides cluster (45). In a recent study comparing M. agalactiae strains using whole-genomic and proteomic approaches, the differential expression of some drp genes was found in association with the membrane fraction. These data suggested that this family may participate in generating surface diversity, with some drp genes presenting features of lipoproteins and being expressed and others serving as sequence reservoirs (34). Interestingly, the drp genes are part of the gene pool which has undergone HGT with members of the phylogenetically distant mycoides cluster. This cluster contains only ruminant-pathogenic mycoplasma species, and in the absence of a cell wall, surface-exposed lipoproteins may play an important role in mediating interactions with the host. Further studies are needed to confirm the role of these CDSs in the M. agalactiae interaction with mammalian cells, but they provide an interesting subset of mutants that can reasonably be analyzed in vivo. Remarkably, a number of integration events occurred in pseudogenes or in NCR that map within a particular 20-kb locus containing a vestige of an integrative conjugative element, ICEA. The implication of an ICEA-related open reading frame (ORF) in the M. agalactiae interaction with HeLa cells remains to be confirmed. However, it is worth noting that best alignments for ICEA products of the PG2 strain were consistently obtained with the ICEC counterparts of M. capricolum subsp. capricolum (45), a member of the mycoides cluster which causes similar symptoms. Defining whether the growth deficiency phenotype observed in selected mutants resulted from the single transposon insertion or from phase variation or spontaneous mutation of other unrelated genes is essential, especially if regions apparently deprived of CDSs are involved. However, this is hampered by the difficulties in the genetic manipulation of these organisms. These difficulties have been overcome so far by complementation with the two mutants that had the most extreme phenotype.
Complementation studies confirmed that key functions conditioning M. agalactiae survival and proliferation under cell culture conditions were encoded by the NIF locus. In M. agalactiae, the locus is composed of two CDSs encoding homologues of nitrogen fixation proteins, NifS and NifU, two proteins involved in iron-sulfur [Fe-S] cluster biosynthesis. Documented in various organisms (14, 26), [Fe-S] cluster assembly systems are poorly understood in Gram-positive bacteria. Recent studies with Enterococcus faecalis identified the SUF machinery as the only [Fe-S] cluster biosynthetic system present in the Firmicutes genome (38). As expected by the taxonomic position of mycoplasmas, sequence features of the SUF machinery were identified in M. agalactiae NifS and NifU proteins. These include the amino acid sequence RSGIFCA surrounding Cys343 of MAG0720 (NifS), which is indicative of a group II (SUF-type) bacterial cysteine desulfurase, whose consensus sequence is RXGHHCA; this sequence clearly distinguishes NifS from group I enzymes of the iron-sulfur cluster, ISC (IscS-type), that display the sequence signature SSGSAC(T/S)S. Similarly, the MAG0730 (NifU) product and SufU scaffold proteins share several features that distinguish them from IscU homologues. They both lack the LPPVK motif present in IscU and contain an 18- to 21-amino-acid insertion between the second and the third conserved cysteine residues. However, despite important sequence homologies with bacterial SUF machineries, mycoplasma NIF proteins exhibit several unique features.
The NIF locus present in mycoplasmas is a simplified version of more complex SUF operons and may encode cysteine desulfurases and scaffold proteins with unique biochemical properties. Its strict conservation among all mycoplasma genomes sequenced so far emphasizes its biological importance in mollicutes. Recent studies with a number of pathogenic bacteria, including Mycobacterium tuberculosis, Shigella flexneri, and the plant pathogen Erwinia chrysanthemi, have established a link between [Fe-S] cluster biosynthesis and virulence (24, 32, 39, 42). The central role played by bacterial SUF machineries in resistance to iron limitation and oxidative stress suggest that the NIF locus might play a similar role in M. agalactiae-host interactions. However, preliminary in vitro studies with M. agalactiae failed to reveal a particular susceptibility of NIF mutants when bacteria were exposed to oxidative stress or iron limitation. In vivo studies are in progress to determine the virulence of the NIF mutants in the animal host. The potential implication of this locus in a broad number of processes involving [Fe-S] proteins considerably increases the functions that can be affected in the NIF mutants. The additional functions that have been attributed to cysteine desulfurases (30), such as the biosynthesis of selenoproteins and multiple cofactors (biotin, lipoic acid, molybdopterin, thiamine, and NAD) as well as iron homeostasis and tRNA modifications, make the situation even more complex.
