Abstract
The gene regulatory mechanism determining the developmental pathway of the temperate bacteriophage TP901-1 is regulated by two phage-encoded proteins, CI and MOR. Functional domains of the CI repressor were investigated by introducing linkers of 15 bp at various positions in cI and by limited proteolysis of purified CI protein. We show that insertions of five amino acids at positions in the N-terminal half of CI resulted in mutant proteins that could no longer repress transcription from the lytic promoter, PL. We confirmed that the N-terminal domain of CI contains the DNA binding site, and we showed that this part of the protein is tightly folded, whereas the central part and the C-terminal part of CI seem to contain more flexible structures. Furthermore, insertions at several different positions in the central part of the CI protein reduced the cooperative binding of CI to the operator sites and possibly altered the interaction with MOR.
Temperate bacteriophages provide a classical example for studies of the choice between two alternative modes of development. Following infection of a sensitive host, a temperate bacteriophage may choose (i) to enter the lysogenic state, in which the viral genome is silenced and integrated into the host genome to be replicated along with the host DNA when the cell divides, or (ii) to enter the lytic state, in which phage replication in the cell leads to production of new phages, cell lysis, and release of phage progeny (4, 21, 25). The choice made by a temperate phage regarding which life cycle to enter is based on a gene regulatory system named the genetic switch (25).
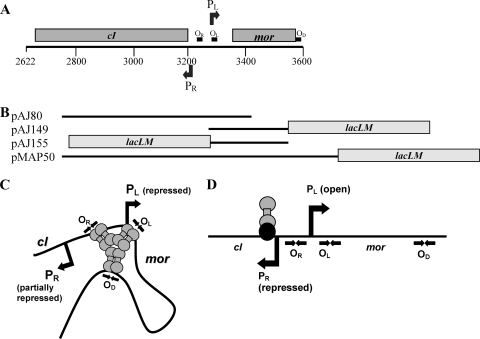
A DNA fragment obtained from the temperate lactococcal phage TP901-1 cloned into a low-copy-number plasmid was shown to exhibit bistable gene expression mimicking the lysis-lysogeny choice when introduced into Lactococcus lactis. The cloned DNA fragment contains the two divergently oriented early promoters PR and PL and the two promoter proximal genes cI and mor (Fig. 1A). PL is the lytic promoter from which MOR, the modulator of repression, is the first gene to be transcribed, and PR is the promoter for CI repressor synthesis. Furthermore, three CI binding sites, OR, OL, and OD, are present on the DNA fragment, which was fused to the reporter genes lacLM in order to measure the activity of either PR or PL (22). Following transformation of such plasmids (pMAP50 in Fig. 1B) into a lactococcal host, two different stable states are obtained; either PL is repressed by CI (immune state, shown in Fig. 1C) or PL is open (anti-immune state, shown in Fig. 1D). The open state of PL requires a large amount of MOR, which apparently counteracts CI binding to the operator sites (17). Knockout mutations introduced in either cI or mor showed that tight repression of PR in the anti-immune state requires the presence of both MOR and CI, and hence this repression of PR was suggested to occur by binding of both proteins to the DNA (Fig. 1D) (22). The nature and the location of the protein complex on the DNA are unknown, but the OR site is not necessary for the repression in the anti-immune state (22). A mathematical model built on a simplified version of the gene regulatory circuit containing an unidentified binding site for the CI:MOR complex and only one of the three CI operator sites, OL, predicted that the two proteins, CI and MOR, form a complex in solution before binding to DNA and thereby inhibit transcription from PR (19). When PL is repressed by CI in the immune state, PR is also repressed by cooperative binding of CI at the three operator sites OR, OL, and OD (Fig. 1C); however, sufficient CI is produced from PR to ensure tight repression of PL. The lytic PL promoter in the TP901-1 prophage may be derepressed by activating the host SOS response with UV light or mitomycin C (17). Since this requires the presence of a functional RecA in the host (17) and a functional MOR protein (17, 22), it is suggested that the CI protein contains regions important for RecA and/or MOR interaction. Previous studies have shown that the TP901-1 repressor exists as a hexamer in solution, most probably a trimer of dimers, which allows it to bind cooperatively at OR, OL, and OD (23). The N-terminal part of the protein contains a helix-turn-helix motif, and site-directed mutagenesis indicates that this motif is involved in DNA interaction, whereas deletion of the C-terminal end of the protein reduces the hexameric form to a dimer and thus the C-terminal end appears to be important for multimerization (23). The ability of the C-terminal half of the protein to form hexamers on its own demonstrates that all the amino acids required for oligomerization are positioned in the C-terminal half of the protein (23).
FIG. 1.
A 979-bp fragment from the TP901-1 genome (positions 2622 to 3600) is known to function as a bistable genetic switch. (A) The gray boxes indicate the positions of the cI and mor genes. The numbers below the line correspond to the location in the TP901-1 genome. The positions and directions of the promoters, PL and PR, are indicated as arrows above and below the line. The CI binding sites, OR, OL, and OD, are shown as black boxes. (B) Schematic representations of the cloned DNA fragments used in this study. Plasmid pAJ80 was used for the transposon mutagenesis, and pAJ149 and pAJ155 were used as reporter plasmids for measuring activity of PL or PR in the presence of wild-type or mutant CI proteins expressed in trans. Plasmid pMAP50 and its derivatives (pMAP131 to pMAP133, pMAP140, and pMAP141) contain a functional mor gene and were used to determine the activity of PL-expressing wild-type or mutant CI protein in cis. The gray boxes indicate the position of the reporter genes lacLM. (C) In the immune state, CI is suggested to bind to the three operator sites, OR, OL, and OD, forming a CI-DNA loop structure. Momentary release of CI from OR, which has the lowest affinity for CI, allows transcription from PR. (D) In the anti-immune state, a CI:MOR complex is suggested to bind DNA and prevent transcription from PR, whereas transcription from PL is allowed. Repression of PR in this state requires the presence of both functional mor and cI genes, whereas the OR site is not required (22). The nature of the protein complex and its location on the DNA are unknown. The head-to-head arrows represent the palindromic sequences of the CI operator sites.
