Abstract
Coxiella burnetii is a Gram-negative, obligate intracellular bacterial pathogen that resides within the harsh, acidic confines of a lysosome-like compartment of the host cell that is termed a parasitophorous vacuole. In this study, we characterized a thiol-specific peroxidase of C. burnetii that belongs to the atypical 2-cysteine subfamily of peroxiredoxins, commonly referred to as bacterioferritin comigratory proteins (BCPs). Coxiella BCP was initially identified as a potential DNA-binding protein by two-dimensional Southwestern (SW) blots of the pathogen's proteome, probed with biotinylated C. burnetii genomic DNA. Confirmation of the identity of the DNA-binding protein as BCP (CBU_0963) was established by matrix-assisted laser desorption ionization-tandem time of flight mass spectrometry (MALDI-TOF/TOF MS). Recombinant Coxiella BCP (rBCP) was generated, and its DNA binding was demonstrated by two independent methods, including SW blotting and electrophoretic mobility shift assays (EMSAs). rBCP also demonstrated peroxidase activity in vitro that required thioredoxin-thioredoxin reductase (Trx-TrxR). Both the DNA-binding and peroxidase activities of rBCP were lost upon heat denaturation (100°C, 10 min). Functional expression of Coxiella bcp was demonstrated by trans-complementation of an Escherichia coli bcp mutant, as evidenced by the strain's ability to grow in an oxidative-stress growth medium containing tert-butyl hydroperoxide to levels that were indistinguishable from, or significantly greater than, those observed with its wild-type parental strain and significantly greater than bcp mutant levels (P < 0.05). rBCP was also found to protect supercoiled plasmid DNA from oxidative damage (i.e., nicking) in vitro. Maximal expression of the bcp gene coincided with the pathogen's early (day 2 to 3) exponential-growth phase in an experiment involving synchronized infection of an epithelial (Vero) host cell line. Taken as a whole, the results show that Coxiella BCP binds DNA and likely serves to detoxify endogenous hydroperoxide byproducts of Coxiella's metabolism during intracellular replication.
Coxiella burnetii is a Gram-negative bacterium that causes Q fever in humans. Growth of the pathogen is restricted to an intracellular lysosome-like compartment, termed a parasitophorous vacuole (PV). The pH (∼4.5) of the PV is ideal for C. burnetii's acidophilic lifestyle (11, 12); however, the effect of concomitant oxidative stress on the bacterium and the defense mechanisms used to counter it have not been well characterized.
Bacteria deal with oxidative stress by employing antioxidants such as glutathione, vitamins A, C, and E, and carotenoids, and they counter reactive oxygen species (ROS) by using a variety of enzymatic effectors, including superoxide dismutase (SOD), catalase, and peroxidase. Previous work suggests that catalase and SOD are potential persistence factors for Coxiella bacteria residing in the intracellular niche (1, 14). In addition, a secreted acid phosphatase possibly reduces the respiratory burst of the host cell by inhibiting NADPH oxidase (4, 23). in silico analysis of the C. burnetii RSA 493 genome (22) revealed predicted Mn- and Cu/Zn-SODs, catalase, and four peroxiredoxins (Prxs), which are antioxidant enzymes (EC 1.11.1.15) that are thiol-containing reductants used to detoxify hydroperoxides. Interestingly, certain C. burnetii strains possess a frame-shifted Cu/Zn-SOD gene (e.g., Dugway) or a markedly truncated catalase gene (Nine Mile or G) (18), suggesting that these strains, likely undergoing reductive evolution in the intracellular niche, must resort to alternative effectors for protection against ROS.
For this report, we characterized the Coxiella BCP (CBU_0963), a representative of a 2-cysteine (2-Cys) Prx subfamily that is widely employed by pathogenic bacteria to deal with potentially toxic H2O2 and organic hydroperoxides. We demonstrate that the bcp gene of Coxiella bacteria is maximally expressed during early exponential-phase growth and that BCP is a DNA-binding protein that exhibits peroxidase activity and can protect supercoiled DNA from oxidative damage in vitro.
MATERIALS AND METHODS
Bacterial strains, cell lines, and growth conditions.
Nine Mile phase II C. burnetii (strain RSA 439, clone 4) was propagated in African green monkey kidney (Vero) epithelial cells (CCL-81; American type Culture Collection [ATCC], Manassas, VA), as previously described (21). C. burnetii small cell variants (SCVs) were prepared and purified as described before (21). The resulting SCVs were used to inoculate Vero cell monolayers to produce synchronized infections (8).
Escherichia coli strains were grown overnight at 37°C in Luria-Bertani (LB) medium. Supplements of chloramphenicol (20 μg/ml), kanamycin (50 μg/ml), or ampicillin (LBamp) (100 μg/ml) were added to the medium, as required. Two E. coli strains (DH5α and JM109) were used for routine cloning and plasmid production, and a third strain (M15 [pREP4]; Qiagen, Valencia, CA) was used for production of recombinant BCP (rBCP) expressed from cloned Coxiella bcp. An E. coli bcp mutant (JS2465-1) and an otherwise isogenic parental strain, BW25113, were obtained from the E. coli Genetic Stock Center (CGCS). An in-frame bcp deletion mutant was generated by transforming (6) JS2465-1 with pCP20 to utilize the encoded Flp recombinase for excision of bcp-728Δ::kan, as previously described (5). The resulting strain, LH001, was transformed with the plasmids pQBCP and pQE30 (Qiagen) to generate strains LH100 and LH200, respectively. BW25113 was transformed with pQE30 (Qiagen) to create strain LH300. A summary of data for these strains, plasmids, and the cell line is provided in Table 1.
TABLE 1.