The growth-deficient phenotype exhibited by the NIF mutants under cell culture conditions and also potentially by other mutants (Fig. 2) revealed the decisive role played by metabolic functions in the adaptation of M. agalactiae to changing environments. A link between carbon metabolism and pathogenicity in mycoplasmas has already been suggested by several groups (5, 21-23, 36). The availability of a carbon source in vivo and its influence on bacterial pathogenicity has reignited interest in using carbon metabolic pathways as viable targets for antibiotic development (7). This might be particularly important for mycoplasmas, which have limited metabolic capacities and are dependent on the host for many nutrients (37).
Attempts to define the minimal amount of genetic information in various mycoplasma species revealed several discrepancies since orthologous genes found to be essential in one organism may be dispensable in another. This is illustrated here by the NIF locus, which was essential for axenic growth of M. genitalium (18, 25) but apparently dispensable in other mycoplasma species such as M. pneumoniae, M. pulmonis, and M. agalactiae (15, 18, 25). A number of situations may account for the occurrence of transposition events in essential genes. The identification of a paralog in M. pulmonis of the cysteine desulfurase-encoding gene provided a simple explanation for the apparent dispensability of the NIF locus in this species (15). Given the central role played by [Fe-S] proteins in a variety of fundamental biological processes, the absence of a paralog in the M. agalactiae genome suggests that essential functions might be performed by unrelated or very distantly related nonorthologous proteins.
The understanding of the basic molecular mechanisms underlying cellular life is of broad interest, and considerable effort has been devoted to establishing candidate minimal genomes. Our study provides a means for addressing this issue at a higher level of complexity, the host-cell context, and new opportunities to decipher mycoplasma-host interaction and virulence.
Acknowledgments
We thank P. Giammarinaro for helpful discussion and critical reading of the manuscript and M. C. Hygonenq and A. Brouillaud for excellent technical assistance. We also thank C. Leroux (INRA, Lyon, France) for providing the TIGMEC and TIGEF cell lines and R. Herrmann for providing pMT85.
This work was supported by a joint grant from INRA and CIRAD and by a doctoral fellowship to A.S. from INRA and DGER.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 1 February 2010.
REFERENCES
- 1.Ambur, O., T. Davidsen, S. Frye, S. Balasingham, K. Lagesen, T. Rognes, and T. Tønjum. 2009. Genome dynamics in major bacterial pathogens. FEMS Microbiol. Rev. 33:453-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barré, A., A. de Daruvar, and A. Blanchard. 2004. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 32:D307-D310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergonier, D., X. Berthelot, and F. Poumarat. 1997. Contagious agalactia of small ruminants: current knowledge concerning epidemiology, diagnosis and control. Rev. Sci. Tech. 16:848-873. [DOI] [PubMed] [Google Scholar]
- 4.Bigot, A., E. Botton, I. Dubail, and A. Charbit. 2006. A homolog of Bacillus subtilis trigger factor in Listeria monocytogenes is involved in stress tolerance and bacterial virulence. Appl. Environ. Microbiol. 72:6623-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischof, D., C. Janis, E. Vilei, G. Bertoni, and J. Frey. 2008. Cytotoxicity of Mycoplasma mycoides subsp. mycoides small colony type to bovine epithelial cells. Infect. Immun. 76:263-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bocs, S., S. Cruveiller, D. Vallenet, G. Nuel, and C. Médigue. 2003. AMIGene: annotation of microbial genes. Nucleic Acids Res. 31:3723-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, S., K. Palmer, and M. Whiteley. 2008. Revisiting the host as a growth medium. Nat. Rev. Microbiol. 6:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chopra-Dewasthaly, R., M. Marenda, R. Rosengarten, W. Jechlinger, and C. Citti. 2005. Construction of the first shuttle vectors for gene cloning and homologous recombination in Mycoplasma agalactiae. FEMS Microbiol. Lett. 253:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chopra-Dewasthaly, R., M. Zimmermann, R. Rosengarten, and C. Citti. 2005. First steps towards the genetic manipulation of Mycoplasma agalactiae and Mycoplasma bovis using the transposon Tn4001mod. Int. J. Med. Microbiol. 294:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citti, C., G. Browning, and R. Rosengarten. 2005. Phenotypic diversity and cell invasion in host subversion by pathogenic mycoplasmas, p. 439-484. In A. Blanchard and G. Browning (ed.), Mycoplasmas: molecular biology, pathogenicity and strategies for control. Horizon Bioscience, Norfolk, United Kingdom.
