Abstract
Der, an essential Escherichia coli tandem GTPase, has been implicated in 50S subunit biogenesis. The rrmJ gene encodes a methyltransferase that modifies the U2552 residue of 23S rRNA, and its deletion causes a severe growth defect. Peculiarly, overexpression of Der suppresses growth impairment. In this study, using an rrmJ-deletion strain, we demonstrated that two GTPase domains of Der regulate its association with 50S subunit via the KH-like domain. We also identified a region of Der that is critical for its specific interaction with 50S subunit.
Emerging evidence indicates that many Escherichia coli GTPases play critical roles in ribosome biogenesis (6). For example, E. coli Era consists of a conventional GTP-binding domain and a KH domain (an RNA-binding domain) with a consensus VIGXXGXXI sequence (9). The direct interaction between Era and 16S rRNA was demonstrated by structural studies of a Thermus thermophilus 30S ribosomal subunit complexed with Era (24). Peculiarly, Era was shown to suppress the cold-sensitive cell growth of the rbfA-deletion strain whose gene product resembles a KH domain and plays an important role in 30S subunit assembly at low temperature (13, 16). A unique GTPase subfamily of Der (double Era-like GTPase; also known as EngA) is conserved only in eubacteria. We have previously demonstrated that Der is cofractionated with 50S subunits in a GTP-dependent manner and that Der plays a critical role in 50S subunit maturation at a later biogenesis step (15).
Interestingly, both GTP-binding domains (G domains) were essential for cell growth; moreover, the two G domains function cooperatively, suggesting that GTP-induced conformational changes and GTPase activity are essential for cell viability as well as function (1, 15). The X-ray crystal structures of two Der orthologs from Thermotoga maritima and Bacillus subtilis revealed that the C-terminal domain has a topology similar to that of a KH domain without a consensus sequence motif and is flanked by two G domains (22, 23). It was suggested that the GTP-bound form of YphC (a Der ortholog in B. subtilis) triggers a dramatic conformational change, which favors an interaction with negatively charged ribonucleic acids by exposing a positively charged KH-like domain with a high pI value (14, 22).
Overexpression of E. coli Der functionally suppressed the slow growth defect of a deletion strain of the rrmJ gene, whose gene product is a methyltransferase, modifying the U2552 residue in the A loop of 23S rRNA in an intact 50S subunit (5, 26). Even though ΔrrmJ (strain HB23) is viable, it causes a serious defect of cell growth by accumulating 50S and 30S ribosomal subunits at the expense of 70S ribosomes. Thus, overexpression of Der seems to overcome its weak interaction with 50S subunits that are unmethylated at U2552. In this study, using an ΔrrmJ strain as a genetic background, we tried to elucidate the nature of the functional regulation of two G domains and the KH-like domain. We further characterized the KH-like domain by random mutagenesis and identified crucial residues for its association with 50S subunits. Our data suggest that the unique C-terminal domain indeed plays a role in rRNA-ribosome recognition.
Both G domains of Der are associated with its suppression of ΔrrmJ.
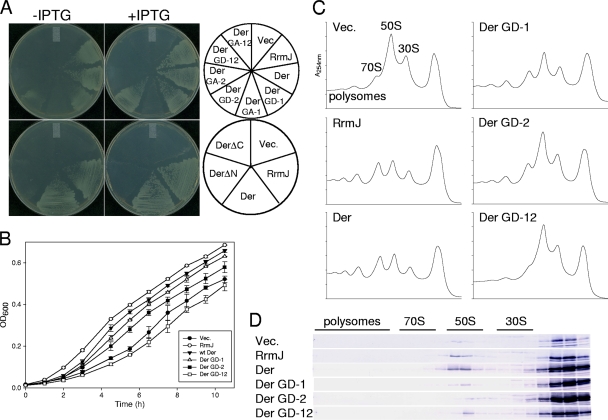
Previously, it was shown that DerN118D, DerN321D, DerS16A, and DerS216A could not support the cell growth of a der-deletion strain (1, 15). However, it is unknown whether suppression of ΔrrmJ (strain HB23) is associated with G domains of Der. Therefore, we introduced seven der alleles, including wild-type and six different G-domain mutants, into ΔrrmJ to test their suppression effect. First, the open reading frames of der and rrmJ were digested with NdeI and EcoRI and cloned into the NdeI-EcoRI site of pINA (an ampicillin-resistant [Ampr], IPTG [isopropyl-β-d-thiogalactopyranoside]-inducible plasmid) (10), yielding pINAEcDer and pINARrmJ, respectively. The mutant plasmids were constructed by site-directed mutagenesis, yielding pINAEcDerGD-1, pINAEcDerGD-2, pINAEcDerGD-12, pINAEcDerGA-1, pINAEcDerGA-2, and