We learn metabolic biochemistry largely as a series of pathways such as glycolysis, the citric acid cycle, the histidine biosynthetic pathway, etc., where each step in the pathway is catalyzed by a specific enzyme. Exceptions to this are known and are increasing. In the work by Kozmin et al. (12) directed at determining the pathway that allows Escherichia coli cells to protect themselves against toxic 6-N-hydroxylaminopurine (HAP), they have demonstrated that only the CysJ component of the sulfite reductase complex (CysJ8CysI4 or α8β4) serves along with YcbX in the reduction of HAP back to nontoxic adenine that can enter back into metabolism via a purine-salvaging pathway. Before this work, YcbX was predicted to be only a [Fe2-S2] cluster-containing protein. Their work not only confirms the function of the ycbX open reading frame in E. coli but shows that this enzyme also requires the molybdenum cofactor (MoCo) to function. The work thus demonstrates that the CysJ portion of the sulfite reductase complex can have multiple roles in cells by supplying reducing equivalents to other enzymes. This raises the possibility that other examples of redox carrier proteins functioning with multiple acceptor proteins are waiting to be discovered. This observation of multiple functions is not to be confused with so-called moonlighting enzymes, where more than one function is found in a single polypeptide chain (9), since here the enzyme is still performing the same function. Another protein implicated in this detoxification of hydroxylamines is YiiM, which the authors show also to be a MoCo-dependent enzyme but one that is not reduced by CysJ. Thus, the work also indicates the presence of two previously unknown MoCo-dependent enzymes.
Role of CysJ in sulfite reductase.
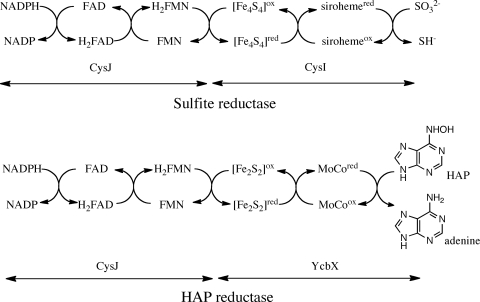
To understand how CysJ could function in the HAP reductase, one must consider how it functions in sulfite reductase. Early work showed that the α-chain of the E. coli sulfite reductase (CysJ) contained the NADPH, flavin adenine dinucleotide (FAD), and flavin mononucleotide (FMN) binding sites and that the β-chain (CysI) contained iron, sulfide, and siroheme. There were indications that the iron in the siroheme was the site at which the sulfite is bound for reduction. The electron flow was proposed early on to proceed as follows: NADPH → FAD → FMN → siroheme → sulfite (18). After the crystal structure of the β-chain CysI was obtained, it was clear that a [Fe4-S4] center was likely used to transfer the electrons from the flavins of the CysJ through a single cysteine thiol to the siroheme, as shown in Fig. 1 (4). Recently the monomeric ferredoxin-dependent sulfite/nitrite reductase structure was determined (17), and despite the fact that it has only 23% sequence similarity to the E. coli enzyme, the structures of the enzyme-coenzyme complexes in the two enzymes are identical and both contain the single cysteine thiol linking the [Fe4-S4] cluster to the siroheme (17). In neither of these structures is the nature of the possible interaction with CysJ revealed, since no CysJ is present.
FIG. 1.
Comparison of electron flow in sulfite dehydrogenase with what may be occurring in the CysJ-YcbX complex. Note that with sulfite reductase, three NADPHs are required to reduce one sulfite, whereas with HAP reductase, only one NADPH is required to reduce one HAP.
Likely electron transfer to MoCo and its function in YcbK.
In its role in protecting against N-hydroxylated base analogs, CysJ functions as a partner with the YcbX molybdoenzyme, where it catalyzes the NADPH-dependent reduction of its contained FAD and FMN cofactors and also very likely facilitates electron transfer to the MoCo center of YcbX, with the electrons localized at the reduced molybdenum center of the enzyme. These electrons are then used for the reduction of the hydroxylated amine back to the functional amine. The essence of this reaction is typical for molybdenum cofactor-containing enzymes that catalyze a net transfer of an oxygen atom from or to a substrate in a two-electron process (7, 8). The atom to which this oxygen is attached can be carbon as in the case of xanthine oxidase, sulfur as in the case of sulfite oxidase, or nitrogen as in the case of nitrate reductase. The reaction catalyzed by YcbX is unique because in this reaction a hydroxylamine is reduced to an amine, unlike the case for nitrate reductase, where a nitrate group is reduced to a nitrite group. That this transfer likely occurs through the formation of a protein complex between CysJ and YcbX has been indicated through the work identifying all the protein complexes in E. coli (2).