- 11.Da Silva Teixeira, M., V. Lambert, L. Mselli-Lakahl, A. Chettab, Y. Chebloune, and J. Mornex. 1997. Immortalization of caprine fibroblasts permissive for replication of small ruminant lentiviruses. Am. J. Vet. Res. 58:579-584. [PubMed] [Google Scholar]
- 12.Dybvig, K., C. Zuhua, P. Lao, D. Jordan, C. French, A. Tu, and A. Loraine. 2008. Genome of Mycoplasma arthritidis. Infect. Immun. 76:4000-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleury, B., D. Bergonier, X. Berthelot, E. Peterhans, J. Frey, and E. Vilei. 2002. Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect. Immun. 70:5612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontecave, M., and S. Ollagnier-de-Choudens. 2008. Iron-sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch. Biochem. Biophys. 474:226-237. [DOI] [PubMed] [Google Scholar]
- 15.French, C., P. Lao, A. Loraine, B. Matthews, H. Yu, and K. Dybvig. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 69:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey, J. 2002. Mycoplasmas of animals, p. 73-90. In S. Razin and R. Herrmann (ed.), Molecular biology and pathogenicity of mycoplasmas. Plenum, New York, NY.
- 17.Gibson, D., G. Benders, C. Andrews-Pfannkoch, E. Denisova, H. Baden-Tillson, J. Zaveri, T. Stockwell, A. Brownley, D. Thomas, M. Algire, C. Merryman, L. Young, V. Noskov, J. Glass, J. Venter, C. r. Hutchison, and H. Smith. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215-1220. [DOI] [PubMed] [Google Scholar]
- 18.Glass, J., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. Lewis, M. Maruf, C. Hutchison, H. Smith, and J. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glew, M., M. Marenda, R. Rosengarten, and C. Citti. 2002. Surface diversity in Mycoplasma agalactiae is driven by site-specific DNA inversions within the vpma multigene locus. J. Bacteriol. 184:5987-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glew, M., L. Papazisi, F. Poumarat, D. Bergonier, R. Rosengarten, and C. Citti. 2000. Characterization of a multigene family undergoing high-frequency DNA rearrangements and coding for abundant variable surface proteins in Mycoplasma agalactiae. Infect. Immun. 68:4539-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halbedel, S., C. Hames, and J. Stülke. 2007. Regulation of carbon metabolism in the mollicutes and its relation to virulence. J. Mol. Microbiol. Biotechnol. 12:147-154. [DOI] [PubMed] [Google Scholar]
- 22.Hames, C., S. Halbedel, M. Hoppert, J. Frey, and J. Stülke. 2009. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson, P., T. Gorton, L. Papazisi, K. Cecchini, S. J. Frasca, and S. Geary. 2006. Identification of a virulence-associated determinant, dihydrolipoamide dehydrogenase (lpd), in Mycoplasma gallisepticum through in vivo screening of transposon mutants. Infect. Immun. 74:931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huet, G., M. Daffé, and I. Saves. 2005. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen's survival. J. Bacteriol. 187:6137-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchison, C., S. Peterson, S. Gill, R. Cline, O. White, C. Fraser, H. Smith, and J. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D., D. Dean, A. Smith, and M. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 27.Kannan, T., and J. Baseman. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 103:6724-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marenda, M., E. Sagné, F. Poumarat, and C. Citti. 2005. Suppression subtractive hybridization as a basis to assess Mycoplasma agalactiae and Mycoplasma bovis genomic diversity and species-specific sequences. Microbiology 151:475-489. [DOI] [PubMed] [Google Scholar]
- 29.McAuliffe, L., R. Ellis, K. Miles, R. Ayling, and R. Nicholas. 2006. Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 152:913-922. [DOI] [PubMed] [Google Scholar]