Bacterial strains, cell line, plasmids, and PCR primers used in the study
| Designation | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| C. burnetii | ||
| RSA 439 | Nine Mile Phase II, clone 4 | R. A. Heinzen |
| E. coli | ||
| DH5α | Host strain for plasmid production | Gibco-BRL |
| JM109 | Host strain for cloning | Promega |
| M15 (pREP4) | Host strain for His6 fusion proteins | Qiagen |
| JS2465-1 | bcp-728Δ::kan | CGSC (3) |
| BW25113 | Parent strain to JS2465-1 | CGSC (3) |
| LH001 | In-frame bcp mutant derived from JS2465-1 | This study |
| LH100 | LH001 containing pQBCP | This study |
| LH200 | LH001 containing pQE30 | This study |
| LH300 | BW25113 containing pQE30 | This study |
| LH400 | M15 (pREP4) containing pQE30 | This study |
| LH500 | M15 (pREP4) containing pQBCP | This study |
| LH600 | M15 (pREP4) containing pQCOM1 | This study |
| LH700 | M15 (pREP4) containing pQHBPA | This study |
| Cell line | ||
| Vero | Green monkey epithelial line | ATCC CCL-81 |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector | 30 |
| pCP20 | flp+, Repts, Apr, Cmra | 5 |
| pQE30, 31 | Expression vectors for His6 fusion proteins | Qiagen |
| pREP4 | lacI+ in trans repressor plasmid | Qiagen |
| pQBCP | pQE30 with cloned Coxiella bcp and sdr | This study |
| pQCOM1 | pQE30 with cloned Coxiella com1 | This study |
| pQHBPA | pQE31 with cloned Bartonella quintana hbpA | This study |
| PCR primers | ||
| BCP_For | GCGCATGCTCCATTGAAGTCGGCCAGAAGG | This study |
| BCP_Rev | TCGCTGCAGGCTTGGTTAGCTGAACATAC | This study |
| BCP2_For | TGAATGATTGATGTGCTTTAACG | This study |
| BCP2_Rev | CGGATGAAGGCGAAATGC | This study |
Ap, ampicillin; Cm, chloramphenicol; ts, temperature sensitive; s, susceptible.
2D SDS-PAGE and Southwestern (SW) blots.
Proteins from a total cell lysate were prepared by adding 500 μl of purified SCVs (∼8 × 1010 genomes) to an equal volume of lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 50 mM MgCl2, 5 mM dithiothreitol [DTT], and a “Complete Mini” protease inhibitor cocktail tablet [Roche Diagnostics, Indianapolis, IN]). Approximately 500 μl of 0.1-mm-diameter glass beads was added to the mixture, cells were vortexed overnight at 4°C using a Disruptor Genie (Scientific Industries, Bohemia, NY), and the resulting lysate was centrifuged (10,000 × g for 10 min, 4°C) to remove cellular debris. The supernatant was collected and the protein concentration determined with a bicinchoninic acid (BCA) protein assay kit per the manufacturer's instructions (Thermo Scientific, Rockford, IL). Two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2D SDS-PAGE) was done using 70 μg of protein and the general methods of O'Farrell (20). For the first dimension, a Protean isoelectric focusing (IEF) cell was used with nonlinear IEF strips (pI, 3 to 10) as instructed by the manufacturer (Bio-Rad, Hercules, CA). The resulting IEF strips were resolved in the second dimension using 12.5% (wt/vol) acrylamide SDS slab gels. 2D gels were stained with SilverQuest as instructed by the manufacturer (Invitrogen, Carlsbad, CA) or immediately blotted.
For SW blotting, 2D gels were incubated for 3 h in renaturing buffer (10 mM Tris-HCl [pH 7.0], 50 mM NaCl, 2 mM EDTA, 4 M urea, and 0.1 mM DTT) to remove SDS with urea. Proteins were subsequently transferred overnight (25 mA, 25°C) onto NitroPure 45 μm membranes (Cole Parmer, Vernon Hills, IL) by the use of transfer buffer (10 mM Tris-HCl [pH 7.0], 0.05 M NaCl, 2 mM EDTA, 0.1 mM DTT) and the general methods of Towbin et al. (26). Blots were washed for 30 min in binding buffer (10 mM Tris-HCl [pH 7.0], 1 mM EDTA, 0.02% bovine serum albumin, 0.02% Ficoll, 0.02% [wt/vol] polyvinylpyrrolidone, and 0.05 M NaCl). A biotin-labeled DNA probe for SW blotting was synthesized in two steps. First, C. burnetii chromosomal DNA was randomly amplified using a GenomiPhi V2 DNA amplification kit as instructed by the manufacturer (GE Healthcare, Piscataway, NJ). Second, a portion of the amplicons was biotin labeled with a NEBlot Phototope kit, as instructed by the manufacturer (New England Biolabs, Ipswich, MA). DNA-binding proteins were identified by incubating blots in 20 ml of binding buffer plus the biotin-labeled DNA probe (60 min at 25°C with rocking). The blots were washed four times with binding buffer by replacing the entire volume at 15-min intervals. The resulting blots were UV cross-linked using a GS gene linker (Bio-Rad) per the manufacturer's instructions. Blots were developed using a chemiluminescent nucleic acid detection module (Thermo Scientific, Rockford, IL) following the manufacturer's instructions but with slight modifications, including the use of 60 ml of blocking buffer (instead of 20 ml), a 1:3,000 dilution of streptavidin-horseradish peroxidase (instead of 1:300), and 60 ml of wash buffer (instead of 20 ml). DNA-binding proteins were visualized using an LAS-3000 imaging system (Fujifilm, Stamford, CT).
Mass spectrometry (MS) and proteomics.
