Abstract
ohrR encodes an organic hydroperoxide sensor and a transcriptional repressor that regulates organic hydroperoxide-inducible expression of a thiol peroxidase gene, ohr, and itself. OhrR binds directly to the operators and represses transcription of these genes. Exposure to an organic hydroperoxide leads to oxidation of OhrR and to subsequent structural changes that result in the loss of the repressor's ability to bind to the operators that allow expression of the target genes. Differential induction of ohrR and ohr by tert-butyl hydroperoxide suggests that factors such as the repressor's dissociation constants for different operators and the chemical nature of the inducer contribute to OhrR-dependent organic hydroperoxide-inducible gene expression. ohrR and ohr mutants show increased and decreased resistance to organic hydroproxides, respectively, compared to a parental strain. Moreover, the ohrR mutant had a reduced-virulence phenotype in the Pseudomonas aeruginosa-Caenorhabditis elegans pathogenicity model.
Pseudomonas aeruginosa is an important opportunistic bacterial pathogen that causes a variety of diseases. The incidence of nosocomial P. aeruginosa infection has increased worldwide (23, 32). The ability of a pathogen to successfully invade the host is often linked to its capacity to overcome host defenses, including the oxidative burst that occurs during host-pathogen interactions (15). Reactive oxygen species produced by host defense mechanisms include hydrogen peroxide (H2O2), superoxide anion, and organic hydroperoxide. In P. aeruginosa, the defense mechanisms against H2O2 have been well studied. Exposure of the bacteria to sublethal levels of H2O2 leads to induction of genes involved in oxidative stress protective pathways, including catalases and alkyl hydroperoxide reductases (5, 28, 31). Peroxide-inducible gene expression is regulated by the peroxide sensor and transcription regulator OxyR (26).
Less is known about the response of P. aeruginosa to organic hydroperoxides. OxyR-regulated genes are upregulated in the presence of organic hydroperoxides (26). In addition, inactivation of alkyl hydroperoxide reductase genes leads to an increase in sensitivity toward both cumene hydroperoxide (CHP) and H2O2 (26). An organic hydroperoxide resistance gene (ohr) encoding a thiol peroxidase is highly expressed in response to exposure to sublethal concentrations of organic hydroperoxides (3, 25). Inactivation of ohr renders the mutant bacteria hypersensitive to organic hydroperoxides (3, 25). In vivo, unlike catalase and alkyl hydroperoxide reductase, Ohr seems to have a very specific role in detoxification of organic hydroperoxides but not of H2O2 (6, 11, 25, 29). In vitro, Ohr also catalyzes H2O2 degradation, albeit at a rate 20 times lower than that for the degradation of an organic hydroperoxide (7).
It has been shown in Bacillus subtilis, Xanthomonas campestris, Agrobacterium tumefaciens, and Streptomyces coelicolor that ohr is regulated by OhrR (6, 11, 27, 29). OhrR in Gram-negative bacteria belongs to the two-cysteine OhrR family. Analysis of OhrR revealed that the conserved N-terminal cysteine residue is essential for the redox sensing mechanism of OhrR (12, 24, 30). The current model for the sensing of organic hydroperoxides by members of the two-cysteine OhrR family involves an initial oxidation of the N-terminal cysteine to an unstable sulfenic acid intermediate by the organic hydroperoxide, followed by disulfide bond formation with another conserved C-terminal cysteine (30). Structural analysis has shown that the oxidized form of OhrR undergoes a major conformational change, including the rotation of the winged helix, resulting in dissociation from DNA (24). For the one-cysteine family of OhrR, oxidation of cysteine leads to cysteine-sulfenic acid and to the formation of a mixed disulfide bond with low-molecular-weight intracellular thiols, for example, cysteines, coenzyme A, and a structurally uncharacterized 398-Da thiol (12, 17). These structural changes and DNA dissociation permit RNA polymerase to bind to the promoter and to commence gene expression.
In several bacteria, ohr has a major role in protection of the cells from organic hydroperoxides. We report here the characterization of OhrR, a transcription regulator of ohr in P. aeruginosa. Expression analysis of ohrR, determination of the DNA binding site, and investigation of the sensing mechanism of OhrR were performed. The physiological and pathological roles of P. aeruginosa ohrR were also investigated.
MATERIALS AND METHODS
Construction of mutant strains.
The pKsΔohr::tet plasmid used to disrupt the ohr gene in P. aeruginosa PAO1 was constructed as follows. Two sets of primers were designed to amplify the predicted Ohr coding sequence plus additional flanking regions from the PAO1 genome. The 797-bp upstream fragment containing the 5′ region of ohr was amplified by PCR with the primers BT512 (5′-TCGGAATTCCGCCGCCCTGGG 3′; EcoRI site underlined) and BT514 (5′-TCACGGCCTCCGGTGGCG-3′). The 720-bp downstream fragment containing the 3′ region of ohr was amplified by PCR with the primers BT1612 (5′-CGCCACCGGAGGCCGTGAACGTGTCGGTCTGAAGCTTCCGAC-3′; BT514 complementary sequence underlined) and BT1613 (5′-GCAGATTGGCTATGATACGC-3′). Primer BT1612 contained sequences complementary to BT514. The PCR products were used in a joining PCR, and the joined product was cloned into the pBluescript II KS(−) (Stratagene) vector between the EcoRV and HincII sites, yielding pKsΔohr. The MluI-digested Tet resistance cassette was excised from the pKNOCK-Tet plasmid (1) and was subcloned into pIC20H (18) at the EcoRV site. This new plasmid, pIC20H-Tet, contained a HindIII site at each end of the cassette. The Tet cassette was then excised using HindIII and was inserted between the upstream and downstream fragments of ohr in pKsΔohr at the HindIII site. The resulting plasmid, pKsΔohr::tet, was linearized and was transformed into PAO1 by electroporation. The expected double recombination event creating a gene knockout was selected using tetracycline-containing medium and was then screened for using carbenicillin sensitivity. Gene replacement in mutant clones was confirmed by PCR and Southern blot analysis.
The unmarked deletion of PAO1 ohrR was constructed using a Cre-lox system (19). A 1,688-bp fragment containing ohrR was amplified with the primers BT1433 (5′-GCAGCTTCATCGACCTGCAG-3′) and 263 (5′ AGTCGGAAGCTTCAGAC 3′) and was cloned into pUC18 at the PvuII sites that had been filled in, yielding pUCohrR. An EcoRI and EcoICRI fragment containing the Gm resistance cassette flanked with lox sites from pCM357 (19) was gap filled using Klenow fragment polymerase. This fragment was cloned into pUCohrR at blunt-end PvuII-BstEII sites, yielding pUCΔohrR::Gm. The PvuII-BstEII digestion deleted 378 bp of the coding region of ohrR; only 54 bp at the 5′ end and 23 bp at the 3′ end of the ohrR gene remained. The pUCΔohrR::Gm plasmid was transformed into PAO1. To select the double crossing-over event, gentamicin resistance colonies were selected and were then screened for carbenicillin-sensitive phenotypes. The ohrR mutant with the antibiotic cassette replacement was then transformed with the pCM157 (19) vector containing the Cre-encoding gene. Cre, a site-specific recombinase, recognizes lox sites, and recombination between these sites deletes the DNA between the two sites. The transformants with the gentamicin cassette deletion or a gentamicin-sensitive phenotype contained the unmarked deletion of ohrR. pCM157 was cured by growing cells under a nonselective condition for several generations. The deletion of ohrR was confirmed by PCR and Southern blot analysis.