pINAEcDerGA-12; these clones express DerN118D, DerN321D, DerN118D/N321D, DerS16A, DerS216A, and DerS16A/S216A, respectively. Both Asn and Ser residues are conserved in the motif sequence of the conventional G domain (14), and their mutations to Asp or Ala effectively inhibit GTPase activity (1, 15, 23, 27). The ΔrrmJ strain was transformed with plasmids as described above, and the transformed cells were plated on LB plates containing ampicillin with or without IPTG followed by incubation at 30°C. On both plates, plasmid pINAEcDer was able to fully suppress the null phenotype of ΔrrmJ as pINARrmJ expressing wild-type RrmJ (Fig. 1A). Plasmids pINAEcDerGD-1 and pINAEcDerGD-2, however, partially suppressed ΔrrmJ by forming smaller colonies. However, cells transformed with pINAEcDerGD-12 did not show any suppression effect at all. Unlike Asn-to-Asp mutations, none of the pINAEcDerGA-1, pINAEcDerGA-2, and pINAEcDerGA-12 plasmids suppressed the growth defect of ΔrrmJ under the described set of conditions, suggesting that the Ser-to-Ala mutation in G domains is more inhibitory than the Asn-to-Asp mutation. Furthermore, we tested whether truncated forms of Der are able to restore the null phenotype of ΔrrmJ. pINAEcDerΔN and pINAEcDerΔC were constructed by truncating the first G domain and the entire C-terminal domain of Der, respectively. Plasmid pINAEcDerΔN or pINAEcDerΔC was transformed into ΔrrmJ, and transformants were incubated at 30°C. Neither of the transformants was able to support the cell growth, suggesting that both the N-terminal G domains and the C-terminal domain are indispensable (Fig. 1A). For the comparisons of suppression effects caused by Der variants, cell growth levels in a liquid medium were measured. In this experiment, IPTG was not added for the following reasons. (i) Hager et al. observed that in case of RrmJ, the leaky expression observed was enough to restore cell growth (12). We observed that the leaky expression of not only RrmJ but also Der was able to restore growth of ΔrrmJ. (ii) We wanted to observe the different effects caused by moderate leaky expression of various mutant proteins (Fig. 1B). Our results clearly demonstrate that plasmids pINAEcDerGD-1 and pINAEcDerGD-2, but not plasmid pINAEcDerGD-12, partially suppressed the phenotype of ΔrrmJ.
FIG. 1.
Phenotypes of ΔrrmJ expressing G domain and mutant Der. (A) The ΔrrmJ (strain HB23) cells transformed with plasmids were grown at 30°C overnight in LB medium containing ampicillin (50 μg/ml), and cultures were diluted. The diluted cultures were streaked on LB plates containing ampicillin with or without 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). GD-1, GD-2, GA-1, and GA-2 indicate N118D, N321D, S16A, and S216A mutations, respectively. GD-12 and GA-12 indicate the double mutations. (B) Growth curves of ΔrrmJ harboring different plasmids. Cells were cultured in LB medium containing ampicillin at 30°C, and cell cultures were diluted five times before measurement of the optical density at 600 nm (OD600). (C) Polysome profiles of wild-type and G-domain mutant Der in ΔrrmJ. The ΔrrmJ cells harboring various plasmids were cultured to the log phase at 30°C in LB medium containing ampicillin. Polysomes were trapped by the addition of chloramphenicol to the culture to achieve a final concentration of 0.1 mg/ml. The cell pellets were resuspended with BP buffer (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM NH4Cl, and 5 mM β-mercaptoethanol). GDPNP was added at a final concentration of 100 μM. Approximately 10 A260 units of each cleared extract was layered onto a sucrose gradient. The preparation of cell lysates and polysome analysis were carried out by ultracentrifugation in a Beckman SW41 rotor for 3.5 h at 210,000 × g. as described previously (15). (D) Association of wild-type and G-domain mutant Der with 50S subunits in ΔrrmJ. Individual gradient fractions (16 μl) prepared as described for panel C were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis followed by Western blotting using anti-Der antisera (15).
Inability of G-domain mutant Der to interact with 50S ribosomal subunits.