Protein spots representing potential DNA-binding proteins on SW blots were excised from three silver-stained 2D SDS-PAGE gels run in parallel with the blots. 2D gel spots were subjected to trypsin digestion and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) peptide mass fingerprinting and MALDI-tandem TOF (MALDI-TOF/TOF) peptide sequencing (Alphalyse, Palo Alto, CA).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were done using a Lightshift chemiluminescence EMSA kit (Pierce, Rockford, IL) as instructed by the manufacturer. The biotinylated-EBNA DNA probe (B-DNA) supplied in the kit was used for all reactions. The EBNA extract (EE) provided in the kit was used as a positive control DNA-binding protein. Heat-denatured EE (hd-EE) was prepared by boiling for 10 min and cooling to 25°C. EMSA reactions were prepared using 10× binding buffer (2 μl), 50% glycerol (1 μl), 0.5 μg/μl φX174 replicative form (RF) HaeIII DNA fragments (Invitrogen) (1 μl), 1% NP-40 (1 μl), and 40 fmol of biotin-EBNA. Native rBCP (1 to 10 μM) or heat-denatured rBCP (hd-rBCP) (1 to 10 μM; boiled 10 min and cooled to 25°C) was added, and the final volume was brought to 20 μl with distilled water (dH2O). Following 20 min of incubation at 25°C, reaction mixtures were loaded onto a nondenaturing polyacrylamide gel (5% [wt/vol] acrylamide) and electrophoretically separated at 25°C using 0.5% Tris-borate-EDTA buffer. Gels were then transferred at 4°C to Immobilon-NY+ (Millipore, Bedford MA) using 0.5% Tris-borate-EDTA and a Mini Trans-Blot cell (Bio-Rad) and then cross-linked with a Bio-Rad cross-linker (program C3; damp membranes) at 150 mJ. Biotin-EBNA probes were visualized using a chemiluminescent nucleic acid detection module (Pierce) as instructed by the manufacturer and an LAS-3000 imaging system (Fujifilm).
Nucleic acid purification and manipulation and recombinant protein production.
Chromosomal DNA was purified with a DNeasy blood and tissue kit as instructed by the manufacturer (Qiagen). RNA was purified using a commercial kit (RiboPure bacteria; Ambion, Austin, TX). Plasmid DNA was prepared with a QIAprep spin miniprep kit (Qiagen). Nucleic acids were quantified by spectrophotometry at 260 nm. The C. burnetii bcp gene (CBU_0963) was amplified using chromosomal DNA and primers containing SphI and PstI sites (BCP_For and BCP_Rev) and PCR following standard protocols (2). The resulting amplicon was subjected to restriction endonuclease digestion with SphI and PstI and cloned into the same sites of pQE30 (Qiagen) by a standard protocol (2) to generate pQBCP, a plasmid encoding an in-frame N-terminal translational fusion of BCP and the vector's His6 tag. Following transformation of E. coli M15 (pREP4) with pQBCP, induction and affinity purification of His6-tagged rBCP (under native conditions) were accomplished using nickel-nitrilotriacetic acid (Ni-NTA) agarose per the protocols of the manufacturer (Qiagen). A list of plasmids and primers used in the study is provided in Table 1.
qRT-PCR.
Quantitative real time-PCR (qRT-PCR) and RNA purification were accomplished as previously described (21). Primer sets were designed using Beacon Designer 7.5 software (Biosoft International, Palo Alto, CA), culminating in primers specific to bcp (BCP2_For and BCP2_Rev; see Table 1). Amplified cDNA was normalized to genome equivalents determined by quantitative PCR (Q-PCR) and a primer set specific to Coxiella's rpoS gene, as previously described by Coleman et al. (8). Q-PCR data were also used to generate a growth curve for Coxiella bacteria, showing genome equivalents as a function of time in infected Vero cell cultures.
trans-complementation of E. coli bcp.
Functional expression of Coxiella's bcp and trans-complementation of an E. coli bcp mutant were analyzed by comparing growth phenotypes of E. coli strains LH100, LH200, and LH300 by using standard growth curves. Briefly, each strain was grown for 16 h in LB broth containing ampicillin (LBamp) (100 μg/ml) at 37°C with shaking. A 2-ml aliquot was subsequently used to inoculate 20 ml of LBamp. The resulting cultures were grown for 2 h at 37°C to an optical density at 600 nm (OD600) of 1.0, and 500 μl was removed and used to inoculate 50 ml of LBamp containing 0 or 0.2 mM tert-butyl hydroperoxide (t-BOOH). Bacterial growth was quantified spectrophotometrically at 600 nm over an 8-h period.
Prx and DNA protection assays.
rBCP was assayed for Prx activity by the methods of Jeong et al. (15). NADPH, thioredoxin (Trx), thioredoxin reductase (TrxR), and t-BOOH were obtained from Sigma Chemical (St. Louis, MO). Reaction volumes of 1 ml were used for reactions performed using various amounts of purified C. burnetii rBCP (1 to 3 μM rBCP in HEPES buffer [pH 7.0]). Typical reactions were done in 50 mM HEPES-NaOH (pH 7.0) containing 8 μM Trx, 0.3 μM TrxR, and 0.25 mM NADPH. Each reaction mixture was mixed and incubated at 25°C for 2 min, prior to addition of t-BOOH to a final concentration of 900 μM. Absorbance was measured spectrophotometrically (340 nm) at 30-s intervals during a 5-min reaction. Control assays were run individually without rBCP, Trx, or TrxR or using 3 μM hd-rBCP.
The ability of rBCP to protect supercoiled plasmid DNA against oxidative damage was assayed essentially by the methods of Imlay et al. (14). Briefly, supercoiled pUC19 (100 ng)-0.8% NaCl (wt/vol)-10 mM ethanol was mixed on ice with rBCP at a final concentration of 0.1 μM. H2O2 was added to achieve a final concentration of 6 mM, and the resulting 20-μl mixture was incubated for 30 min at 37°C. Reactions were immediately analyzed by electrophoresis through ethidium bromide-stained 1.5% (wt/vol) agarose gels. Control reaction experiments using hd-rBCP (prepared as described above), or in the absence of rBCP or H2O2, were also conducted. Densitometry analysis (performed by measuring the intensity of pixels per square millimeter, corrected for background) of the resulting DNA bands was done using Quantity One 4.6.2 software (Bio-Rad).