Complementation plasmids.
A 457-bp DNA fragment containing full-length ohr and a ribosome binding site was generated by PCR using the primers 262 (5′-TCAGACAGGTGACTCTC-3′) and 263 (5′-AGTCGGAAGCTTCAGAC-3′). This fragment was cloned into a broad-host-range vector, pBBR1MCS-4 (16), at the SmaI site to yield the plasmid pohr. For construction of a full-length ohrR gene and its ribosome binding site, 489-bp PCR fragments generated from the primers 260 (5′-CTTGGAAGACAACCATG-3′) and 261 (5′-GACAGGTAGGTATAAGCCCCT-3′) were cloned into pBBR1MCS-4 at the SmaI site. This plasmid was named pohrR. The direction of fusion was checked by PCR to ensure that the gene was fused to the Escherichia coli lacUV5 promoter present in the pBBR1MCS-4 vector. The insertions were then checked by sequencing. pohr or pohrR was introduced into PAO1 strains by electroporation and selected for ampicillin resistance colonies.
Promoter activity analysis.
In order to clone the promoter regions of the ohr and ohrR genes, the corresponding DNA fragments were amplified using genomic DNA from PAO1 as a template. The oligonucleotides used for PCR amplification of the ohr and ohrR promoters were BT513 (5′-ACCGAATTCAGGGGCTTATAC-3′; EcoRI site underlined) and BT514 (5′-TCACGGCCTCCGGTGGCG-3′) and BT512 (5′-TCGGAATTCCGCCGCCCTGGG-3′; EcoRI site underlined) and BT601 (5′-ATGCAGGCGGGAATGCTC-3′), respectively. The 210-bp PCR product generated from BT513 and BT514 contained a 201-bp ohr promoter fragment covering the −87 to +114 region of ohr. The 515-bp PCR product generated from BT512 and BT601 contained a 506-bp ohrR promoter fragment covering the −112 to +394 region of ohrR. The PCR fragments were cloned into the pDrive cloning vector (Qiagen). pDrive with the ohr or ohrR promoter inserted in the opposite direction to the lacZ cassette was chosen. The ohr or ohrR promoter was excised from pDrive with BamHI and EcoICRI and then directionally cloned 5′ of the lacZ cassette in pUC18Sfi lacZ, which was digested with SmaI and BamHI. The HindIII fragment from pUC18Sfi Psohr-lacZ containing the ohr-lacZ fragment or the HindIII fragment from pUC18Sfi PsohrR-lacZ containing the ohrR-lacZ fragment was then cloned into a broad-host-range expression vector, p027Ery, to give p027Psohr-lacZ and p027PsohrR-lacZ, respectively. The promoter fragment was checked by sequencing. The plasmid was then introduced into P. aeruginosa strains by electroporation and selected for erythromycin resistance colonies. pUC18Sfi lacZ and p027Ery were constructed for this study as followed. An SfiI-digested trp′-lacZ cassette from pUTmini-Tn5 lacZ1 (10) was cloned into pUC18Sfi (13) at the SfiI sites to yield pUC18Sfi lacZ. The first step for construction of p027Ery was the insertion of an EcoRI-digested erythromycin cassette from pIM13 (22) into a pKK223-3 derivative, containing multiple cloning sites from pIC20H (18), at the EcoRI site. The erythromycin cassette was then digested with a BamHI-EcoRV fragment and cloned into pIC20H at BamHI-SmaI. The PstI-BglII fragment of the erythromycin cassette from pIC20HEry was further inserted into p027tet (9) at PstI-BamHI, to yield p027Ery. The p027Ery plasmid contains the pSa origin of DNA replication, the partition locus parA from the Agrobacterium plasmid pTAR, an erythromycin resistance selection marker, and a lacZ alpha cassette with cloning sites (9).
Semiquantitative analysis of oxidant sensitivity.
Approximately 2 × 109 washed cells taken from an exponentially growing culture were mixed with 10 ml of Luria-Bertani (LB) semisoft agar (0.75%) and were then poured onto plates containing 50 ml 1.5% LB agar. Paper discs (6-mm diameter) soaked with 10 μl of the indicated oxidant were placed on top of the plates. Plates were incubated overnight at 37°C, and the diameters of the cleared zones were measured. Alternatively, logarithmically growing cells were serially diluted, and 10 μl of each dilution was spotted onto solid LB medium containing oxidants. The ability of bacteria to grow on an oxidant-containing medium was recorded after an overnight incubation at 37°C.
Northern blot analysis.
Total RNA was extracted from exponential-phase cells (optical density at 600 nm [OD600] of ∼0.4) cultured in Luria-Bertani broth using the hot acid phenol method (21). Expression of specific transcripts was measured by Northern blot analysis. Briefly, 10 μg of each RNA sample was subjected to electrophoresis on a 1.2% formaldehyde gel and was then transferred to a Hybond-XL membrane (Amersham Biosciences). Membranes were prehybridized for 30 min at 55°C in hybridization buffer (0.5 M phosphate buffer [pH 7.0], 7% sodium dodecyl sulfate [SDS], 1 mM EDTA, and 1% bovine serum albumin [BSA]) and were then hybridized at 55°C overnight to a randomly primed [α-32P]dCTP labeling probe. Membranes were washed twice at 25°C for 5 min under low-stringency conditions (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.1% SDS) and once at 55°C under high-stringency conditions (0.5× SSC plus 0.1% SDS). Washed membranes were exposed to X-ray film overnight. DNA probes containing the internal regions of ohr (oligonucleotides 262 and 263) and ohrR (oligonucleotides 260 and 261) were used.
RT-PCR.
Total RNA samples were prepared as described for Northern blot analysis experiments except that contaminating DNA in the total RNA samples was hydrolyzed by addition of DNase using the Ambion Turbo DNA-free kit (Applied Biosystems) according to the manufacturer's instructions. The quality of the RNA was assessed at this step by using agarose gel electrophoresis. To control for DNA contamination, each DNase-treated RNA sample was used as a template in a standard PCR assay as a control. The absence of PCR products in this control reaction indicated that the reverse transcription-PCR (RT-PCR) products were not derived from contaminating DNA. The reverse transcription reaction was performed using a RevertAid Moloney murine leukemia virus (M-MuLV) reverse transcriptase kit (Fermentas) according to the manufacturer's instructions using a random hexaprimer. The PCR was performed using gene-specific primers. For proof of the operonic structure, the primers BT468 (5′-TCCGAGGATACCGCACGG-3′) and BT484 (5′-AGGGGATCCTACCTACCTGTCTGG-3′; BamHI site underlined) were used. The expected PCR product was 222 bp in size. For expression of the ohr gene, BT1554 (5′-GCACTCCGCGCGAACTGG-3′) and BT1555 (5′-CGGCAGGTTGATGTGCAG-3′) were used with the expected product size of 218 bp. The amount of template used in each reaction was controlled by a PCR using BT2828 (5′-AACTGGAGGAAGGTGGGGAT-3′) and BT2829 (5′-AGGAGGTGATCCAACCGCA-3′), which are specific to the 16S rRNA gene, with an expected product size of 371 bp. A 100-bp marker (Fermentas) was used as the DNA size marker.