Next, we examined the GTP analog-dependent 50S subunit association of Der in those partially suppressed cells. Cells transformed with wild-type or mutant plasmids were grown in LB medium at 30°C, and cell lysates were prepared in the presence of GDPNP, a GTP analog. First, polysomes from each strain were separated by 5-to-40% sucrose density gradient sedimentation. Cells harboring a control vector accumulated a substantial amount of ribosomal subunits, with concomitant reductions of polysomes and 70S ribosomes (approximately 12% 70S, 55% 50S, and 33% 30S; Table 1), while the suppressor strain containing pINAEcDer recovered a normal ribosome profile (36% 70S, 42% 50S, and 22% 30S), as demonstrated by Tan et al. (26). The value for 70S ribosomes increased to 37.2% with pINRrmJ in the absence of IPTG. When RrmJ was induced by the addition of IPTG, the value further increased to 51.6% (Table 1). Notably, partially suppressed cells harboring either pINAEcDerGD-1 or pINAEcDerGD-2 accumulated less ribosome subunits than pINA transformants (Table 1). The cells expressing the double mutant DerN118D/N321D showed the same polysome defect as the ΔrrmJ cells harboring pINA (Fig. 1C).
TABLE 1.
The amounts of free 30S, 50S, and 70S ribosomes in ΔrrmJ cells expressing RrmJ or Der variants
| Plasmid | Ribosomea (%) |
||
|---|---|---|---|
| 70S | 50S | 30S | |
| pINA | 12.3 | 55.2 | 32.5 |
| pINARrmJ | 37.2 | 39.6 | 23.3 |
| pINARrmJ + 1 mM IPTG | 51.6 | 23.9 | 24.4 |
| pINAEcDer | 36.2 | 41.6 | 22.2 |
| pINAEcDerGD-1 | 24.9 | 45.5 | 29.6 |
| pINAEcDerGD-2 | 16.6 | 54.8 | 28.6 |
| pINAEcDerGD-12 | NAb | NA | NA |
| pINAEcDerG414R | 23.9 | 49.6 | 26.4 |
| pINAEcDerG424D | NA | NA | NA |
| pINAEcDerN469K | 24.5 | 48.7 | 26.9 |
| pINAEcDerT472A | 26.8 | 48.2 | 25.1 |
Values represent the average amounts of free 30S, 50S, and 70S ribosomes determined in duplicate experiments.
NA, the value for 70S was not available.
Subsequently, using fractions of each sucrose gradient, Western blot analysis of Der proteins in ΔrrmJ was carried out to detect the association of Der proteins with the 50S subunit. In two strains harboring vector only or pINARrmJ, endogenous Der colocalized with the 50S subunit. Note that more Der proteins are associated with 50S subunits in cells harboring pINAEcDer than in those harboring pINARrmJ. The experiment also revealed that the associations of DerN118D, DerN321D, and DerN118D/N321D with the 50S subunit were significantly diminished compared to that of wild-type Der (Fig. 1D), even though a large amount of 50S subunits accumulated in the polysome profile (Fig. 1C). It seems that only endogenous Der was detected with 50S subunit fractions of ΔrrmJ harboring pINAEcDerGD-1, pINAEcDerGD-2, or pINAEcDerGD-12, suggesting very weak or limited binding of G-domain mutant Der proteins to the 50S subunit. These data indicate that ΔrrmJ suppression by Der requires two intact G domains.
Isolation of Der KH mutants by random mutagenesis.
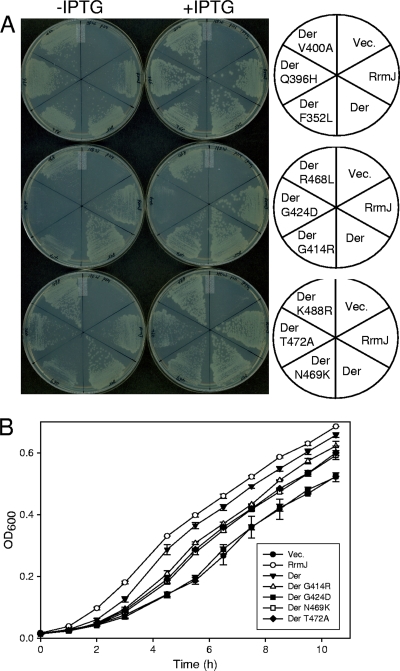
As described briefly above, the binding of GTP to Der may trigger substantial conformational changes, exposing KH-like domains to solution, which in turn promotes its ribosome interaction (22). Thus, in order to explore the possibility that the C-terminal domain of Der is responsible for ribosome interactions, we used the ΔrrmJ genetic backgrounds to screen KH-domain mutants. We created random mutations by PCR specifically targeting the C-terminal region of der by incorporating dITP. The mutated PCR fragments and pINAEcDer were digested with ClaI-EcoRI, and the doubly digested PCR fragments were ligated into the ClaI-EcoRI site of pINAEcDer. Then, the resulting ligation mixture was transformed into