In this work we further investigated the domain structure of the CI repressor by linker insertion mutagenesis. Linkers of 15 bp were randomly introduced into the cI gene, the resulting CI mutant proteins were screened for their ability to repress the lytic PL promoter in vivo, and selected mutants were tested for (i) the ability to repress the lysogenic PR promoter and (ii) the ability to establish two different stable states in the presence of a functional mor gene. In addition, biochemical analysis of purified CI was performed using limited proteolysis to gain insights into its tertiary folding. It was shown that the N-terminal half of the protein containing the DNA binding domain was quite resistant to proteolysis, implying a tightly folded conformation, whereas the distal C-terminal part containing the oligomerization domain was sensitive to proteolysis, suggesting the presence of partially unfolded or flexible regions.
MATERIALS AND METHODS
Bacterial strains, transformation, and growth conditions.
The bacterial strains used in this work are listed in Table 1. Lactococcus lactis subsp. cremoris MG1363 (6) was propagated at 30°C in GM17 medium (M17 broth from Oxoid Limited supplemented with 0.5% glucose) (28). Erythromycin (Erm) at 5 μg/ml and chloramphenicol (Cam) at 5 μg/ml were added to the medium when appropriate. MG1363 was transformed by electroporation (10). Screening on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates was performed with an X-Gal concentration of 90 μg/ml. Escherichia coli was grown at 37°C with agitation in Luria-Bertani (LB) medium (26). Bacto Agar was used at 1.5% (wt/vol) in solid media, and 150 μg/ml Erm, 15 μg/ml Cam, or 20 μg/m kanamycin (Kan) was added to the media when appropriate. Electrocompetent E. coli was obtained by growth in LB medium to an optical density at 450 nm (OD450) of 0.8 followed by washing in 10% glycerol. The cells were stored at −80°C, and DNA was transformed by electroporation (200 Ω, 25 μF, 2.0 kV).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Description or sequence | Antibiotic resistance | Reference or source |
|---|---|---|---|
| L. lactis subsp. cremoris strains | |||
| MG1363 | Plasmid-free derivative of NCDO712 | 6 | |
| MP401 | MG1363 containing pMAP50 | Ermr | 23 |
| AJ189 | MG1363 containing pAJ149 (PL-lacLM) | Ermr | 13 |
| AJ218 | MG1363 containing pAJ155 (PR-lacLM) | Ermr | 13 |
| Escherichia coli strains | |||
| DH5α | supE44 hsdR17 deoR recA1 endA1 lacZΔM15 | Stratagene | |
| M15[pREP4] | Expression host for pQE vectors, contains pREP4 | Kanr | Qiagen |
| Plasmids | |||
| pCI372 | Shuttle vector | Camr | 8 |
| pAJ149 | pAK80::P3 (3142-3458)a PL-lacLM fusion, containing OR and OL | Ermr | 13 |
| pAJ155 | pAK80::P3 (3142-3458)a PR-lacLM fusion, containing OR and OL | Ermr | 13 |
| pAJ80 | pCI372::PRORcI (2622-3332) | Camr | 16 |
| pMAP50 | pAK80::cI+OR+OL+mor+OD+, PL-lacLM | Ermr | 23 |
| pMAP131 | pMAP50;15 base pairs inserted at codon 120 in cI | Ermr | This study |
| pMAP132 | pMAP50;15 base pairs inserted at codon 98 in cI | Ermr | This study |
| pMAP133 | pMAP50;15 base pairs inserted at codon 135 in cI | Ermr | This study |
| pMAP140 | pMAP50;15 base pairs inserted at codon 78 in cI | Ermr | This study |
| pMAP141 | pMAP50;15 base pairs inserted at codon 89 in cI | Ermr | This study |
| pMAP150 | pCI372::PRcI; 15 base pairs inserted at codon 78 | Camr | This study |
| pMAP151 | pCI372::PRcI; 15 base pairs inserted at codon 89 | Camr | This study |
| Oligonucleotides | |||
| MLN6.rev | 5′-GGGGGGGTTTAAACATTTTAATTGAATTTTTGCAGTATC-3′ | ||
| morBamHI.rev | 5′-GGGGGATCCATGAGTTATGATTATTCGTCAC-3′ | ||
| MLN6.for | 5′-GGGGGGGTTTAAACATAAAAGAACAAGATGAGCAAAATAAAGTTA-3′ | ||
| pAK80erm | 5′-CTTTCAACTGCCTGGCAC-3′ | ||
| MLN8.rev | 5′-GGGGGGGTTTAAACAAACTGTAATAGTTTCTTCTATATT-3′ | ||
| MLN8.for | 5′-GGGGGGGTTTAAACACAGTTATGAAAAAATTGGAAGAGCCAAGGC-3′ | ||
| MLN27.rev | 5′-GGGGGGGTTTAAACATCAGACGGTAATCTTCTATTTGTT-3′ | ||
| MLN27.for | 5′-GGGGGGGTTTAAACATCTGAGTGATGAATATCTTGAAGAACAAAT-3 | ||
| MLN2.rev | 5′-GGGGGGGTTTAAACATACATCAAAGCCCATTAACCACGC-3′ | ||
| MLN2.for | 5′-GGGGGGGTTTAAACACAGTTCCTATGGTAGAATCTTCTAAAATA-3 | ||
| MLN3.rev | 5′-GGGGGGGTTTAAACAATCATTTTCTATTTTAGAAGATTC-3′ | ||
| MLN3.for | 5′-GGGGGGGTTRAAACACAGTTTCTGAAAATATAGAAGAAACTATT-3 | ||
| CIBamHI | 5′-GGGGGGGATCCGCTCTCTAAGTTTACTCATAAATCG-3 | ||
| erm2BamH | 5′-CGGGATCCCGCACGTTTCATGAACTTT-3′ |
Numbers indicate nucleotide positions in the TP901-1 genome.
DNA techniques.
Primers were supplied by TAG Copenhagen A/S, Denmark. The GFX PCR DNA and gel band purification kit were supplied by Amersham Biosciences. T4 DNA ligase, shrimp alkaline phosphatase, restriction enzymes, and buffer systems were supplied by Fermentas Life Sciences and used as recommended by the supplier. Plasmid DNA from E. coli was isolated using the Qiagen plasmid kit. Lactococcus strains were treated with 10 mg/ml lysozyme for 20 min at 37°C prior to plasmid purification using the Qiagen plasmid kit. Cloning using the TOPO TA cloning kit (Invitrogen) was performed as recommended by the supplier (Invitrogen). DNA sequencing was performed by MWG Biotech AG, Germany.
Construction of plasmids.