Primer extension.
The primers used to detect the start sites of the ohr and ohrR mRNAs were BT468 (5′-TCCGAGGATACCGCACGG-3′) and BT467 (5′-ATACAGGGCGAAGCACAG-3′), respectively. A primer, BT467, complementary to nucleotide sequences between +52 and +69 relative to the start codon of OhrR and a BT468 primer located between +60 and +79 of ohr were used in the extension reactions. Two picomoles of primer was annealed to 10 μg of total RNA preparation. The primers were previously labeled at their 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase. Primer extension reactions were carried out by incubating the annealing mixture with Superscript III reverse transcriptase (Invitrogen). The products were analyzed by 6% polyacrylamide gel electrophoresis (PAGE). A sequence ladder was generated using the fmol DNA cycle sequencing system (Promega) with pGEM-3zf(+) as a DNA template and M13 forward (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) as a primer.
Site-directed mutagenesis of C19 and C121 of OhrR.
Site directed mutagenesis was done by the PCR mutagenesis method. Primers containing mutated sites at C19 and C121 were designed. The primers used were as follows: BT859 (5′-ACCCAGCAGTCCTTCGCCCTG-3′) and BT860 (5′-CAGGGCGAAGGACAGCTGGTT-3′) for mutation of cysteine 19 into serine and BT861 (5′-ATTCCCGCCTCCATCGTCGAG-3′) and BT862 (5′-CTCCAGGATGGAGGCGGGAAT-3′) for mutation of cysteine 121 into serine. Changed bases are underlined, and members of each pair are complementary to each other. To construct OhrR containing C19S, BT859 and M13, forward primers, and BT860 and M13, reverse primers, were used in two separate amplification reactions with pohrR as a template. The second round of amplification reaction contained the two DNA fragments produced above without template and primers. This reaction allows the product to join. After 5 cycles, M13 forward and reverse primers were added to the reaction, and the product was amplified. The desired fragment from the second-round PCR was digested with EcoRI and SacI and cloned into pBBRMCS-4. The resultant plasmid was sequenced and designated pohrR-C19S. pohrR-C121S was generated using the same strategy except that the PCR primers BT861 and BT862 were used in the PCR.
Purification of OhrR.
Based on the genome sequence of PAO1, the primers BT602 (5′-TGGAAGACAACCATGGCCCG-3′, NcoI site underlined) and 261 (5′-GACAGGTAGGTATAAGCCCCT-3′) were designed and used in a PCR with PAO1 genomic DNA, pohrR-C19S, or pohrR-C121S as a template. The purified OhrR fragment was digested with NcoI and ligated to the pETBlue2 expression vector at NcoI and HincII to give a recombinant plasmid, pET-PsOhrR. The mutated ohrR(C19S) and ohrR(C121S) genes were similarly produced using PCR with plasmid templates before PCR fragments cloned into the expression vector. ohrR in the expression vector was sequenced to assess the accuracy of the gene sequence. pET-PsOhrR, pET-PsOhrR-C19S, and pET-PsOhrR-C121S were transformed into the BL21(DE3) strain. These transformants were used for protein production. Essentially, BL21(DE3) containing pET-PsOhrR was grown in 200 ml Luria-Bertani broth containing 100 μg/ml ampicillin at 37°C on a rotary shaker until the cell density reached an OD600 of about 0.6 to 0.8. Then, isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to the culture, and growth was continued for 2 h. Cells were harvested with centrifugation, washed, and resuspended in KEG buffer (25 mM KPO4, 1 mM EDTA, and 5% glycerol, pH 8.0) containing 1 mM dithiothreitol (DTT). Cells were disrupted by sonication at 0°C. The lysates were clarified by centrifugation and loaded onto a column of DEAE previously equilibrated with the same resuspension buffer. Nonabsorbed materials were removed by passage of this buffer through the column. OhrR was eluted with KEG buffer containing 100 mM KPO4 and 1 mM DTT. After dialysis in resuspension buffer, this fraction was loaded onto a heparin Sepharose column preequilibrated with the resuspension buffer. After the column was washed with resuspension buffer, OhrR was eluted with KEG buffer containing 200 mM KPO4 and 1 mM DTT. Then, this fraction was dialyzed, concentrated, and stored in KEG buffer containing 20% glycerol and 1 mM DTT. The purity of OhrR was estimated to be at about 90% on a Coomassie blue-stained SDS-polyacrylamide gel.
DNA binding assays.
Promoter fragments were end labeled with [γ-32P]ATP using T4 polynucleotide kinase. The ohrR promoter fragment was 194 bp long and was produced by PCR using 32P-labeled BT483 (5′-CTGGGATCCGGCCCTGCAGCA-3′; BamHI site underlined) and BT467 (5′-ATACAGGGCGAAGCACAG-3′), covering the region of ohrR from −87 to +98. The ohr promoter fragment was produced by PCR using 32P-labeled BT484 (5′-AGGGGATCCTACCTACCTGTCTGG-3′; BamHI site underlined) and BT514 (5′-TCACGGCCTCCGGTGGCG-3′) covering the region of the ohr gene from −87 to +114. DNA binding reactions were carried out at room temperature for 15 min in a mixture containing 20 mM Tris-Cl (pH 7.0), 50 mM KCl, 0.5 mM dithiothreitol, 1 mM EDTA, 5% glycerol, 50 μg/ml BSA, 5 μg/ml calf thymus DNA, 0.025 U poly(dI-dC) (Amersham Biosciences), γ-32P-labeled promoter fragment, and various amounts of OhrR. Samples were then analyzed on a 5% polyacrylamide gel in 0.25× Tris borate-EDTA buffer containing 2.5% glycerol. The gel was dried and analyzed by autoradiography. For DNase I footprinting analysis, the binding reactions were treated with DNase I, and the DNA fragments were analyzed on a 6% denaturing polyacrylamide gel (12). A sequence ladder was generated using an fmol DNA cycle sequencing system (Promega). The template for the sequencing ladder was pGEM-3zf(+) or the promoter fragment itself.
Pathogenicity test (8).
Brain heart infusion medium (BHI) was used to analyze the fast killing mechanism. Freshly streaked bacterial strains on BHI agar (Becton, Dickinson and Company) were resuspended in BHI broth and adjusted to an OD660 of 0.1. A 150-μl aliquot of this suspension was spread on a 5.5-cm-diameter BHI agar plate in triplicate, and the plates were then incubated at 37°C overnight to form lawns of bacteria. Approximately 100 to 200 synchronized adult worms (Caenorhabditis elegans Bristol N2 type strain, a gift from B. P. Braeckman, Biology Department, Ghent University, Belgium) were added to each plate. The numbers of live and dead/paralyzed worms were recorded over time under a microscope at room temperature (22°C).