ΔrrmJ. Transformants with a slower growth rate at 30°C were screened as mutant Der, and pINA and pINAEcDer were used as negative and positive controls, respectively, to compare the growth rates. Three candidates were initially screened as defective mutants, and all three clones expressed the full-length Der proteins, as confirmed by Western blot analysis (data not shown). Each plasmid contained a combination of three mutations; the first plasmid contained R468L, N469K, and K488R, the second contained Q396H, V400A, and G414R, and the third contained F352L, G424D, and T472A. To determine which mutation was responsible for slow cell growth, site-directed mutagenesis for each of the nine mutations was carried out and nine mutant plasmids were transformed and tested for growth phenotypes with ΔrrmJ. ΔrrmJ harboring pINAEcDer or pINARrmJ formed colonies normally on LB plates with or without IPTG; among nine mutations, F352L, Q396H, V400A, R468L, and K488R did not affect colony formation. However, cell growth of ΔrrmJ was not supported by pINAEcDerG414R, pINAEcDerG424D, pINAEcDerN469K, or pINAEcDerT472A (Fig. 2A). Cell growth in a liquid medium was further monitored to quantitate the growth rate of each transformant. Consistently, ΔrrmJ cells harboring pINAEcDerG414R, pINAEcDerG424D, pINAEcDerN469K, or pINAEcDerT472A grew slowly; in particular, pINAEcDerG424D showed the same growth phenotype as ΔrrmJ (Fig. 2B). These data indicate that those four residues (Gly414, Gly424, Asn469, and Thr472) may play important roles in the function of the C-terminal domain.
FIG. 2.
(A) Screening of KH mutant Der proteins and their phenotypes in ΔrrmJ. The C-terminal part of the der gene was randomly mutagenized by PCR as described by Lerner et al. (18) and Spee et al. (25). The four deoxynucleoside triphosphates (dNTPs) (dATP, dCTP, dGTP, and dTTP) were depleted one at a time in PCR. MnCl2 (0.5 mM) and dITP (200 μM) were also included in the reaction, and error-prone Taq polymerase was used. Plasmids with mutations were transformed into ΔrrmJ, and transformants were streaked onto LB plates containing ampicillin with or without 1 mM IPTG. (B) Growth curves of ΔrrmJ transformed with KH mutant plasmids. Cell culture and optical density measurement were carried out as described for Fig. 1B.
KH mutant Der proteins are impaired in polysome recovery.
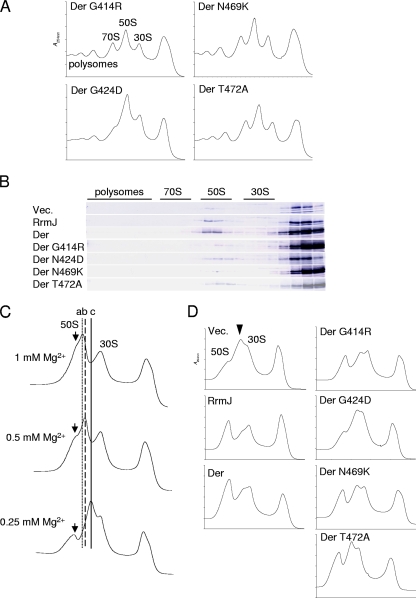
In order to examine whether these four residues are directly involved in the association of the KH-like domain with the 50S subunit, we prepared cell lysate samples in the presence of GDPNP and analyzed the polysome and ribosomal subunit profiles. Sucrose gradient centrifugation was carried out at a 10 mM Mg2+ concentration as described in the Fig. 1C legend. All the ΔrrmJ cells expressing KH mutant Der proteins accumulated both 50S and 30S subunits with a reduced amount of 70S and polysomes (Fig. 3A and Table 1). The ΔrrmJ cells expressing wild-type RrmJ or Der recovered the normal polysome profiles as described in the Fig. 1C legend. The DerG424D mutant showed the most severe polysome impairment compared to DerG414R, DerN469K, and DerT472A proteins. Subsequently, sample fractions of sucrose gradients were subjected to Western blot analysis as shown in Fig. 3B. The analysis revealed that the DerG414R, DerG424D, DerN469K, and DerT472A mutants substantially diminished the association with highly accumulated 50S subunits, suggesting that each mutation interrupted the associations of Der to 50S subunits, which in turn caused the accumulations of ribosomal subunits in ΔrrmJ. Note that since all four mutant proteins have two intact G domains, it is the KH-like domain that plays a major role in the ribosome binding of Der.