Statistical analysis.
At least three independent determinations were used to calculate means and standard deviations for all numerical data. Statistics and graphs were analyzed and produced using InStat3 (GraphPad, La Jolla, CA) and/or Excel 2003 (Microsoft, Redmond, WA) software. Statistical significance was determined using Student's t test; a P value of < 0.05 was considered significant.
RESULTS AND DISCUSSION
Oxidative stress resistance is essential to the viability of aerobic organisms and helps prevent membrane and macromolecular damage from endogenous ROS byproducts of metabolism, such as superoxide anions, H2O2, and hydroxyl radicals. In addition, an ability to resist host-derived, exogenous ROS is a property shared by many bacterial pathogens (25), especially those that reside within professional phagocytes, such as C. burnetii. Bacteria deal with ROS by employing SOD, catalase, and peroxidase or by nonenzymatic means involving glutathione, vitamins A, C, and E, and carotenoids. Prx's are members of a biologically ubiquitous family of thiol-dependent peroxidases that can be divided into two main groups that contain either a single, N-terminal cysteine (i.e., 1-Cys Prx's with a peroxidatic Cys) or an N-terminal peroxidatic Cys plus a second Cys residue (2-Cys Prx's with a resolving Cys) for reduction of H2O2 and organic hydroperoxides (29). Unlike many peroxidases, Prx's do not require redox cofactors (metals or prosthetic groups) for activity, and reducing equivalents are typically provided by NADPH through the Trx-TrxR pathway.
Bacterioferritin comigratory proteins (BCPs) were originally named for their propensity to comigrate with bacterioferritin proteins when subjected to SDS-PAGE, prior to elucidation of their function (19). BCPs are atypical 2-Cys Prx's whose redox partner has been identified as Trx (15, 29). BCPs constitute a subfamily of the thiol-specific-antioxidant (TSA)-alkyl hydroperoxidase C (AhpC) Prx family (15, 27) and are also referred to as PrxQ-like (13, 16) or Class 1 Prx's (9). BCPs are common to several bacterial pathogens (15); however, there is little information regarding their role in conferring ROS resistance during pathogenesis. Previous work has shown that Helicobacter pylori BCP helps the pathogen establish long-term colonization of the stomach in a murine model of infection (28).
Discovery of BCP during SW blotting.
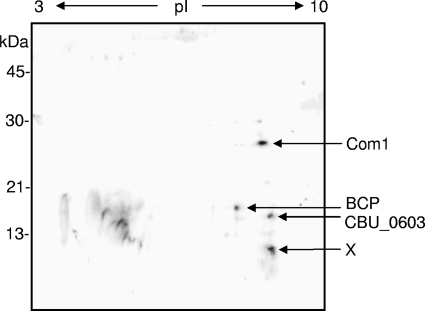
In an effort to discover DNA-binding proteins that are possibly involved in C. burnetii's developmental cycle, SCVs were prepared and proteomes from a SCV lysate were resolved by 2D SDS-PAGE, followed by SW blotting. SW blots of 2D SCV protein profiles probed with random, biotinylated genomic Coxiella DNA fragments revealed four putative DNA-binding proteins, plus several minor species (Fig. 1). Analysis of the protein spots obtained from three individual 2D-SDS-PAGE gels was done by MS (MALDI-TOF peptide mass fingerprinting and MALDI-TOF/TOF peptide sequencing of tryptic digests). The results of the MS consistently identified BCP (CBU_0963), Com1 (CBU_1910), and a hypothetical protein (CBU_0603). MS analysis of BCP generated four peptides with high-confidence identification that represented ∼25% coverage of BCP's amino acid sequence. A prominent, low-molecular-mass spot of ∼10 kDa and basic pI (marked “X” in Fig. 1) could not be identified by MS, despite numerous attempts.
FIG. 1.
Representative 2D SW blot prepared from a total cell lysate of purified Coxiella SCVs (70 μg of total protein). IEF (pI, 3 to 10) was employed in the first dimension and a 12.5% (wt/vol) acrylamide SDS-PAGE gel was used in the second dimension, prior to blotting to nitrocellulose. The SW blot was probed with biotin-labeled, C. burnetii genomic DNA and visualized by streptavidin-horseradish peroxidase (see text for details). Potential DNA-binding proteins from the SW blot were identified by MALDI-TOF peptide mass fingerprinting and MALDI-TOF/TOF peptide sequencing and are listed on the right. The spot marked “X” could not be fingerprinted, despite numerous attempts.