The plasmids and primers used in this study are listed in Table 1. For construction of plasmid pMAP131, primers MLN6.rev and morBamHI.rev were used to amplify mor along with cI containing an insertion of 15 base pairs. Primers MLN6.for and pAK80erm were used to amplify cI containing an insertion of 15 base pairs. Plasmid pMAP50 was used as the template. The two PCR products were purified, digested with restriction enzyme PmeI, ligated, and cloned using the TOPO TA cloning kit (Invitrogen). The insert was transferred as a HindIII-BamHI fragment to pAK80 and verified by sequencing. Plasmids pMAP132, pMAP133, pMAP140, and pMAP141, were constructed in a similar way using primers MLN8.rev and MLN8.for, MLN27.rev and MLN27.for, MLN2.rev and MLN2.for, and MLN3.rev and MLN3.for, respectively. Plasmid pMAP150 and pMAP151, expressing CI with insertions at codons 78 and 89, respectively, were constructed by amplification of the cI genes in plasmid pMAP140 and pMAP141, respectively, using primers CIBamHI and erm2BamHI. The resulting PCR products were digested with BamHI, cloned into pCI372 digested with BamHI, and verified by sequencing.
Proteolysis of CI.
Purified CI protein (23) was diluted to 1.5 mg/ml in 50 mM Tris (pH 8) and 0.2 M NaCl. Subtilisin was added to a final concentration of 4 μg/ml, and samples were incubated at 37°C for 2, 5, 10, 15, 30, or 60 min before addition of 6× SDS sample buffer followed by boiling for 5 min. Polypeptides were separated by SDS-PAGE (14) using 15% gels.
N-terminal sequencing.
Protein samples were resolved by SDS-PAGE and blotted onto a polyvinylidene difluoride (PVDF) membrane. Following electrotransfer, the protein of interest on the blot was cut out. N-terminal sequencing was performed by Anne Blicher, Center for Systems Microbiology, Technical University of Denmark, by Edman degradation on an Applied Biosystems Procise 494 sequencer.
Mass spectrometry.
Peptide samples were taken from the gel, digested with trypsin or Asp-N, and loaded on the AnchorChip target using the sample/matrix/wash (SMW) method (29). Mass spectrometry was kindly performed by Birgit Andersen, Center for Systems Microbiology, Technical University of Denmark.
GPS-LS mutagenesis and selection of transformants.
GPS-LS reactions were performed according to the manufacturer's recommendations (New England Biolabs) using 0.08 μg of pAJ80 (Fig. 1B). Following in vitro mutagenesis, plasmid pAJ80 was transformed into E. coli DH5α (Stratagene), and plasmids containing the 1.7-kbp transprimer were selected on LB plates containing 15 μg/ml Cam and 20 μg/ml Kan. Colonies originating from eight independent transformations were pooled, and plasmid was extracted from each of the eight independent pools. The purified plasmid DNA was transformed into AJ189 containing pAJ149 (PL-lacLM) (Fig. 1B) (13), and bacteria which did not express CI proteins able to repress transcription from PL were selected as blue colonies on GM17 plates containing X-Gal. In general, approximately 50% blue colonies were observed. Thirty-nine colonies were randomly chosen for plasmid purification. Plasmids were transformed into E. coli DH5α and new plasmid preparations made. The inserted 1.7-kbp transposon was removed from the purified plasmids by restriction with enzyme PmeI followed by ligation, leaving a 15-base-pair linker inserted in the plasmid. Plasmids lacking the kanamycin marker gene and hence the 1.7-kbp transposon were purified, and the cI gene, including the upstream region, was sequenced. The phenotypes of the plasmids were determined by transformation into the lactococcal strain AJ189 containing plasmid pAJ149 (PL-lacLM) (Fig. 1B). Selected plasmids were also transformed into strain AJ218 containing the plasmid pAJ155 (PR-lacLM) (Fig. 1B).
Enzyme assays.
β-Galactosidase activity was determined by permeabilizing cells with sodium dodecyl sulfate (0.1%) and chloroform, and the assay was performed as described previously (18). The specific activities are reported in Miller units [A420/(min·ml·A600)]. The activities listed in Table 2 are average values determined from at least three independent experiments.
TABLE 2.
Isolated plasmids containing linker insertions
| No. isolated | Plasmid | Linker position in cI | Position of stop codon or linker insertion in CI | Mean promoter activity (Miller units) ± SDa |
|
|---|---|---|---|---|---|
| PL | PR | ||||
| 1 | pML37 | 30 | TAA at 12 | ND | ND |
| 1 | pML2 | 36 | TAA at 14 | ND | ND |
| 1 | pML10 | 48 | TAA at 18 | ND | ND |
| 1 | pML11 | 49 | TAA at 18 | ND | ND |
| 1 | pML15 | 162 | TAA at 56 | ND | ND |
| 1 | pML5 | 192 | TAA at 66 | ND | ND |
| 1 | pML26 | 227 | TAA at 78 | ND | ND |
| 1 | pML3 | 285 | TAA at 96 | ND | ND |
| 1 | pML25 | 317 | TAA at 108 | ND | ND |
| 1 | pML39 | 351 | TAA at 119 | ND | ND |
| 1 | pML30 | 8 | 3 | 141 ± 15 | ND |
| 1 | pML21 | 19 | 6 | 0.7 ± 0.2 | ND |
| 1 | pML19 | 31 | 11 | 778 ± 25 | ND |
| 1 | pML7 | 41 | 14 | 892 ± 40 | ND |
| 2 | pML17/pML18 | 44 | 15 | 1,095 ± 28 | ND |
| 1 | pML28 | 62 | 20 | 1,081 ± 154 | ND |
| 1 | pML34 | 124 | 41 | 861 ± 163 | ND |
| 2 | pML13/pML14 | 227 | 75 | 243 ± 19 | ND |
| —b | pMAP150 | 233 | 78 | 1.3 ± 0.4 | 2.0 ± 0.1 |
| —b | pMAP151 | 257 | 89 | 0.8 ± 0.3 | 1.5 ± 0.2 |
| 1 | pML8 | 295 | 98 | 0.6 ± 0.2 | 1.4 ± 0.1 |
| 2 | pML9/pML38 | 307 | 101 | 1.4 ± 0.1 | 10.7 ± 1 |
| 1 | pML35 | 340 | 113 | 1.1 ± 0.1 | 6.6 ± 2 |
| 1 | pML6 | 358 | 120 | 1.1 ± 0.3 | 7.7 ± 0.4 |
| 1 | pML32 | 362 | 121 | 1.3 ± 0.1 | 5.2 ± 0.3 |
| 1 | pML33 | 368 | 122 | 0.6 ± 0.1 | 2.1 ± 0.2 |
| 2 | pML27/pML36 | 406/407 | 135 | 0.3 ± 0.1 | 1.2 ± 0.1 |
| None | pAJ80 | 1.0 ± 0.3 | 1.3 ± 0.1 | ||
| None | pCI372 | No cI | 1,023 ± 38 | 34 ± 5 | |
A420/(ml·min·OD600). ND, not determined.