RESULTS
Transcription organization of the ohrR-ohr operon.
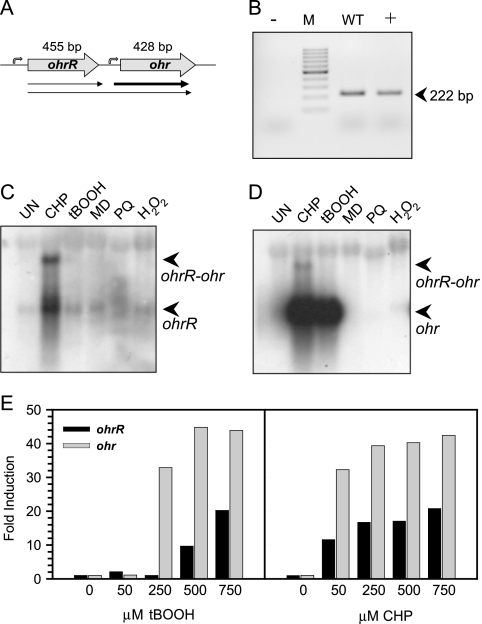
The organic hydroperoxide-inducible expression of ohr in P. aeruginosa has been reported previously (25). However, the regulator of ohr expression has not been identified. In most bacteria, the regulators of ohr are located in close proximity to the ohr gene (6, 11, 27, 36). Hence, we searched for a regulator in the vicinity of ohr. An annotated putative regulatory gene, PA2849 (35), located upstream of ohr was identified in P. aeruginosa. The amino acid sequence of the PA2849 protein shares 38 to 50% identity with those of previously identified OhrRs (6, 11, 27, 36). Hence, we designated the PA2849 gene P. aeruginosa ohrR. ohrR is located upstream of ohr, and the two genes are oriented in a head-to-tail manner (Fig. 1A) (25). The transcription organization of these genes was determined using RT-PCR with primers covering the ohrR-ohr intergenic region. The results shown in Fig. 1B show the expected 222-bp PCR products, suggesting that ohrR and ohr were transcribed in an operon. The RT-PCR results also supported subsequent Northern blot analysis of ohrR and ohr expression, in which 1.1-kb transcripts corresponding to the expected size of the two-gene operon were detected (Fig. 1C and D). However, the transcription organization of the genes was more complex. Additional smaller transcripts approximately 0.5 kb in length, which is equivalent to the expected size for monocistronic ohrR or ohr mRNAs, were also detected using either ohrR or ohr probes (Fig. 1C and D). The hybridization signals using ohrR as the probe revealed that ohrR-ohr and ohrR transcripts had similar hybridization intensities (Fig. 1C). On the other hand, the ohr hybridization signals for bicistronic and monocistronic ohr transcripts differed significantly. The shorter monocistronic transcripts of ohr were more than 50-fold more abundant than the longer bicistronic ohrR-ohr transcripts (Fig. 1D).
FIG. 1.
Expression analysis of ohrR and ohr. (A) Genome organization of the ohr-ohrR operon. (B) RT-PCR of the intergenic region between the ohr and ohrR genes. The primers BT468 and BT484 were used, giving the expected 222-bp PCR products. +, positive control; −, negative control; M, molecular weight marker (GeneRuler 100-bp DNA ladder; Fermentas); WT, PAO1 DNA as the template. (C and D) Phosphor images of Northern blots of total RNA extracted from wild-type P. aeruginosa cultures treated with various oxidants for 15 min. Northern blots were hybridized with radiolabeled probes specific to ohrR (C) or ohr (D). Positively hybridizing bands are indicated by arrows. UN, uninduced; CHP, 250 μM CHP; tBOOH, 250 μM tBOOH; MD, 500 μM menadione; PQ, 500 μM paraquat; H2O2, 500 μM H2O2. (E) Quantitative analysis of Northern blot analysis of wild-type P. aeruginosa cultures treated with various concentrations of CHP and tBOOH for 30 min.
Expression analysis of ohrR and ohr.
The expression profiles of P. aeruginosa ohrR in response to organic hydroperoxides were determined. The results of the Northern blot analysis show that ohrR was maximally induced by treatment of PAO1 cultures with 250 μM CHP, a synthetic hydrophobic organic hydroperoxide, while exposure to 250 μM tert-butyl hydroperoxide (tBOOH), a simple synthetic organic hydroperoxide, did not significantly induce ohrR expression (Fig. 1C, D, and E). Nonetheless, induction of ohrR expression could be detected when higher concentrations (500 and 750 μM) of tBOOH were used (Fig. 1E). Other oxidants and oxidant-generating compounds, such as H2O2 (1 mM), menadione (500 μM), and paraquat (500 μM), failed to induce ohrR expression (Fig. 1C). The expression profile of ohr in response to oxidants shows that both 250 μM CHP and 250 μM tBOOH but not the other oxidants tested induced maximal expression of ohr (Fig. 1D). The ohr induction profile is similar to the results from a previous study (25). The expression profiles show that ohrR transcripts were efficiently induced by CHP and were less efficiently induced by tBOOH, whereas the ohr transcripts were induced by both CHP and tBOOH (Fig. 1C and D). The ability of 250 μM tBOOH to induce expression of ohr but not ohrR suggested a complex differential regulation mechanism for these genes.
Primer extension analysis of ohrR and ohr genes.
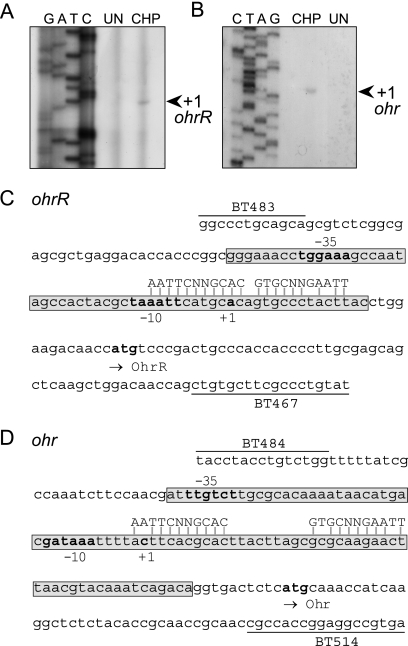
The transcription start sites of P. aeruginosa ohrR and ohr were identified by primer extension analysis using RNA samples prepared from uninduced and 250 μM CHP-induced cultures. For ohrR, 98-bp primer extension products were detected in the CHP-induced RNA sample. This placed the ohrR transcription start site 29 bp upstream from the ohrR translation initiation codon (Fig. 2A). This information allowed prediction of the putative −10 (TAAATT) and −35 (TGGAAA) boxes of the ohrR promoter (Fig. 2C). Primer extension analysis of ohr gave 137-bp products in the CHP-induced RNA sample (Fig. 2B). This placed the ohr transcription start site at −58 bp from the ohr translation start site (Fig. 2D), with the predicted putative ohr promoter motifs GATAAA and TTGTCT at the −10 and −35 regions, respectively. The ohr promoter is located within the ohrR-ohr intergenic region. The quantification of ohrR and ohr primer extension products revealed that there were more than 50-fold more products detected in RNA samples prepared from the CHP-induced culture than in the RNA samples from the uninduced culture. These findings confirmed the Northern blot results, which showed that CHP induced expression of ohrR and ohr. The organic hydroperoxide-inducible expression of these genes most likely results from increased transcription from their promoters.