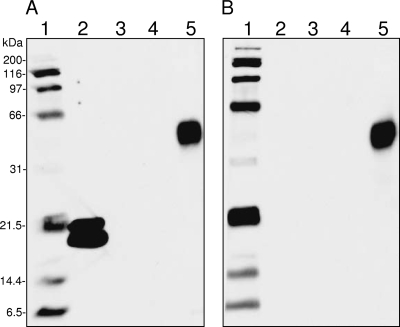
To verify the novel DNA-binding activity of BCP, Coxiella's bcp gene was cloned into pQE30 to create pQBCP, a plasmid containing an in-frame translational fusion of BCP to an N-terminal His6 tag. pQBCP was also used to synthesize rBCP for purification via nickel affinity chromatography. SW blots were prepared using rBCP and probed with biotinylated Coxiella genomic DNA as for Fig. 1. Control blots done without a DNA probe were run in parallel. The resulting SW blots clearly show that rBCP binds DNA (Fig. 2A, lane 2) and that the signal is dependent on the DNA probe (Fig. 2A versus 2B, lanes 2). SW blots consistently showed two molecular mass forms of rBCP (i.e., ∼19 kDa and 21.5 kDa). The unexpected observation that Com1 may be a DNA-binding protein (Fig. 1) was reexamined by using rCom1, prepared from E. coli strain LH600, on the SW blot. The results show that rCom1 does not bind DNA or give a signal under the conditions employed (Fig. 2A and 2B, lanes 3). One explanation for the apparent Com1 discrepancy between Fig. 1 and 2 is that the proteins represented in Fig. 1 were prepared for 2D SDS-PAGE using urea, while those in Fig. 2 were subjected to heat denaturing in Laemmli sample buffer for the 1D SDS-PAGE gel. Com1, a predicted oxidoreductase (thioredoxin), may have retained activity following urea denaturation (Fig. 1; Com1 signal), whereas it was lost upon heat denaturation (Fig. 2; no Com1 signal). Interestingly, carbonic anyhdrase in the molecular mass standard (Fig. 2, lanes 5) was clearly unaffected by heat denaturation and gave a peroxidase signal that was not dependent on the presence of a DNA probe. To ensure that reactivity in SW blots did not arise from the presence of the His6 tag, an irrelevant fusion protein, rHbpA (recombinant Bartonella quintana heme-binding protein A), was purified from E. coli strain LH700 and included on the SW blot. The results clearly show that rHbpA did not give a signal after being subjected to probing with the genomic DNA probe (Fig. 2A, lane 4).
FIG. 2.
One-dimensional SW blot demonstrating DNA binding by rBCP. Blots were prepared from 1D SDS-PAGE gels (12.5% [wt/vol] acrylamide) with 5 μg of protein per well. (A) SW blot probed with biotin-labeled, C. burnetii genomic DNA. (B) SW blot duplicating that shown in panel A but done without a biotin-labeled DNA probe. Lanes: 1, biotinylated SDS-PAGE standards (Bio-Rad); 2, rBCP; 3, rCom1; 4, rHbpA; 5, Kaleidoscope prestained standards (Bio-Rad). The carbonic anyhdrase standard in lanes 5 (∼40.3 kDa) demonstrates intrinsic peroxidase activity and generates a signal on the SW blot regardless of whether or not a DNA probe is employed. Blot signals were detected using enhanced chemiluminescence (ECL) reagents and a 3-min exposure.
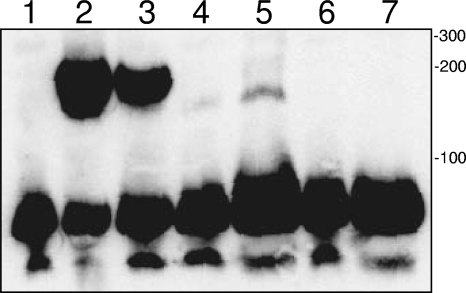
Since DNA binding has not been described for any 2-Cys Prx to date, we wanted to verify the activity by a different method. To that end, EMSAs were done using rBCP and a LightShift chemiluminescence EMSA kit (Pierce). EMSAs clearly demonstrated that rBCP bound to DNA in a concentration-dependent fashion, as shown by the protein's ability to retard electrophoretic migration of the kit's biotin-EBNA probe compared to the negative control (Fig. 3, lanes 4 and 5 versus lane 1). Retardation of the biotin-EBNA probe's mobility was also observed with a positive-control DNA-binding protein (EBNA extract) (Fig. 3, lane 2). Interestingly, DNA-binding activity of EBNA extract was resistant to heat (Fig. 3, lane 3) whereas rBCP activity was sensitive to heat denaturation at either of the concentrations tested (Fig. 3, lanes 6 to 7). Taken as a whole, the results of the two SW blot analyses (Fig. 1 and 2) and the EMSA (Fig. 3) clearly demonstrate that BCP bound DNA. To our knowledge, this is the first report of DNA-binding activity by a 2-Cys Prx.
FIG. 3.
EMSA demonstrating DNA binding by rBCP. Components of a LightShift chemiluminescence EMSA kit were used as instructed by the manufacturer (Pierce). A comparison of mobilities for the biotin-EBNA DNA probe following acrylamide gel electrophoresis, blotting to nylon, and visualization of the signal is shown (see text for details). Lanes show the results of reactions performed with a biotin-EBNA DNA probe. Lanes: 1, no DNA-binding protein (negative control); 2, EBNA extract (positive control containing known DNA-binding protein); 3, heat-denatured (hd) EBNA extract; 4, 2 μM rBCP; 5, 10 μM rBCP; 6, 2 μM hd-rBCP; 7, 10 μM hd-rBCP. Sizes (indicated to the right in base pairs) were determined by the use of a GeneRuler biotinylated DNA ladder (KPL, Gaithersburg, MD).
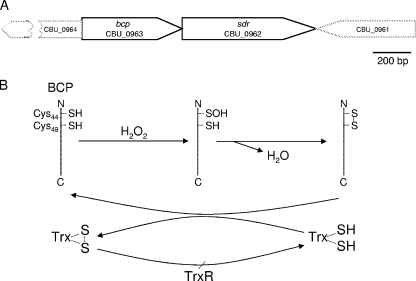
Because C. burnetii resides in a lysosome-like PV that is undoubtedly rich in ROS, we focused our efforts on characterizing the putative BCP Prx (CBU_0963), with the underlying hypothesis that it may serve as an antioxidant effector for the pathogen. in silico analysis of the bcp locus in the RSA493 genome (GenBank number AE016828) revealed several noteworthy characteristics (Fig. 4A). First, the last nucleotide of the bcp stop codon also serves as the “A” in the ATG start codon of an sdr gene located downstream (CBU_0962), encoding a putative short-chain dehydrogenase (Cluster of Orthologous Group COG4221; short-chain alcohol dehydrogenase). Second, a GenBank search revealed that the bcp-sdr linkage is unique to C. burnetii and conserved in all five sequenced genomes of the bacterium. Of note is that sdr is annotated as a pseudogene in CbuG_Q212 due to a frameshift (CbuG_1043; GenBank number NC_011527). Third, the CBU_0964 open reading frame (ORF) that occurs upstream of bcp is in the opposite orientation and is separated by only three nucleotides from bcp. The CBU_0961 ORF that lies downstream of sdr is in the opposite orientation and separated by only 10 nucleotides from sdr (Fig. 4A). These observations lead to several questions regarding regulation of the polygenic locus and suggest that bcp and sdr are cotranscribed due to their overlapping linkages. It is tempting to speculate that the predicted oxidoreductase encoded by the sdr gene immediately downstream of bcp also serves as an antioxidant effector.