—, the 15-base-pair linker was obtained not by using the GPS-LS linker scanning system but by site-directed mutagenesis.
Induction by mitomycin C.
Induction with mitomycin C was performed mainly as described previously (17). Bacteria were grown exponentially in GM17 liquid medium containing 5 μg/ml Erm and then diluted to an OD600 of 0.05 and grown to an OD600 of 0.3 before the cell culture was divided in two. Mitomycin C was added to a final concentration of 2.5 μg/ml to one of the cultures, while the other served as a control. Samples were removed at different time points, harvested, and analyzed by Western blotting using CI-specific antibodies. The total amount of protein was determined by measuring the OD280, and the same amount of total protein was loaded in each lane on the gel.
Western blotting.
Cell cultures were grown exponentially to an OD600 of 0.5. Twenty milliliters of culture was harvested, and the cell pellet was washed in phosphate-buffered saline (PBS), pH 7.4. The cell pellet was resuspended in 0.5 ml PBS, 200 μl glass beads (Sigma) was added, and the cells were lysed using the FastPrep FP120 at speed 4 three times for 30 s. Samples were centrifuged at 17,000 × g for 10 min at 4°C, and the supernatant was transferred to new Eppendorf tubes. The protein concentration was determined by measuring the OD280 (1 OD280 unit = 1 mg/ml). Generally, samples containing 150 μg protein were prepared for analysis by SDS-PAGE (14) using 15% gels. Following separation, the proteins were transferred to Hybond P (PVDF) membranes (GE Healthcare Life Sciences) by electroblotting using a semidry blotter (Bio-Rad) or by wet transfer. Western blotting was performed using the RPN5783 Western blotting reagent packet (GE Healthcare Life Sciences). Polyclonal antibodies against CI were made at the Department for Biochemistry and Nutrition, Technical University of Denmark, by immunization of a rabbit with purified CI extract from AJ157 (12). Antibodies against CI were diluted 25,000-fold for use in the Western blot assay. The primary antibodies were detected using an anti-rabbit secondary antibody conjugated with alkaline phosphatase (GE Healthcare Life Sciences RPN5783), the blots were scanned using a Storm 860 imager, and the relative amount of CI was estimated using the ImageQuant TL software (Amersham Biosciences).
Incubation of CI at alkaline pH.
CI was purified as described previously (13). The CI protein was diluted in buffer to final concentrations of 100 mM Tris (pH 7.5), 100 mM Tris (pH 8), 100 mM Tris (pH 8.8), or 100 mM glycine-NaOH (pH 9.5) with 10 mM CaCl2 and 100 mM NaCl. Samples were incubated at 30°C for 1 h and 15 h before being analyzed by SDS-PAGE (14).
RESULTS
Limited proteolysis identifies a tightly folded N-terminal domain in CI.
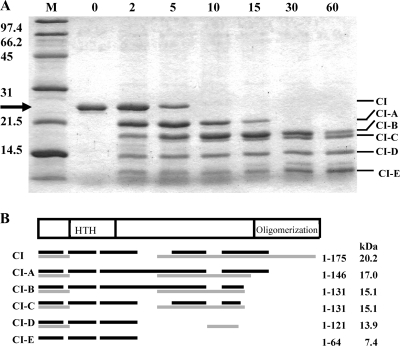
Limited proteolysis of a polypeptide chain with proteases exhibiting broad substrate specificity can be used to identify sites or regions characterized by high flexibility or local unfolding (5, 11). To identify such flexible regions of the protein, purified CI protein was digested with the protease subtilisin, and the resulting peptides were resolved by SDS-PAGE (Fig. 2). The intact CI polypeptide with a molecular mass of 21 kDa was readily cleaved to a metastable product with a molecular mass of 18 kDa, which was subsequently digested to polypeptides of approximately 16, 15, 12, and 9 kDa. For precise localization of the proteolytically susceptible regions in CI, the intact protein and its fragments were subjected to N-terminal sequencing and mass spectrometry. The minimal molecular masses of the five peptide bands were calculated as 17, 15, 15, 14, and 7 kDa, respectively (Fig. 2), based on the sequences identified by mass spectrometry. N-terminal sequencing of the polypeptides identified that the first five residues of CI, MKTDT, were retained in all the polypeptides, indicating that proteolysis must occur in the C-terminal region of CI. The digestion pattern shows that the C terminus of CI is rapidly degraded, since full-length CI is no longer observed following 10 minutes of incubation with the protease. CI-A is formed first and then CI-B, which seems to be quite stable for more than 30 min and is still present after 60 min (Fig. 2A). These results demonstrate chain flexibility in the C-terminal part of CI, which includes the domain responsible for hexamer formation from intact dimers (23). A rather short peptide formed early, CI-E, shows that CI also has a flexible region downstream of amino acid 64. This fragment was also clearly seen after 60 min, implying a tightly folded conformation of the N-terminal region, the domain responsible for DNA binding.
FIG. 2.
Limited proteolysis of CI using subtilisin. (A) SDS-PAGE of purified CI protein treated with the protease subtilisin. First lane, molecular weight standard. The numbers to the left of the gel show the sizes of the molecular weight markers. Second lane, CI before addition of subtilisin. Third to eighth lanes, samples of CI treated with subtilisin for 2, 5, 10, 15, 30, and 60 min. The arrows at the left indicate the positions of the full-length CI protein. The lines to the right show polypeptides appearing following digestion with subtilisin. Each polypeptide band was analyzed by N-terminal sequencing and mass spectrometry. (B) The individual peptide bands were purified from the gel, treated with trypsin or Asp-N, and analyzed by mass spectrometry. The black lines indicate the sequences identified following trypsin digestion and mass spectrometry analysis, and the gray lines indicate the sequences identified following Asp-N digestion and mass spectrometry analysis. The amino acid numbers at the right of the figure indicate the minimal lengths of the peptides produced after subtilisin digestion.