Nature of the possible interaction between CysJ and YcbX.
From what has been presented here and in the paper by Kozmin et al. (12), one could infer that electron transfer from the FMN of the CysJ directly to the [Mo-S2(3)] center of YcbX could occur. This type of transfer has never been observed before. However, when one looks at homologs of YcbX, it is clear, due to conserved cysteine residues in their sequences, that they all have the potential to contain multiple [Fe-S] clusters. This being the case, the electrons from FMN can be transferred through the [Fe-S] cluster(s) on their way to MoCo. What is really interesting is that the enzyme has had to evolve the ability to donate electrons not only to YcbX but also to CysI and maybe even other enzymes. This aspect of the enzyme is thus like that seen with ferridoxins, thioredoxin, and glutaredoxin, which can donate electrons to a wide variety of different proteins, and is thus not that special (13). Each of these redox carriers is placed in a reduced state by a separate oxidoreductase such as ferridoxin NADPH oxidoreductase or thioredoxin reductase. The CysJ is then like a thioredoxin reductase and thioredoxin combination, where a single enzyme extracts reducing power from NADPH, transferring it to FMN for transfer to the acceptor protein. The redox transfer reactions are also different from those of these enzymes, since they use thiols or [Fe4-S4] clusters as their redox carriers. CysJ is different, since the flavin transfers the electrons to the acceptor. Transfer of electrons from flavins to [Fe-S] clusters is well known in many different proteins. An excellent example of this is found in the dihydropyrimidine dehydrogenases, where both FAD and FMN transfer electrons in and out of a string of [Fe4-S4] clusters in the same enzyme (16). What is different here is the apparent transfer of electrons from flavins in one enzyme to MoCo in another enzyme through [Fe2-S2] centers. The MoCo can be oriented in such a way that it can participate directly in electron transfer to and from different cofactors (10).
Possible electron flow in the HAP reductase.
The native protein is either an α8β4 or α8β8 complex (18, 20), and no structure is currently known for the intact complex. As a result we do not know how the flavin in the CysJ may be located in the CysJ-YcbX complex next to the [Fe-S] cluster in YcbX. The flavoprotein component of the enzyme contains two prosthetic groups, one FAD binding site and one FMN binding site (5, 6), with each binding site in a different domain that evolved from different proteins (15). The FAD is associated with a ferredoxin-NADP+ binding site, and the FAD binding domain is homologous to bacterial flavodoxins. The enzyme belongs to a family of electron transfer flavoproteins that includes NADPH-cytochrome P450 reductase, nitric oxide synthase, cytochrome P450, and methionine reductase.
Source and toxicity of N-hydroxylated compounds.
N-hydroxylated base analogs are stated in the paper by Kozmin et al. (12) to be “formed in vivo as a consequence of normal cellular metabolism or produced by chemical and physical factors, such as alkylating agents or ionizing radiation.” I could not find any examples where this has been shown for HAP production. This work was thus done with a compound that appears to have never been identified as a natural product. The compound can, however, be generated by the microsomal N-hydroxylation of adenine (3) and has been used extensively as a very strong mutagen in bacterial, fungal, and mammalian cells (1, 11, 14). Considering the apparently small amount of HAP that may be present in natural systems, it is likely that HAP is not the natural substrate for this enzyme.
The paper by Kozmin et al. (12) also establishes a possible function for the CysJ-YcbX “hybrid” protein encoded by the genomes of two Vibrio species and demonstrates once again that the functional assignment of genomes always requires experiments (19).