FIG. 4.
(A) Linkage map showing C. burnetii bcp with proximal genes, derived from genomic sequence data for Nine Mile strain RSA 493 (22). Note the overlap of sdr with the 3′ end of bcp and the inverted but closely linked CBU_0964 and CBU_0961 ORFs. (B) A possible mechanism whereby BCP serves as an atypical 2-Cys Prx enzyme for hydroperoxides, based on the model of Kong et al. (16). A predicted role of Trx as an electron donor that reduces BCP through a cysteine thiol-disulfide exchange (plus TrxR for reduction of the Trx disulfide bond) is shown.
The results of a BLASTP search indicate that BCP is a Prx (a thioredoxin-dependent thiol peroxidase). BCP enzymes are widely utilized by pathogenic bacteria to detoxify hydroperoxides and/or organic peroxides. The predicted amino acid sequence of BCP possesses the conserved catalytic triad common to the BCP subfamily of Prx's (i.e., Thr42, Cys45, Arg121), wherein deprotonation of the catalytic (peroxidatic) Cys45 thiol would presumably be facilitated by H-bonding with the hydroxyl group of Thr42 and the positively charged Arg121 sidechain, as previously described (10, 29). The activity of the Coxiella BCP is predicted to conform to the process represented by the diagram shown in Fig. 4B. This scheme is based upon a model first proposed for a 2-Cys BCP ortholog of Sedum lineare (16) that shares many features with the Coxiella BCP, including two Cys residues (Cys45 and Cys50) in the N-terminal region that are separated by four amino acids and a dependency on Trx for activity (see Fig. 7).
FIG. 7.
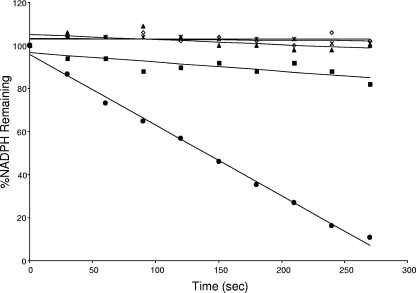
Assay demonstrating in vitro peroxidase activity of rBCP. Percentages of NADPH substrate remaining as a function of incubation time are shown. Reactions were done as described in the text but with reaction mixtures containing 3 μM rBCP (filled circle), 0 μM rBCP (open diamond), 3 μM heat-denatured rBCP (× with vertical line), 0 μM Trx (filled square), or 0 μM TrxR (filled triangle). The assay is a representative from three independent experiments.
Expression of bcp during infection of host cells.
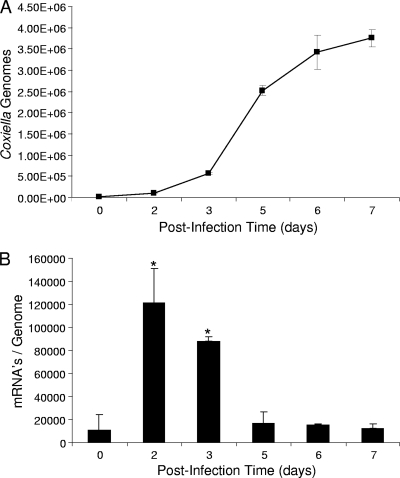
To better understand the role of BCP in Coxiella growth and metabolism, we quantified Coxiella genomic DNA and bcp mRNA transcript levels over the course of synchronous 7-day infections of cultured Vero cells. The results showed that bcp was maximally expressed in Vero cells at days 2 to 3, coinciding with the pathogen's early exponential-phase growth (Fig. 5A) and with the period when the large-cell-variant (LCV) morphotype predominates (8). With these data in mind, BCP likely functions as an antioxidant for endogenous peroxides generated during active replication. The qRT-PCR results (Fig. 5B) of this experiment are in good agreement with a previous proteomic analysis of C. burnetii developmental stage morphotypes, which showed that BCP, while present in the SCV proteome, was considerably more abundant (>2-fold greater abundance) in the LCV proteome (7).
FIG. 5.
bcp gene expression is maximal during early log-phase growth of C. burnetii in synchronized infections of cultured Vero cells. (A) Coxiella growth curve showing numbers of genome equivalents as a function of postinfection time, as determined by Q-PCR of the same source cultures used in qRT-PCR. (B) Corresponding qRT-PCR results, showing bcp mRNA transcript values normalized to the Coxiella genome equivalents. The qRT-PCR and Q-PCR values represent the means ± standard deviations of the results of 6 independent experiments. Asterisks denote a statistically significant increase in bcp mRNA relative to the day 0 postinfection value (P < 0.05).
trans-complementation of E. coli bcp by using cloned Coxiella bcp.