Identification of functional domains in CI using linker insertion mutagenesis.
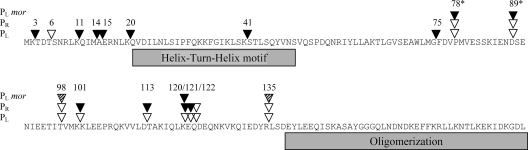
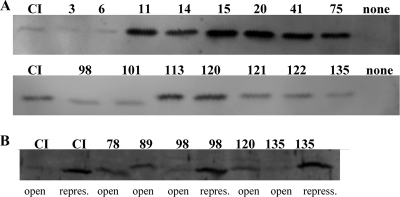
A genetic analysis of the domain structure of the cI gene product was performed using the GPS-LS linker scanning system (3, 7). cI mutants in which a 15-base pair linker was inserted randomly into the gene were constructed. Insertion of five additional amino acids in a surface loop with no specific target function, in a connector region, or in a relatively unstructured region is expected to have a minor effect on the function of the protein, whereas insertion within tightly folded regions with secondary structures or functionally important loops should have a greater effect (1, 2, 9). In this study we tested the ability of the mutated cI genes to produce proteins able to repress transcription from PL in vivo. First we mutagenized in vitro the plasmid pAJ80 containing the lysogenic PR promoter, the promoter proximal gene, cI, OR, and part of the OL site (Fig. 1B). Insertion of the 1.7-kb Tn7-based transposon within the PR promoter or cI was expected to abolish CI repression. Such mutants were identified by introduction of the mutagenized plasmids into lactococcal cells containing the reporter plasmid pAJ149 carrying the two CI operator sites, OR and OL, and the lytic PL promoter fused to the reporter genes lacLM (Fig. 1B) (13). Insertions that affected the ability of CI to repress transcription from PL were selected and the transposon removed, leaving a 15-bp insertion in the plasmid. The position of the inserted sequence was identified by sequencing (Table 2), and the ability of the resulting CI mutant protein to repress PL was tested. In 11 of the plasmids, the inserted linker introduced a TAA translational stop codon in cI, resulting in truncated versions of CI. All of these truncated CI proteins were shorter than the Δ58CI protein of 122 amino acids known to express a nonfunctional repressor protein in vivo (23), and hence these CI mutant proteins were not studied further. Fifteen different linker insertion positions in cI were obtained from a total of 19 plasmids. In addition to these 15 cI mutants, we introduced 15 bp at codon 78 or 89 in cI within the pAJ80 plasmid using site-specific mutagenesis. The PL-directed β-galactosidase activity was determined for each of the mutant repressors (Table 2). In the absence of CI, 1,023 ± 38 Miller units of β-galactosidase was produced, while wild-type CI repressed PL more than 1,000-fold (1.0 ± 0.3 units). The positions of the inserted linkers in CI are illustrated in Fig. 3. Seven plasmids, containing insertions in the 5′-terminal half of cI at codon 3, 11, 14, 15, 20, 41 or 75, expressed CI proteins that were unable to repress PL (Table 2) or repressed it maximally 7-fold (insert at codon 3), compared with the 1,000-fold repression by the wild-type pAJ80 plasmid. This supports the data obtained from previous mutant studies, which show that the N-terminal part contains the DNA binding domain (23). To confirm that CI mutant proteins were present in the cells and not degraded due to incorrect folding of the protein, Western blot analyses were performed using specific antibodies against CI. The expression levels of repressor proteins having five amino acids inserted in the N-terminal part of the protein at codon 3 or 6 showed CI levels comparable to those of the wild-type protein, whereas insertion at codon 11, 14, 15, 20, 41, or 75 caused more than a 10-fold elevation compared to wild-type CI (Fig. 4A). Thus, the inability of the CI mutants to repress PL was not due to lack of repressor protein. Furthermore, the increased levels of these CI mutant proteins verified that expression of CI from PR in plasmid pAJ80 is autoregulated by CI even though this plasmid contains only the OR site and part of the OL site. In contrast to the dramatic effect of linker insertions near the N terminus, insertions in the middle of CI at codon 98, 101, 113, 120, 121, 122, or 135 had only minor effects on CI levels (Fig. 4B) (less than 2-fold variation), and all of these modified CI proteins were able to repress PL in vivo.
FIG. 3.
Amino acid sequences of the TP901-1 CI repressor protein. The gray boxes represent regions involved in DNA binding (helix-turn-helix motif) and oligomerization (residues 138 to 180) (23). The positions where five amino acids were inserted by transposon mutagenesis are shown above the sequences as triangles, and the numbers identify the positions of the inserts in CI. At two positions (78 and 89) marked with asterisks, five amino acids were inserted by site-specific mutagenesis. Three lines to the left of the sequence are marked with PL, PR, or PLmor, and the triangles in each line represent the state of PL, PR, or PL in the presence of a functional mor gene, resulting from the given CI mutant, respectively. ▾, positions where linker insertion in CI abolish repression of PL or PR; ▿, positions where linker insertion still allows repression of PL or PR. Wild-type CI protein represses transcription from both PL and PR (white triangles) in the absence of mor. In the presence of a functional mor gene and wild-type CI, each transformant has a choice between two states, i.e., PL repressed or PL open (hatched triangle). Insertion of linkers at positions 98 and 135 showed wild-type phenotypes, but the repressed state of PL was not inducible.
FIG. 4.
Western blot analysis using specific antibodies against TP901-1 CI. (A) Protein extracts were obtained from cells expressing cI, cI3, cI6, cI11, cI14, cI15, cI20, cI41, cI75, cI98, cI101, cI113, cI120, cI121, cI122, or cI135 from the lysogenic promoter, PR, cloned in the pCI372 plasmid. Cells containing plasmid pCI372 without a cI insert was used as a negative control. The relative amount of CI was estimated using the ImageQuant TL software. (B) Protein extracts were obtained from cells containing a functional mor gene in concert with cI, cI78, cI89, cI98, cI120, or cI135. The state of the lytic PL promoter (open or repressed) is shown below each lane.