FIG. 2.
Primer extension analysis of ohrR (A) or ohr (B) using total RNA extracted from P. aeruginosa cultures with and without cumene hydroperoxide treatment. Arrows indicate the transcription start site. The Sanger sequencing ladder to the left of the primer extension lanes was generated using pGEM-3zf(+) as the template and M13 forward as a primer. UN, uninduced; CHP, 250 μM CHP. (C and D) Promoter sequences of ohrR (C) or ohr (D). The putative −10 and −35 promoter regions, the transcription start site (+1), and the putative ohrR and ohr translation start codons are shown in bold. Locations and names of oligonucleotides (BT483, BT467, BT484, and BT514) used in this study are shown. The DNase I-protected regions for OhrR binding are boxed. The putative OhrR binding sites on each promoter are aligned with the predicted conserved sequence of the OhrR box.
Effect of OhrR on expression of ohrR and ohr.
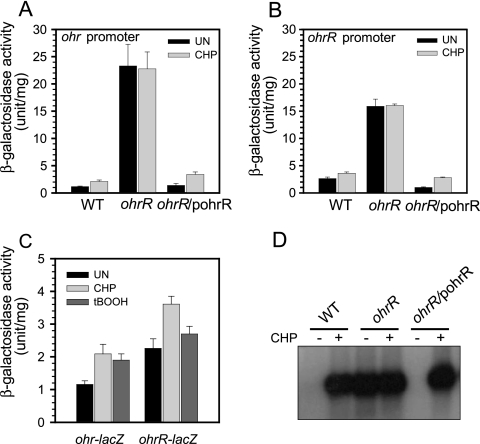
The role of OhrR as a transcription regulator of the ohrR-ohr operon was investigated using in vivo promoter fusion analysis. An ohrR mutant was constructed as described in Materials and Methods. The in vivo transcription activities from the ohrR and ohr promoters were monitored using promoter-lacZ fusion plasmids constructed as described in Materials and Methods. The p027Psohr-lacZ and p027PsohrR-lacZ plasmids were transformed into wild-type PAO1 and into an ohrR mutant. The levels of β-galactosidase produced by p027Psohr-lacZ and p027PsohrR-lacZ in wild-type PAO1 were 1.2 ± 0.1 and 2.6 ± 0.3 U/mg, respectively (Fig. 3A and B). Treatment of the wild-type strain containing p027Psohr-lacZ and p027PsohrR-lacZ with 1 mM CHP resulted in an increase in β-galactosidase activity to 2.1 ± 0.3 and 3.6 ± 0.2 U/mg, respectively. The experiments were repeated with an ohrR mutant harboring these plasmids. In the ohrR mutant, p027Psohr-lacZ and p027PsohrR-lacZ produced high levels of β-galactosidase, 23.3 ± 4.0 U/mg protein and 15.9 ± 1.3 U/mg protein, respectively. CHP treatment of these strains did not further increase the β-galactosidase levels (Fig. 3A and B). When an expression vector containing ohrR was introduced into the ohrR mutant harboring either p027Psohr-lacZ or p027PsohrR-lacZ, the β-galactosidase activity in these strains dropped to 1.4 ± 0.3 U/mg protein and 1.0 ± 0.1 U/mg protein, respectively, in uninduced cultures and to 3.4 ± 0.5 U/mg protein and 2.8 ± 0.1 U/mg protein, respectively, in 1 mM CHP-induced cultures. These values were comparable to those observed for a wild-type strain harboring the promoter/reporter plasmids (Fig. 3A and B). Using a promoter-lacZ fusion assay, we also confirmed the aforementioned data showing that at a specific concentration, tBOOH induced ohr expression but could not induce ohrR expression (Fig. 3C).
FIG. 3.
Effect of an ohrR mutation on ohrR and ohr expression. (A) β-Galactosidase activities of strains containing an ohr-lacZ fusion. (B) β-Galactosidase activities of strains containing an ohrR-lacZ fusion. (C) β-Galactosidase activities of strains containing ohrR-lacZ and ohr-lacZ fusions in the wild type induced with either 1 mM CHP or 1 mM tBOOH. (D) Northern blot analysis of ohr. Overnight cultures of strains were grown to mid-log phase and collected for β-galactosidase analysis or for mRNA extraction for Northern blot analysis. Experiments were performed three times; error bars represent the standard errors of the means. UN, control sample; CHP, 1 mM CHP; tBOOH, 1 mM tBOOH.
The role of OhrR in the regulation of ohr was independently confirmed by monitoring the expression of ohr in wild-type, ohrR mutant, and ohrR mutant/pohrR strains by Northern blot analysis. The results clearly showed that ohr expression was highly induced by the CHP treatment in an ohrR-dependent manner (Fig. 3D). Lack of CHP induction of ohr in the ohrR mutant could be restored by expression of a functional ohrR in trans (Fig. 3D). These data are strongly indicative of the negative regulatory role of OhrR in the expression of ohrR and ohr at the transcriptional level. The results support the idea that there is independent regulation of the ohrR and ohr promoters. These promoters responded disparately to different organic hydroperoxides.
OhrR binding to ohrR and ohr promoter fragments.
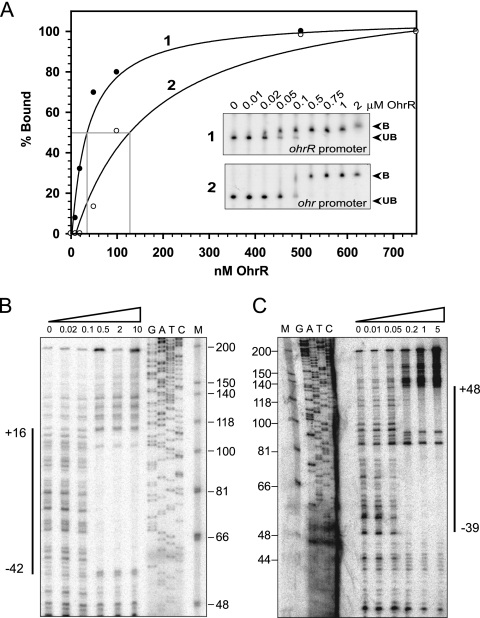
Previous studies have shown that OhrR regulates its target genes by directly binding to the operator site located in the vicinity of its target promoters. Hence, the ability of purified OhrR to bind to its target promoter in vitro was investigated using a gel shift assay. The addition of various concentrations of purified P. aeruginosa OhrR to either ohrR or ohr promoter fragments resulted in slower migration of the promoter fragments in a native acrylamide gel (Fig. 4A). This indicates that OhrR binds directly to its target sites. The OhrR dissociation constants (Kd) for the ohrR and ohr operators were estimated to be 35 nM and 125 nM, respectively (Fig. 4A). The OhrR binding sites on these promoter/operator fragments were determined by DNase I footprinting. Binding of OhrR protected DNA regions on both the ohrR and ohr operators/promoters from DNase I digestion. The protected region on the ohrR operator/promoter fragment extended from nucleotide position −42 to position +16 on the upper strand (Fig. 4B). On the bottom strand of the ohrR operator/promoter fragment, OhrR also bound to the same region (data not shown). On the ohr operator/promoter fragment, OhrR binding protected the ohr operator/promoter fragment from nucleotide position −39 to position +48 on the upper strand and nucleotide position −43 to position +75 on the lower strand (Fig. 4C and data not shown). Taken together, the gel shift and DNase I footprinting results clearly demonstrated that purified OhrR directly bound to the ohrR and ohr operators located in the vicinity of the respective promoter regions.