- 30.Mihara, H., and N. Esaki. 2002. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 60:12-23. [DOI] [PubMed] [Google Scholar]
- 31.Mselli-Lakhal, L., F. Guiguen, C. Fornazero, C. Favier, J. Durand, D. Grezel, A. Moussa, J. Mornex, and Y. Chebloune. 2001. Immortalized goat milk epithelial cell lines replicate CAEV at high level. Vet. Res. 32:429-440. [DOI] [PubMed] [Google Scholar]
- 32.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 33.Nouvel, L., M. Marenda, P. Sirand-Pugnet, E. Sagné, M. Glew, S. Mangenot, V. Barbe, A. Barré, S. Claverol, and C. Citti. 2009. Occurrence, plasticity and evolution of the vpma gene family, a genetic system devoted to high-frequency surface variation in Mycoplasma agalactiae. J. Bacteriol. 191:4111-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nouvel, L., P. Sirand-Pugnet, M. Marenda, E. Sagné, V. Barbe, S. Mangenot, C. Schenowitz, D. Jacob, A. Barré, S. Claverol, A. Blanchard, and C. Citti. 2 February 2010. Comparative genomic and proteomic analyses of two Mycoplasma agalactiae strains: clues to the macro- and micro-events that are shaping mycoplasma diversity. BMC Genomics 11:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, S., and C. Fraser. 2001. The complexity of simplicity. Genome Biol. 2:COMMENT2002. http://genomebiology.com/2001/2/2/comment/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilo, P., J. Frey, and E. Vilei. 2007. Molecular mechanisms of pathogenicity of Mycoplasma mycoides subsp. mycoides SC. Vet. J. 174:513-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riboldi, G., H. Verli, and J. Frazzon. 2009. Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem. 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rincon-Enriquez, G., P. Crété, F. Barras, and B. Py. 2008. Biogenesis of Fe/S proteins and pathogenicity: IscR plays a key role in allowing Erwinia chrysanthemi to adapt to hostile conditions. Mol. Microbiol. 67:1257-1273. [DOI] [PubMed] [Google Scholar]
- 40.Rosati, S., S. Pozzi, P. Robino, B. Montinaro, A. Conti, M. Fadda, and M. Pittau. 1999. P48 major surface antigen of Mycoplasma agalactiae is homologous to a malp product of Mycoplasma fermentans and belongs to a selected family of bacterial lipoproteins. Infect. Immun. 67:6213-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rottem, S. 2003. Interaction of mycoplasmas with host cells. Physiol. Rev. 83:417-432. [DOI] [PubMed] [Google Scholar]
- 42.Runyen-Janecky, L., A. Daugherty, B. Lloyd, C. Wellington, H. Eskandarian, and M. Sagransky. 2008. Role and regulation of iron-sulfur cluster biosynthesis genes in Shigella flexneri virulence. Infect. Immun. 76:1083-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, A., L. Guidry, K. Narasimhulu, D. Mai, J. Trombley, K. Redding, G. Giles, J. J. Lancaster, and A. Steyn. 2007. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U. S. A. 104:11562-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirand-Pugnet, P., C. Citti, A. Barré, and A. Blanchard. 2007. Evolution of mollicutes: down a bumpy road with twists and turns. Res. Microbiol. 158:754-766. [DOI] [PubMed] [Google Scholar]
- 45.Sirand-Pugnet, P., C. Lartigue, M. Marenda, D. Jacob, A. Barré, V. Barbe, C. Schenowitz, S. Mangenot, A. Couloux, B. Segurens, A. de Daruvar, A. Blanchard, and C. Citti. 2007. Being pathogenic, plastic, and sexual while living with a nearly minimal bacterial genome. PLoS Genet. 3:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tully, J. 1995. Culture medium formulation for primary isolation and maintenance of mollicutes, p. 33-39. In S. Razin and J. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology. Academic Press, San Diego, CA.
- 47.Wen, Z., P. Suntharaligham, D. Cvitkovitch, and R. Burne. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmerman, C., and R. Herrmann. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253:315-321. [DOI] [PubMed] [Google Scholar]