Repression of PR in vivo occurs by binding of CI at OR facilitated by cooperative binding at OL and/or OD (22). To test if the CI mutant proteins were able to bind cooperatively to two operator sites, we expressed the mutant proteins in a strain containing plasmid pAJ155 (Fig. 1B), which holds the two CI operator sites, OR and OL, and the divergently oriented promoters, PR and PL, with PR fused to the reporter genes lacLM. Plasmids expressing CI101, CI113, CI120, or CI121 repressed PR less efficiently than the wild-type CI (Table 2). Repression of PR requires cooperative binding of CI at OR and OL, since CI does not bind at OR on its own (13, 22). Furthermore, since repression of PL by these mutant proteins is as efficient as that by wild-type CI, it may be concluded that the inability of the mutant proteins to repress PR to the same level as wild-type CI is not caused by reduced binding of these proteins at OL, which indicates that the dimer-dimer interaction is affected in these mutant proteins.
Note that the screening method used did not identify transposons inserted in the C-terminal quarter of the CI protein, as insertions in this region apparently produce functional proteins (23).
PL repression by CI78, CI89, CI98, CI120, and CI135 in the presence of MOR.
The MOR protein encoded by phage TP901-1 functions as a modulator of repression by inhibiting CI repression of the lytic PL promoter, possibly by interacting with CI (13, 23). In order to examine if the region between the DNA binding domain and the oligomerization domain needed for hexamer formation in CI is involved in interaction with MOR, 15-bp linkers were introduced in cI at codon 78, 89, 98, 120, or 135, in a DNA segment also containing the two divergent promoters PR and PL, the promoter proximal gene mor, and the three CI operator sites OR, OL, and OD.
When the wild-type construct, pMAP50 (Fig. 1B), was transformed into the lactococcal host, two different stable states were obtained; i.e., in some clones the lytic PL promoter was open, and in other clones PL was repressed. Of 1,887 transformants, 23 (approximately 1%) contained the lytic promoter in the repressed state. If mor is knocked out in pMAP50, the plasmid no longer exhibits bistability when introduced into Lactococcus. Only one stable state is observed, namely, the repressed state, since CI now always represses PL CI (22). In the case of the mutant CI proteins, bistability is lost when five amino acids are introduced at position 78, 89, or 120 in CI in the presence of a functional mor gene, since PL is always found in the open state (more than 5,000 colony transformants were screened). These results show that in the presence of MOR, the mutant CI proteins are no longer able to establish repression of PL, even though they are able to repress PL in the absence of MOR (Table 2). The levels of CI78, CI89, and CI120 protein were shown by Western blot analysis to be higher than the wild-type CI level in cells where PL is in the open state (Fig. 4B) and slightly lower than the CI level in cells where PL is repressed (Fig. 4B). This shows that although repression of PL by CI78, CI89, or CI120 is lost in the presence of MOR, CI protein is still present in the cells and PR is considerably repressed, presumably by the CI:MOR complex.
When five amino acids were inserted at codon 98 or 135 in cI and expressed in the presence of a functional mor gene, both the open and the repressed states of PL were obtained. The frequency of finding PL in the repressed state was 1% when using these plasmids, which is comparable to the frequency obtained when using the wild-type construct. This frequency of obtaining PL in the repressed state was also observed in previous studies (22). Repression of PL by wild-type CI in the presence of functional mor and recA genes may be released by activating the host SOS response, for instance, by treating the host cells with mitomycin C (17). However, repression of PL by CI98 or CI135 could not be relieved following treatment of the cells with mitomycin C (data not shown). The lack of PL induction was apparently not caused by elevated levels of CI98 or CI120 in the cells (Fig. 4B).
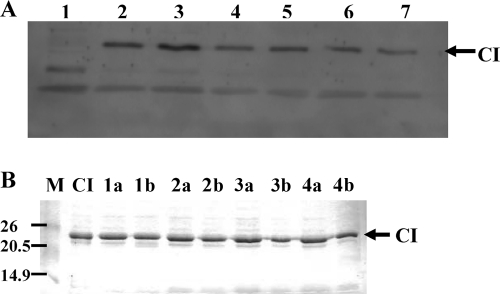
In order to examine if wild-type CI is cleaved following activation of the host SOS response and released from the DNA, allowing transcription from PL, the lactococcal host cells containing the genetic switch of phage TP901-1 holding PL in the repressed state were treated with mitomycin C. Protein samples were analyzed by Western blotting before and after addition of mitomycin C using CI-specific anti-bodies (Fig. 5A). No cleavage products were observed following PL induction. Furthermore, to determine if CI may be autocleaved at alkaline pH as is the case for CI encoded by phage lambda (15), purified CI was incubated at 30°C at pH 7.5, 8, 8.8, or 9.5 for 1h and 15 h and analyzed by SDS-PAGE (Fig. 5B). No cleavage products of CI were observed following incubation at alkaline pH in vitro. The possibility still remains that the cleavage product may be quickly degraded or that only a small fraction of CI is cleaved in vivo and in vitro and therefore not detected, but the results suggest that the inactivation of CI by the SOS response may occur by a mechanism different from that of phage lambda.
FIG. 5.
No cleavage product of CI is observed in vivo following PL induction or following incubation at alkaline pH in vitro. (A) In vivo induction of the repressed PL promoter analyzed by Western blotting using CI-specific antibodies. The cloned bistable switch from phage TP901-1 carrying PL in the repressed state was treated with mitomycin C, which activates the host SOS response, thereby allowing increased transcription from PL. Lane 1, protein sample from the host strain Lactococcus lactis subsp. cremoris MG1363 without the genetic switch from phage TP901-1. Lanes 2 and 3, protein samples collected at 10 and 2 min, respectively, before treatment with mitomycin C. At time zero the culture was divided into two, and one of the cultures was treated with mitomycin C. Lanes 4 and 5, are protein samples collected at 15 and 60 min, respectively, after time zero (no mitomycin C was added). Lanes 6 and 7, protein samples collected 15 and 60 min, respectively, after addition of mitomycin C. (B) Purified CI protein incubated at alkaline pH. First lane, protein marker. Numbers to the left of the gel represent molecular weights in thousands. Second lane, CI protein at pH 7.5 without incubation. Lanes 1a and b, CI incubated at pH 7.5 for 1 and 15 h, respectively. Lanes 2a and b, CI incubated at pH 8 for 1 and 15 h, respectively. Lanes 3a and b, CI incubated at pH 8.8 for 1 and 15 h, respectively. Lanes 4a and b, CI incubated at pH 9.5 for 1 and 15 h, respectively. The arrows indicate the position of CI.