FIG. 4.
(A) Binding of OhrR to the ohrR promoter fragment. The percentage of OhrR bound to the ohrR operator fragment was determined using a gel shift assay with the ohrR promoter and purified OhrR. The various concentrations of OhrR added to the binding reactions are indicated above each lane. The unbound promoter fragment is designated UB, and the protein-DNA complex is designated B. (B and C) Mapping of the OhrR binding sites on the P. aeruginosa ohrR (B) and ohr (C) promoter fragments by DNase I footprinting. PCR-generated probe fragments were labeled on one strand by end labeling one of the primers with 32P prior to amplification. The sequencing ladder used to localize the binding sites on the ohrR promoter (B) was generated using pGEM-3zf(+) as a template and M13 forward as a primer. The sequencing ladder (G, A, T, C) used to localize the binding sites on the ohr promoter (C) was generated using the promoter fragment itself as a template and the same labeled oligonucleotide as was used to generate the probe as a primer. “M” indicates molecular weight marker, φx174/HinfI (Promega). Numbers above each lane indicate amounts of the OhrR protein (μM) used in each reaction.
Role of conserved cysteine residues in organic hydroperoxide sensing.
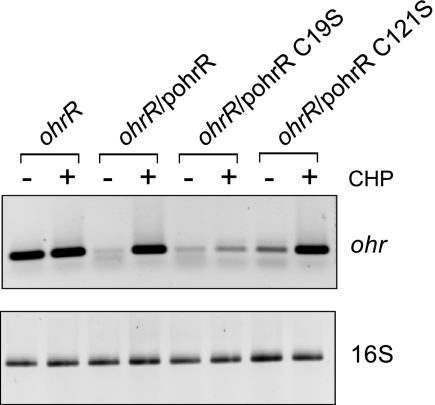
OhrR of P. aeruginosa belongs to the two-cysteine family of OhrRs. P. aeruginosa OhrR contains the conserved oxidant-sensing cysteine residue located near the N terminus at amino acid position 19 (C19) and the conserved C-terminal cysteine residue located at position 121 (C121). The importance of these two cysteine residues in organic hydroperoxide sensing by P. aeruginosa OhrR was investigated. We performed site-directed mutagenesis at C19 and C121, changing these cysteines to serines. The mutated ohrR gene was cloned into an expression vector, pBBR1MCS-4, and the resultant plasmids were introduced into a P. aeruginosa ohrR mutant. The levels of ohr mRNA in these strains were measured by RT-PCR under uninduced and CHP-induced growth conditions. In the absence of OhrR, ohr expression became constitutively high regardless of whether or not CHP was present (Fig. 5). CHP-inducible expression of ohr could be achieved by introduction of a plasmid containing wild-type ohrR into the mutant (Fig. 5). These results confirmed the expression pattern of ohr shown by the Northern blot analysis and the transcription fusion assays (Fig. 1 and 3). The system also allowed us to test the effects of cysteine mutations in OhrR on ohr expression. PAO1 ohrR/pohrRC19S showed low-level expression of ohr compared to the level attained in the ohrR mutant strain, indicating that OhrRC19S could function as a transcription repressor on the promoter (Fig. 5). However, the derepression of ohr expression by CHP treatment was not observed (Fig. 5). Introduction of OhrRC121S into an ohrR mutant resulted in the wild-type pattern of ohr expression under uninduced and CHP-induced conditions (Fig. 5). These in vivo data indicate that C19 has an important role in organic hydroperoxide sensing.
FIG. 5.
Effect of OhrR cysteine mutations on expression of ohr. Expression of ohr was monitored by RT-PCR. The first panel shows the level of ohr expression. The bottom panel shows the expression of a housekeeping gene, the 16S rRNA gene. CHP, 250 μM CHP.
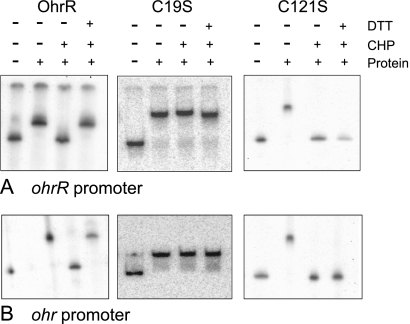
We further tested the DNA binding and oxidant sensing abilities of mutated and wild-type OhrRs in vitro. OhrRC19S and OhrRC121S were purified using the same procedure that was used for wild-type OhrR. In the gel shift assay, purified OhrR bound to the ohr promoter/operator fragment (Fig. 6A). The binding of OhrR to the fragment was abolished in the presence of 1 mM CHP. This reaction to CHP could be reversed by addition of 1 mM DTT, a reducing agent, to the binding reaction (Fig. 6A). OhrRC19S also bound to the ohr operator/promoter fragment with a similar binding affinity (data not shown), although addition of CHP did not affect the binding activity (Fig. 6A). In contrast, OhrRC121S could bind to the promoter/operator fragment, and the addition of CHP also abolished the binding activity (Fig. 6A). Surprisingly, addition of 1 mM DTT to the binding reaction could not restore the binding of OhrRC121S to the promoter fragment. OhrR and various OhrR cysteine mutants bound to the ohrR promoter fragment produced patterns similar to those observed using the ohr promoter fragment (Fig. 6B).
FIG. 6.
Binding of OhrR, OhrRC19S, and OhrRC121S on the ohrR promoter (A) or the ohr promoter (B). DTT, 1 mM DTT; CHP, 1 mM CHP. Protein: OhrR, OhrRC19S, or OhrRC121S, as indicated on the top of each panel.
Role of ohrR and ohr in oxidative defense and pathogenicity.