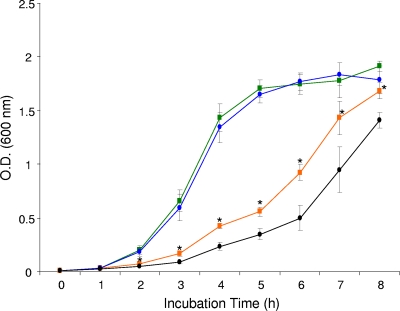
For functional expression of C. burnetii bcp and verification of BCP's role as an antioxidant in vivo, standard growth curves were determined in triplicate experiments using otherwise isogenic E. coli strains, including LH300 (wild-type E. coli transformed with pQE30; not shown), LH200 (bcp mutant transformed with pQE30), and LH100 (bpc mutant trans-complemented with pQBCP). Growth in LBamp showed OD600 values that were not significantly different between strains (P ≥ 0.05), although OD600 values for LH200 were consistently lower during exponential-phase growth (Fig. 6). In contrast, growth of these strains in an oxidative-stress growth medium (LBamp containing 0.2 mM t-BOOH) showed statistically significant differences. Specifically, the trans-complemented strain (LH100) produced significantly higher OD600 values at 2 to 8 h than the bcp mutant (LH200) (Fig. 6). In addition, LH100 showed significantly higher OD600 values than the wild type (LH300) at 2 to 4 h but was not significantly different from the wild type at 5 to 8 h (wild-type data not shown). The significantly greater growth of LH100 relative to the LH200 strain from 2 to 8 h was undoubtedly due to LH100's enhanced ability to deal with the exogenous t-BOOH through rBCP, especially considering the isogenic background of these strains.
FIG. 6.
Growth assays demonstrating functional expression of cloned Coxiella bcp and its ability to trans-complement E. coli bcp. Growth curves for an E. coli bcp mutant (LH200; •) and a trans-complemented daughter strain (LH100; ▪) are shown; the curve for the wild-type strain (LH300) was omitted for clarity. The upper lines (blue and green) depict growth in LBamp, while the lower lines (black and orange) show growth in an oxidative-stress growth medium (LBamp containing 0.2 mM t-BOOH). OD600 values represent the means of the results of 3 independent experiments ± standard deviations. Asterisks indicate a statistically significant (P < 0.05) difference between OD600 values at the indicated time points.
BCP's Prx activity and protection of DNA from oxidative damage.
The Prx activity of rBCP was assayed using various concentrations (0 to 3 μM) of enzyme and t-BOOH as a substrate, per several previous reports on Prx's. A representative experiment is shown in Fig. 7. Regardless of the enzyme concentration (1 to 3 μM), results consistently showed that rBCP is a heat-sensitive Prx that requires Trx and TrxR for in vitro activity. Trx is maintained in a reduced state by TrxR in an NADPH-dependent reaction in this assay (see Fig. 4B). The NADPH oxidation rate also directly correlated with the concentration of rBCP employed in these assays (data not shown).
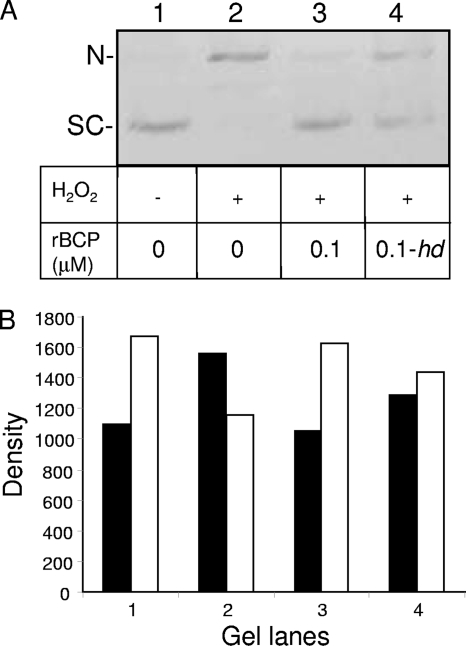
Given the DNA-binding capability of Coxiella BCP (Fig. 1 to 3), its ability to confer resistance to t-BOOH in vivo (Fig. 6), and its Prx activity in vitro (Fig. 7), we wanted to determine whether rBCP would protect supercoiled DNA from oxidative nicking by hydroxyl radicals, as previously reported for BCPs of Sulfolobus solfataricus (17). To this end, supercoiled pUC19 plasmid DNA was prepared and incubated with 0 or 6 mM H2O2 for 30 min at 37°C, in the presence of 0 or 0.1 μM rBCP. The results of the assay showed that supercoiled DNA was the predominant species in reactions lacking both H2O2 and rBCP (Fig. 8, lane 1). When H2O2 was added, the amount of the nicked form of the plasmid increased by ∼42% relative to the untreated control amount (Fig. 8, lane 2 versus lane 1) as a result of oxidative damage. Addition of 0.1 μM rBCP to the reactions decreased the amount of nicked DNA by ∼32% compared to reactions lacking enzyme (lane 3 versus lane 2). Interestingly, hd-rBCP was able to reduce oxidative damage by ∼17.5% when included in reactions (Fig. 8, lane 4 versus lane 2). Since heat denaturation destroys rBCP's DNA binding (Fig. 3) and Prx activity (Fig. 7), the residual oxidative protection conferred by hd-rBCP to DNA (Fig. 8) may have been due to the denatured protein's physical shielding or exclusion of H2O2 from the nucleic acid. Alternatively, hd-rBCP may have chelated trace iron ions present in the reaction buffer, thereby reducing the Fenton reaction and consequential oxidative damage to the DNA. When combined with other results of this study, the data suggest that BCP protects DNA through its DNA binding and Prx activity and, to a lesser extent, in an unknown manner that does not involve binding or enzyme activities. These results are in keeping with previous work showing that 1-Cys and 2-Cys Prx's can protect DNA from oxidative damage (17, 24).
FIG. 8.
rBCP protects supercoiled plasmid DNA from nicking by H2O2. (A) Supercoiled (SC) pUC19 DNA (100 ng) (lane 1) was treated with 6 mM H2O2 for 30 min, resulting in nicked (N) pUC19 (lane 2). Reactions were also done in the presence of 0.1 μM rBCP in its native (lane 3) or heat-denatured “hd” (lane 4) state. Control reactions (lanes 1 and 2) were used as comparators for densitometry. (B) Densitometric analysis of the gel lanes in panel A, where black bars represent the nicked form of pUC19 and white bars depict supercoiled DNA. The assay is a representative from three independent experiments.