DISCUSSION
The phage-encoded repressor protein is a key player in determining the life cycle of a temperate phage following infection of a sensitive host. In the temperate lactococcal phage TP901-1, the repressor protein, CI, represses transcription from both the lytic and the lysogenic promoters by binding to multiple operator sites on the phage DNA. Mutations in the helix-turn-helix DNA binding motif located in the N-terminal part of CI were shown to destroy DNA binding (23). In this study, we show that the DNA binding N-terminal part of CI is tightly folded, since this part of the protein is resistant to proteolysis. Our new results also further support the location of the DNA binding site in the N-terminal part of CI, since insertion of additional amino acids in this region resulted in mutant proteins that could not repress transcription from the lytic promoter in vivo.
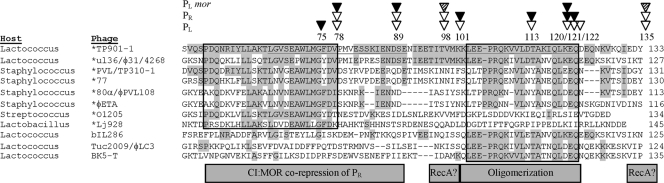
The C-terminal part of CI is required for oligomerization of the protein, and the C-terminal half was previously shown to contain all the amino acids required for oligomerization (amino acids 92 to 180) (23). In this work, we show that amino acids 121 to 180 in the C-terminal part of CI are accessible for proteases, which strongly suggests that parts of the peptide chain in this region of the protein are flexible and not tightly folded. This applies in particular for the region distal to amino acid 146. Although nine mutant proteins containing insertions in the central region of CI were able to repress transcription from the lytic PL promoter, none of these mutants functioned like the wild type. In four mutants the insertion of additional amino acids between positions 101 and 121 reduced the ability of the CI protein to repress the lysogenic PR promoter, suggesting that this region is important for efficient oligomerization of the protein (dimer to hexamer) or that the additional amino acids affect the dimer-to-hexamer formation at a distance. Sequence alignments showed that this region was highly conserved in a number of repressor proteins encoded by phages infecting Gram-positive bacteria (Fig. 6), supporting the notion that this region may be of general importance for the structure of CI and/or for interaction with host- or phage-encoded proteins conserved among the species.
FIG. 6.
Amino acid sequences of the CI repressor protein. Eighty-six amino acids from TP901-1 CI (residues 49 to 133) were searched for sequence identity to other repressor proteins. Phages encoding proteins with sequence similarity to TP901-1 CI and the corresponding host bacteria are shown to the left of the sequences. Bacteriophages that encode MOR homologous are marked with an asterisk. CI encoded by phages ul36, φ31, and 4268 are 98% identical, whereas CI proteins encoded by phages 80α and φPVL180 are 94% identical. Amino acids shown in gray are identical to amino acids in CI of phage TP901-1. Numbers to the right of the sequences indicate the amino acid numbers in the CI proteins. ▿, positions where linker insertion in CI allows repression of PL or PR; ▾, positions where linker insertion abolishes repression of PL or PR; hatched triangle, positions where linker insertion in CI in the presence of MOR allows the choice between the two different states. Two conserved regions are marked with black lines. The gray box below the sequences at the left represents a region conserved only in phages encoding MOR-homologous proteins. Insertion of five amino acids at positions 78 and 89 in this region abolished repression of PL in the presence of MOR, suggesting that this region is important for CI:MOR interactions. The second and the fourth gray boxes represent regions where insertion of five amino acids produced CI mutant proteins that can no longer be derepressed following activation of the host SOS response, suggesting that interaction with RecA may be abolished or that the multimeric form of the protein may be changed. The third gray box represents a conserved region in phage infection in Lactococcus. Insertion of five amino acids in this region abolishes repression of PR, suggesting that the cooperative binding of CI at OL-OR is reduced and hence this region is involved in oligomerization of CI.
Insertion of additional amino acids at position 98 or 135 in CI showed wild-type phenotypes repressing transcription from both the lytic and lysogenic promoters. Also, in the presence of a functional mor gene, a wild-type phenotype was observed, since two stable states of PL (repressed or open) were obtained. However, PL repressed by CI98 or CI135 in the presence of a functional mor gene could not be derepressed when activating the host SOS response as is the case when PL is repressed by wild-type CI. The inability to induce PL following activation of the host SOS response could not be explained by elevated levels of CI (Fig. 4B), suggesting that the introduced amino acids could (i) alter regions required for interaction with RecA or (ii) alter the oligomeric form of the repressor, which could be important for the induction process. The TP901-1 repressor protein does not contain an Ala-Gly cleavage site or any other signature sequence known to be involved in autocleavage and RecA interaction (15, 27). Furthermore, no cleavage products of CI were observed in vitro following incubation at alkaline pH or following PL induction in vivo (Fig. 5A and B), suggesting that induction of the lytic promoter in phage TP901-1 occurs by a mechanism different from that of phage lambda, but may use a mechanism somewhat similar to the switch of Vibrio cholerae phage CTXΦ, where the lysogenic promoter also is controlled by two DNA binding proteins, the phage-encoded RstR and the host-encoded SOS response regulator LexA (20). However, is should be noted that no lexA homologue is found in the genome sequence of the Lactococcus lactis MG1363 strain used in our studies.
Two CI mutants with insertions at position 78 or 89 repressed both the lytic and the lysogenic promoters as efficiently as wild-type CI. However, in the presence of a functional mor gene, only the open state of PL was obtained, showing that the lytic promoter is not repressed by CI78 or CI89 in the presence of MOR. This phenotype might be due to a stronger interaction between MOR and the mutant CI proteins, thereby preventing sufficient CI accumulation to repress PL after transformation. Western blot analysis shows that the open state of PL is not a result of a lack of CI protein in the cell (Fig. 4C). Furthermore, the levels of CI78 and CI89 observed were comparable to those found in the immune state where PR (and PL) is repressed by wild-type CI alone (Fig. 4C). A change in the oligomeric state or the strength of the subunit-subunit interaction of CI probably also is important for the interaction with MOR. Although repression of PR by CI78 and CI89 seems to be like that for the wild-type protein, repression of PR by CI120 is not as efficient as for the wild-type CI protein. A change in the oligomeric state of CI120 might therefore explain the defect in repression of PL in the presence of MOR. All temperate phages encoding MOR homologues also encode a CI protein containing a conserved patch (amino acids 53 to 78) (Fig. 6), a patch which is not conserved in CI proteins from phages that do not encode a MOR homologue protein. This finding supports the possibility that this region is involved in interaction with MOR. Five-amino-acid insertions at position 79 or 89 may thus interfere with the MOR interaction.