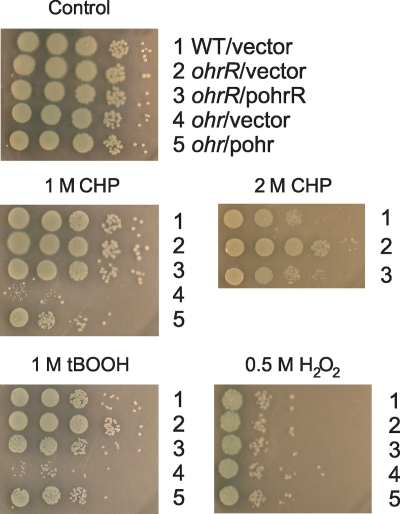
Previous reports have indicated that ohr has important physiological roles in protecting P. aeruginosa from organic peroxide stress. Here we extended these findings by examining the physiological roles of ohrR and ohr in protecting P. aeruginosa from peroxide stresses, particularly organic hydroperoxide stress, and we also investigated the roles of these genes in the pathogenicity of P. aeruginosa. First, we determined the oxidant resistance levels of ohrR, ohr mutants, and a wild-type strain. As expected, the ohr mutant was more sensitive to the organic hydroperoxides CHP and tBOOH than the wild-type strain, and the increased sensitivity of the mutant could be restored to almost wild-type levels by a plasmid containing a functional ohr gene (Fig. 7). The results confirmed the data from a previous study (25). Unexpectedly, the ohrR mutant was more resistant to both CHP and tBOOH (Fig. 7). This phenotype could be complemented by a functional ohrR gene (Fig. 7). Neither mutant showed significant alterations in the level of resistance to H2O2 (Fig. 7).
FIG. 7.
Sensitivities of wild-type P. aeruginosa and ohrR and ohr mutants to oxidants. Exponential-phase cells grown in LB medium were serially diluted (10-fold dilutions), and 10 μl of each dilution was spotted onto LB agar containing either 500 μM H2O2, 1 M CHP, 2 M CHP, or 1 M tBOOH. After incubation at 28°C for approximately 24 h, growth of the bacteria was observed. The experiment was repeated three times with similar results.
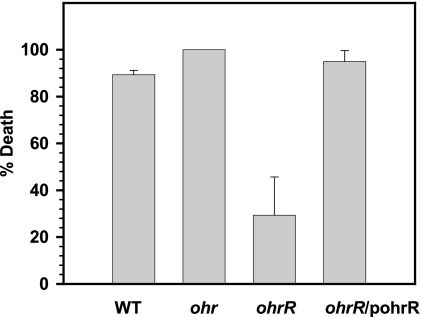
The roles of ohr and ohrR in P. aeruginosa pathogenicity were examined using the C. elegans/P. aeruginosa pathogenicity model. The percent mortality of C. elegans caused by a wild-type strain and an ohr mutant were similar at all time points (Fig. 8), whereas an ohrR mutant strain showed 29.3% ± 16.4% mortality of C. elegans, compared to 89.3% ± 1.9% mortality of C. elegans caused by the wild-type strain at 5 h (Fig. 8). The ability to kill C. elegans was restored to a wild-type level when a functional copy of OhrR was introduced into the ohrR mutant (Fig. 8).
FIG. 8.
Caenorhabditis elegans/P. aeruginosa pathogenicity tests. After incubation of P. aeruginosa with C. elegans, the numbers of live and dead/paralyzed worms were recorded over time. Percentage death was then calculated. The data show percentage death at 5 h. These results are representative of at least three independent experiments. Error bars indicate the standard errors for the technical replicates.
DISCUSSION
Here we identified ohrR, the regulatory gene for ohr, and we elucidated the mechanisms governing the organic hydroperoxide sensing and the regulation of inducible expression of ohr. P. aeruginosa ohrR and ohr show atypical transcription organization. Both genes are transcribed as a bicistronic mRNA and as monocistronic transcripts. These two transcripts are produced in equal proportion for ohrR but are produced in greatly different quantities for ohr. Primer extension and in vivo promoter analysis confirmed the presence of promoters located upstream of ohrR and ohr (Fig. 2). These promoters are likely responsible for the monocistronic transcripts, while the bicistronic transcripts could arise from transcription readthrough from ohrR into ohr. The significance of different sizes of the transcripts is not known. The monocistronic ohr transcripts could be more stable than the bicistronic transcripts and the monocistronic ohrR transcripts, similar to the previously observed situation for Xanthomonas ohrR-ohr (20). This would enhance the overall expression of ohr.
Experimental evidence supports the role of OhrR as a transcription repressor. OhrR binds with high affinity to the ohr and ohrR operators (Fig. 4). There are many potential OhrR binding sites in the ohr and ohrR promoter regions. The OhrR binding sites vary in different bacteria (6, 14, 27, 36). The OhrR binding site is thought to involve an inverted repeat with the core sequence of AATT-N-AATT (6). There are many putative AATT-N-AATT boxes in the vicinity of the OhrR-protected regions of the ohr and ohrR promoters. We found one box in each promoter region for which the surrounding nucleotides are similar, AATTCNNGCAC-N-GtGCNNgAATT (Fig. 2C and D). These boxes are likely the OhrR operators.
In vivo and in vitro analyses using OhrR and OhrRC19S confirmed the role of C19 as a redox-sensing residue in P. aeruginosa OhrR. Addition of CHP to the reaction inactivated the DNA binding properties of OhrRC121S in a manner similar to that for the wild-type protein (Fig. 6); however, addition of 1 mM DTT could not reestablish OhrRC121S DNA binding activity. The results are consistent with the recent hypothesis that the conserved cysteine at the C terminus plays a role in prevention of the overoxidation of the redox-sensing cysteine at the N terminus (33, 34). The overoxidized forms of cysteine are sulfinic acid and sulfonic acid, which cannot be reduced by DTT (17). The carboxyl-terminal C121 residue also has an important role in the overall sensing process of OhrR.
The degree to which an organic hydroperoxide can induce the ohrR-ohr system correlates with the degree of hydrophobicity of the inducer. CHP, a more hydrophobic hydroperoxide, is a better inducer than tBOOH in the ohrR-ohr system. These observations could be clarified by structural analysis of OhrR that could reveal the presence of a hydrophobic patch and hydrophobic amino acid residues in the vicinity of the sensing cysteine (2, 4). This would facilitate the entry of a hydrophobic organic hydroperoxide into the channel, allowing efficient oxidation of the sensing cysteine.
A current model of OhrR sensing and response to organic hydroperoxides involves oxidation of the sensing cysteine residue by an organic hydroperoxide and subsequent disulfide bond formation with C121. The formation of this disulfide bond then results in the loss of the DNA-binding activity of OhrR. This allows RNA polymerase to bind to the promoter and activate transcription of the target genes. tBOOH (250 μM) highly induced ohr expression and yet barely induced ohrR expression (Fig. 1). This suggests involvement of factors in addition to the oxidation of OhrR that govern the induction of these OhrR-regulated genes. These factors could be the OhrR dissociation constants (Kd) for the operators and the chemical nature of the organic hydroperoxide inducer. A model has been postulated to explain the tBOOH induction of ohr and ohrR that takes into account these additional factors. The binding of either OhrR or RNA polymerase to the operator or the promoter is mutually exclusive. Depending on whether OhrR or RNA polymerase successfully binds to its target site, the subsequent events lead to either repression or induction of gene expression. Thus, exposure of reduced OhrR to tBOOH, a poor inducer, leads to oxidation of only a fraction of reduced OhrR. This decreases the overall concentration of reduced OhrR, while the oxidized OhrR dissociates from the operator. Under such conditions, the concentration of the remaining reduced OhrR is sufficient for it to compete with RNA polymerase for binding to the ohrR operator and to prevent the RNA polymerase from binding to the promoter. However, the concentration of reduced OhrR is not sufficient for the repressor to bind to the ohr operator due to the higher Kd of the ohr operator. This allows RNA polymerase to bind to the ohr promoter to activate transcription of this gene. This model has taken into account the observations that OhrR has a Kd for the ohrR operator that is 3.5 times lower (35 nM) than that for the ohr operator (125 nM). In addition, the differential induction could be observed only with a poor organic hydroperoxide inducer.