Continued research on BCP and the other antioxidant cohorts enlisted by C. burnetii (e.g., SodB, SodC, AhpC, AhpD, 2-Cys Prx, and KatE) will provide additional clues regarding effectors required to parasitize professional phagocytes, establish the parasitophorous vacuole, and allow for replication in the harsh confines of this unusual intracellular niche.
Acknowledgments
We are grateful to Kate Sappington and Sherry Coleman for technical assistance and Bob Heinzen for the C. burnetii RSA 439 stock culture.
This work was supported by NIH Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Disease grant U54 AI065357-040023 to M.F.M.
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Akporiaye, E. T., and O. G. Baca. 1983. Superoxide anion production and superoxide dismutase and catalase activities in Coxiella burnetii. J. Bacteriol. 154:520-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley and Sons Inc., New York, NY.
- 3.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.008. [DOI] [PMC free article] [PubMed]
- 4.Baca, O. G., M. J. Roman, R. H. Glew, R. F. Christner, J. E. Buhler, and A. S. Aragon. 1993. Acid phosphatase activity in Coxiella burnetii: a possible virulence factor. Infect. Immun. 61:4232-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 6.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman, S. A., E. R. Fischer, D. C. Cockrell, D. E. Voth, D. Howe, D. J. Mead, J. E. Samuel, and R. A. Heinzen. 2007. Proteome and antigen profiling of Coxiella burnetii developmental forms. Infect. Immun. 75:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, S. A., E. R. Fischer, D. Howe, D. J. Mead, and R. A. Heinzen. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copley, S. D., W. R. P. Novak, and P. C. Babbitt. 2004. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry 43:13981-13995. [DOI] [PubMed] [Google Scholar]
- 10.Flohé, L., H. Budde, K. Bruns, H. Castro, J. Clos, B. Hofmann, S. Kansal-Kalavar, D. Krumme, U. Menge, K. Plank-Schumacher, H. Sztajer, J. Wissing, C. Wylegalla, and H. J. Hecht. 2002. Tryparedoxin peroxidase of Leishmania donovani: molecular cloning, heterologous expression, specificity, and catalytic mechanism. Arch. Biochem. Biophys. 397:324-335. [DOI] [PubMed] [Google Scholar]
- 11.Hackstadt, T. 1983. Estimation of the cytoplasmic pH of Coxiella burnetii and effect of substrate oxidation on proton motive force. J. Bacteriol. 154:591-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hackstadt, T., and J. C. Williams. 1981. Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 78:3240-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoya-Matsuda, N., K. Motohashi, H. Yoshimura, A. Nozaki, K. Inoue, M. Ohmori, and T. Hisabori. 2005. Anti-oxidative stress system in cyanobacteria. Significance of type II peroxiredoxin and the role of 1-Cys peroxiredoxin in Synechocystis sp. strain PCC 6803. J. Biol. Chem. 280:840-846. [DOI] [PubMed] [Google Scholar]
- 14.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 15.Jeong, W., M. K. Cha, and I. H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 16.Kong, W., S. Shiota, Y. Shi, H. Nakayama, and K. Nakayama. 2000. A novel peroxiredoxin of the plant Sedum lineare is a homologue of Escherichia coli bacterioferritin co-migratory protein (Bcp). Biochem. J. 351:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limauro, D., E. Pedone, I. Galdi, and S. Bartolucci. 2008. Peroxiredoxins as cellular guardians in Sulfolobus solfataricus: characterization of Bcp1, Bcp3 and Bcp4. FEBS J. 275:2067-2077. [DOI] [PubMed] [Google Scholar]
- 18.Mertens, K., J. Berg, and J. E. Samuel. 2009. Complementation of a natural katE mutation in Coxiella burnetii Nine Mile enhances growth in murine macrophages. 23rd Meet. Amer. Soc. Rickettsiol., abstr. 115.
- 19.Neidhardt, F. C., V. Vaughn, T. A. Phillips, and P. L. Bloch. 1983. Gene-protein index of Escherichia coli K-12. Microbiol. Rev. 47:231-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 21.Raghavan, R., L. D. Hicks, and M. F. Minnick. 2008. Toxic introns and parasitic intein in Coxiella burnetii: legacies of a promiscuous past. J. Bacteriol. 190:5934-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seshadri, R., I. T. Paulsen, J. A. Eisen, T. D. Read, K. E. Nelson, W. C. Nelson, N. L. Ward, H. Tettelin, T. M. Davidsen, M. J. Beanan, R. T. Deboy, S. C. Daugherty, L. M. Brinkac, R. Madupu, R. J. Dodson, H. M. Khouri, K. H. Lee, H. A. Carty, D. Scanlan, R. A. Heinzen, H. A. Thompson, J. E. Samuel, C. M. Fraser, and J. F. Heidelberg. 2003. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 100:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siemsen, D. W., L. N. Kirpotina, M. A. Jutila, and M. T. Quinn. 2009. Inhibition of the human neutrophil NADPH oxidase by Coxiella burnetii. Microbes Infect. 11:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacy, R. A., E. Munthe, T. Steinum, B. Sharma, and R. B. Aalen. 1996. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol. Biol. 31:1205-1216. [DOI] [PubMed] [Google Scholar]
- 25.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-60. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, DC.
- 26.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdoucq, L., F. Vignols, J. P. Jacquot, Y. Chartier, and Y. Meyer. 1999. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 274:19714-19722. [DOI] [PubMed] [Google Scholar]
- 28.Wang, G., A. A. Olczak, J. P. Walton, and R. J. Maier. 2005. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect. Immun. 73:378-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood, Z. A., E. Schröder, J. R. Harris, and L. B. Poole. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]
- 30.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]