To summarize, the conformational designs of the repressor proteins encoded by phages TP901-1 and -186 have previously been reported to be similar in shape, with both forming flat circular molecules (23, 24). However, the conformation of the monomeric subunits may be different, since the 186 repressor has a stably folded C-terminal region and an N-terminal region which is sensitive to proteases. This is in clear contrast to the data presented here for the TP901-1 repressor. We showed that the N-terminal domain of CI is stably folded except for a region downstream of amino acid 64, whereas the very C-terminal region is much more susceptible to proteolysis and hence contains sites with flexible peptide backbones. In the C-terminal half, insertion of amino acids without loss of repressor function of the dimer form of CI is allowed, suggesting a more flexible structure. The region located between amino acids 138 and 180 was previously shown to be needed for dimer-hexamer formation. Here, we show that the region between amino acids 101 and 121 is also important for oligomerization of the protein. We show that positions 98 and 135 in CI are important for derepression of PL following activation of the SOS response, possibly due to interaction with RecA, and that positions 78 and 89 may be important for interaction with MOR.
Acknowledgments
We thank Vibeke Barkholt and Anne Blicher, BioCentrum-DTU, Technical University of Denmark, for performing the amino acid analysis. We acknowledge Birgit Andersen, who performed the mass spectrometry analysis and helped us with the analysis. We are grateful to Tonny Dedenroth Hansen and Marianne Knudsen for technical support with the Western blotting and to Sine Lo Svenningsen for helpful comments on the manuscript.
This work was supported by the Danish Council for Technology and Innovation (STVF) and by the Danish National Research Foundation through the Center for Models of Life.
Footnotes
Published ahead of print on 29 January 2010.
REFERENCES
- 1.Anton, B. P., and E. A. Raleigh. 2004. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J. Bacteriol. 186:5699-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballicora, M. A., E. D. Erben, T. Yazaki, A. L. Bertolo, A. M. Demonte, J. R. Schmidt, M. Aleanzi, C. M. Bejar, C. M. Figueroa, C. M. Fusari, A. A. Iglesias, and J. Preiss. 2007. Identification of regions critically affecting kinetics and allosteric regulation of the Escherichia coli ADP-glucose pyrophosphorylase by modeling and pentapeptide-scanning mutagenesis. J. Bacteriol. 189:5325-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brussow, H. 2001. Phages of dairy bacteria. Annu. Rev. Microbiol. 55:283-303. [DOI] [PubMed] [Google Scholar]
- 5.Fontana, A., P. P. de Laureto, B. Spolaore, E. Frare, P. Picotti, and M. Zambonin. 2004. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 51:299-321. [PubMed] [Google Scholar]
- 6.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goff, S. P., and V. R. Prasad. 1991. Linker insertion mutagenesis as probe of structure-function relationships. Methods Enzymol. 208:586-603. [DOI] [PubMed] [Google Scholar]
- 8.Hayes, F., C. Daly, and G. F. Fitzgerald. 1990. Identification of the minimal replicon of Lactococcus lactis subsp. lactis UC317 plasmid pCI305. Appl. Environ. Microbiol. 56:202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes, F., and B. Hallet. 2000. Pentapeptide scanning mutagenesis: encouraging old proteins to execute unusual tricks. Trends Microbiol. 8:571-577. [DOI] [PubMed] [Google Scholar]
- 10.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 11.Hubbard, S. J. 1998. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382:191-206. [DOI] [PubMed] [Google Scholar]
- 12.Johansen, A. H. 2000. The genetic switch of the temperate lactococcal bacteriophage TP901-1. Ph.D. thesis.Technical University of Denmark, Lyngby, Denmark.
- 13.Johansen, A. H., L. Brøndsted, and K. Hammer. 2003. Identification of operator sites of the CI repressor of phage TP901-1: evolutionary link to other phages. Virology 311:144-156. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Little, J. W. 1984. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. U. S. A. 81:1375-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madsen, P. L., and K. Hammer. 1998. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology 144:2203-2215. [DOI] [PubMed] [Google Scholar]
- 17.Madsen, P. L., A. H. Johansen, K. Hammer, and L. Brøndsted. 1999. The genetic switch regulating activity of early promoters of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 181:7430-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Nakanishi, H., M. Pedersen, A. K. Alsing, and K. Sneppen. 2009. Modeling of the genetic switch of bacteriophage TP901-1: a heteromer of CI and MOR ensures robust bistability. J. Mol. Biol. 394:15-28. [DOI] [PubMed] [Google Scholar]
- 20.Nickels, B. E. 2009. A new twist on a classic paradigm: illumination of a genetic switch in Vibrio cholerae phage CTX phi. J. Bacteriol. 191:6779-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oppenheim, A. B., O. Kobiler, J. Stavans, D. L. Court, and S. Adhya. 2005. Switches in bacteriophage lambda development. Annu. Rev. Genet. 39:409-429. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, M., and K. Hammer. 2008. The role of MOR and the CI operator sites on the genetic switch of the temperate bacteriophage TP901-1. J. Mol. Biol. 384:577-589. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen, M., L. Lo Leggio, J. G. Grossmann, S. Larsen, and K. Hammer. 2008. Identification of quaternary structure and functional domains of the CI repressor from bacteriophage TP901-1. J. Mol. Biol. 376:983-996. [DOI] [PubMed] [Google Scholar]
- 24.Pinkett, H. W., K. E. Shearwin, S. Stayrook, I. B. Dodd, T. Burr, A. Hochschild, J. B. Egan, and M. Lewis. 2006. The structural basis of cooperative regulation at an alternate genetic switch. Mol. Cell 21:605-615. [DOI] [PubMed] [Google Scholar]
- 25.Ptashne, M. 2004. A genetic switch. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Sauer, R. T., M. J. Ross, and M. Ptashne. 1982. Cleavage of the lambda and P22 repressors by recA protein. J. Biol. Chem. 257:4458-4462. [PubMed] [Google Scholar]
- 28.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, X., L. Shi, S. Shu, Y. Wang, K. Zhao, N. Xu, S. Liu, and P. Roepstorff. 2007. An improved method of sample preparation on AnchorChip targets for MALDI-MS and MS/MS and its application in the liver proteome project. Proteomics 7:2340-2349. [DOI] [PubMed] [Google Scholar]