Further support for this model came from observations that treatment with either higher concentrations of tBOOH (500 and 750 μM) or an efficient inducer, such as CHP (250 μM), could oxidize more reduced OhrR and could lower the concentration of reduced OhrR to a level where it could no longer bind to the ohrR operator. This results in the observed induction of both ohrR and ohr expression by CHP and tBOOH. The repressor binding affinity and the hydrophobicity of the inducer allow fine-tuning and differential induction of expression of OhrR target genes. This situation, in which the nature of the inducer contributes to a measured response, is rare in bacteria. The model is also a departure from a simple on-off model and permits selective expression of genes regulated by the same sensor/transcription regulator under the same stress condition.
Physiological analysis of ohrR and ohr mutants confirms the role that these genes play in protecting bacteria from organic hydroperoxide toxicity (Fig. 7) (25). Inactivation of ohr did not seem to have a significant effect on the pathogenicity of the bacteria, even though the mutant was hypersensitive to organic hydroperoxide-induced death. Surprisingly, the ohrR mutant showed a reduced ability to kill nematodes compared to wild-type PAO1. This phenotype could be complemented by expression of functional ohrR from a plasmid (Fig. 8). This observation could not be accounted for by increased ohr expression in the ohrR mutant. Thus, it is likely that OhrR regulates other genes in addition to ohr and that correct levels of expression of these genes are required for pathogenicity of P. aeruginosa. Transcriptome analysis of an OhrR mutant and a wild-type strain is being done to identify novel OhrR target genes. Nonetheless, the observations presented here provide support for the concept of targeting transcription factors for therapeutic control of P. aeruginosa infection.
Acknowledgments
This work was supported by grants from the National Center for Genetic Engineering and Biotechnology (BTB-01-PG-14-5112), Chulabhorn Research Institute, and Mahidol University.
We are grateful to B. P. Braeckman for his assistance in C. elegans pathogenicity testing and his gift of C. elegans. We also thank Weerachai Tanboon and Aingporn Pagakayai for their aid in the β-galactosidase assays.
Footnotes
Published ahead of print on 5 February 2010.
REFERENCES
- 1.Alexeyev, M. F. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824-826, 828. [DOI] [PubMed] [Google Scholar]
- 2.Arun Kumar, C., and U. N. Das. 1999. Lipid peroxides, nitric oxide and essential fatty acids in patients with Plasmodium falciparum malaria. Prostaglandins Leukot. Essent. Fatty Acids 61:255-258. [DOI] [PubMed] [Google Scholar]
- 3.Atichartpongkul, S., S. Loprasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 4.Azenabor, A. A., and J. B. Mahony. 2000. Generation of reactive oxygen species and formation and membrane lipid peroxides in cells infected with Chlamydia trachomatis. Int. J. Infect. Dis. 4:46-50. [DOI] [PubMed] [Google Scholar]
- 5.Chang, W., D. A. Small, F. Toghrol, and W. E. Bentley. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuchue, T., W. Tanboon, B. Prapagdee, J. M. Dubbs, P. Vattanaviboon, and S. Mongkolsuk. 2006. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J. Bacteriol. 188:842-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cussiol, J. R. R., S. V. Alves, M. A. Oliveira, and L. E. S. Netto. 2008. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 28:11570-11578. [DOI] [PubMed] [Google Scholar]
- 8.Darby, C., C. L. Cosma, J. H. Thomas, and C. Manoil. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:15202-15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuangthong, M., and J. D. Helmann. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U. S. A. 99:6690-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong, M., M. Fuangthong, J. D. Helmann, and R. G. Brennan. 2005. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20:131-141. [DOI] [PubMed] [Google Scholar]
- 15.Jesaitis, A. J., M. J. Franklin, D. Berglund, M. Sasaki, C. I. Lord, J. B. Bleazard, J. E. Duffy, H. Beyenal, and Z. Lewandowski. 2003. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J. Immunol. 171:4329-4339. [DOI] [PubMed] [Google Scholar]
- 16.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 17.Lee, J.-W., S. Soonsanga, and J. D. Helmann. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U. S. A. 104:8743-8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 19.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 20.Mongkolsuk, S., W. Panmanee, S. Atichartpongkul, P. Vattanaviboon, W. Whangsuk, M. Fuangthong, W. Eiamphungporn, R. Sukchawalit, and S. Utamapongchai. 2002. The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol. Microbiol. 44:793-802. [DOI] [PubMed] [Google Scholar]
- 21.Mongkolsuk, S., W. Praituan, S. Loprasert, M. Fuangthong, and S. Chamnongpol. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monod, M., C. Denoya, and D. Dubnau. 1986. Sequence and properties of pIM13, a macrolide-lincosamide-streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol. 167:138-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Nosocomial Infections Surveillance. 2004. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32:470-485. [DOI] [PubMed] [Google Scholar]
- 24.Newberry, K. J., M. Fuangthong, W. Panmanee, S. Mongkolsuk, and R. G. Brennan. 2007. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28:652-664. [DOI] [PubMed] [Google Scholar]
- 25.Ochsner, U. A., D. J. Hassett, and M. L. Vasil. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh, S.-Y., J.-H. Shin, and J.-H. Roe. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189:6284-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma, M., D. DeLuca, S. Worgall, and L. E. N. Quadri. 2004. Transcriptome analysis of the response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 186:248-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panmanee, W., P. Vattanaviboon, W. Eiamphungporn, W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45:1647-1654. [DOI] [PubMed] [Google Scholar]
- 30.Panmanee, W., P. Vattanaviboon, L. B. Poole, and S. Mongkolsuk. 2006. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188:1389-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salunkhe, P., T. Topfer, J. Buer, and B. Tummler. 2005. Genome-wide transcriptional profiling of the steady-state response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 187:2565-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slama, T. 2008. Gram-negative antibiotic resistance: there is a price to pay. Critical Care 12:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soonsanga, S., J. W. Lee, and J. D. Helmann. 2008. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol. Microbiol. 68:978-986. [DOI] [PubMed] [Google Scholar]
- 34.Soonsanga, S., J.-W. Lee, and J. D. Helmann. 2008. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J. Bacteriol. 190:5738-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 36.Sukchawalit, R., S. Loprasert, S. Atichartpongkul, and S. Mongkolsuk. 2001. Complex regulation of the organic hydroperoxide resistance gene (ohr) from Xanthomonas involves OhrR, a novel organic peroxide-inducible negative regulator, and posttranscriptional modifications. J. Bacteriol. 183:4405-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]