Acknowledgments
The National Science Foundation (grant MCB0722787) supported this work.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Barrett, J. C. 1981. Induction of gene mutation in and cell transformation of mammalian cells by modified purines: 2-aminopurine and 6-N-hydroxylaminopurine. Proc. Natl. Acad. Sci. U. S. A. 78:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. H. Yang, X. C. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 3.Clement, B., and T. Kunze. 1990. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem. Pharmacol. 39:925-933. [DOI] [PubMed] [Google Scholar]
- 4.Crane, B. R., L. M. Siegel, and E. D. Getzoff. 1995. Sulfite reductase structure at 1.6 A: evolution and catalysis for reduction of inorganic anions. Science 270:59-67. [DOI] [PubMed] [Google Scholar]
- 5.Eschenbrenner, M., J. Coves, and M. Fontecave. 1995. The flavin reductase-activity of the flavoprotein component of sulfite reductase from Escherichia coli—a new model for the protein-structure. J. Biol. Chem. 270:20550-20555. [DOI] [PubMed] [Google Scholar]
- 6.Eschenbrenner, M., J. Coves, and M. Fontecave. 1995. NADPH-sulfite reductase flavoprotein from Escherichia coli: contribution to the flavin content and subunit interaction. FEBS Lett. 374:82-84. [DOI] [PubMed] [Google Scholar]
- 7.Hille, R. 1996. The mononuclear molybdenum enzymes. Chem. Rev. 96:2757-2816. [DOI] [PubMed] [Google Scholar]
- 8.Hille, R. 1994. The reaction mechanism of oxomolybdenum enzymes. Biochim. Biophys. Acta 1184:143-169. [DOI] [PubMed] [Google Scholar]
- 9.Jeffery, C. J. 2009. Moonlighting proteins—an update. Mol. Biosyst. 5:345-350. [DOI] [PubMed] [Google Scholar]
- 10.Kisker, C., H. Schindelin, D. Baas, J. Retey, R. U. Meckenstock, and P. M. H. Kroneck. 1998. A structural comparison of molybdenum cofactor-containing enzymes. FEMS Microbiol. Rev. 22:503-521. [DOI] [PubMed] [Google Scholar]
- 11.Kozmin, S. G., R. M. Schaaper, P. V. Shcherbakova, V. N. Kulikov, V. N. Noskov, M. L. Guetsova, V. V. Alenin, I. B. Rogozin, K. S. Makarova, and Y. I. Pavlov. 1998. Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 402:41-50. [DOI] [PubMed] [Google Scholar]
- 12.Kozmin, S. G., J. Wang, and R. M. Schaaper. 2010. A role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-hydroxylaminopurine. J. Bacteriol. 192:2026-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer, Y., B. B. Buchanan, F. Vignols, and J. P. Reichheld. 2009. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet 43:335-367. [DOI] [PubMed] [Google Scholar]
- 14.Pavlov, Y. I., V. N. Noskov, E. K. Lange, E. V. Moiseeva, M. R. Pshenichnov, and N. N. Khromov-Borisov. 1991. The genetic activity of N-6-hydroxyadenine and 2-amino-N-6-hydroxyadenine in Escherichia coli, Salmonella typhimurium and Saccharomyces cerevisiae. Mutat. Res. 253:33-46. [DOI] [PubMed] [Google Scholar]
- 15.Porter, T. D., and C. B. Kasper. 1986. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry 25:1682-1687. [DOI] [PubMed] [Google Scholar]
- 16.Schnackerz, K. D., D. Dobritzsch, Y. Lindqvist, and P. F. Cook. 2004. Dihydropyrimidine dehydrogenase: a flavoprotein with four iron-sulfur clusters. Biochim. Biophys. Acta 1701:61-74. [DOI] [PubMed] [Google Scholar]
- 17.Schnell, R., T. Sandalova, U. Hellman, Y. Lindqvist, and G. Schneider. 2005. Siroheme- and [Fe4-S4]-dependent NirA from Mycobacterium tuberculosis is a sulfite reductase with a covalent Cys-Tyr bond in the active site. J. Biol. Chem. 280:27319-27328. [DOI] [PubMed] [Google Scholar]
- 18.Siegel, L. M., and P. S. Davis. 1974. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. IV. The Escherichia coli hemoflavoprotein: subunit structure and dissociation into hemoprotein and flavoprotein components. J. Biol. Chem. 249:1587-1598. [PubMed] [Google Scholar]
- 19.White, R. H. 2006. The difficult road from sequence to function. J. Bacteriol. 188:3431-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeghouf, M., M. Fontecave, and J. Coves. 2000. A simplifed functional version of the Escherichia coli sulfite reductase. J. Biol. Chem. 275:37651-37656. [DOI] [PubMed] [Google Scholar]