FIG. 3.
Ribosome profiles of ΔrrmJ expressing KH mutant Der. (A) The ΔrrmJ cells harboring the indicated plasmids were cultured at 30°C in LB medium containing ampicillin, and polysomes were resolved by ultracentrifugation carried out as described for Fig. 1. (B) Cofractionation of mutant Der with 50S subunits in ΔrrmJ. SDS-PAGE analysis and Western blotting were carried out as described for Fig. 1D. Western blots of three controls were taken from the experiment represented in Fig. 1D. (C) Ribosomal subunit profiles of ΔrrmJ. The cell pellets were resuspended with BP buffer containing 0.25, 0.5, or 1 mM MgCl2. Polysomes were resolved by ultracentrifugation for 3.5 h at 230,000 × g. Three lines (a, b, and c) indicate aberrant 50S subunits with three different migration patterns. Arrows indicate 50S subunits. (D) Ribosomal subunit patterns of ΔrrmJ expressing RrmJ, Der, and the Der KH mutant. The cell pellets were prepared as described for panel A and resuspended in BP buffer containing 0.25 mM MgCl2. Subunits were resolved by ultracentrifugation. An arrowhead indicates the position of aberrant 50S subunits.
Accumulation of aberrant 50S ribosomal subunits in ΔrrmJ.
Der-depleted cells accumulated aberrant 50S subunits in vitro at different Mg2+ concentrations as described previously (15), and the ΔrrmJ cells also accumulated ∼40S ribosomal subunits at a low Mg2+ concentration (3). Next, we tested whether ΔrrmJ accumulates aberrant 50S subunits at various Mg2+ concentrations. The ΔrrmJ cells were grown in LB medium, and cell pellets were resuspended in a buffer containing 0.25, 0.5, or 1 mM Mg2+. Cell lysates were then loaded onto 5-to-25% sucrose gradients containing 0.25, 0.5, or 1 mM Mg2+ followed by ultracentrifugation. 50S subunits of ΔrrmJ migrated at four different positions, depending on the Mg2+ concentration (Fig. 3C). At 1 mM Mg2+, normal 50S and abnormal 50S (line a) appeared, and at the lower Mg2+ concentrations (0.5 and 0.25 mM), two aberrant 50S subunits (lines b and c) with a substantially reduced amount of normal 50S subunits were detected. Note that the 30S subunits remained unaffected under the conditions used. We subsequently analyzed ribosomal subunit profiles of ΔrrmJ expressing DerG414R, DerG424D, DerN469K, and DerT472A. Expression of RrmJ or wild-type Der suppressed the abnormality of 50S subunits, while those four KH mutant Der proteins could not suppress the accumulation of aberrant 50S subunits, as shown in Fig. 3D. These results suggest that DerG414R, DerG424D, DerN469K, and DerT472A proteins cannot stabilize 50S subunits of ΔrrmJ cells at low Mg2+ concentrations. Therefore, it is likely that both Der and RrmJ contribute to the integrity of 50S subunits, likely through a common mechanism.
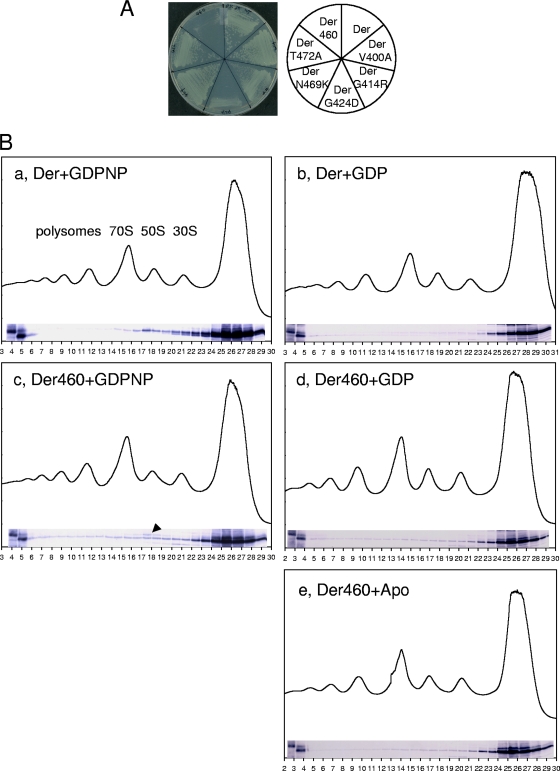
Since the overexpressed DerG414R, DerG424D, DerN469K, and DerT472A proteins significantly reduced binding to the unmethylated U2552 of 23S rRNA, we next tested whether the methylation at U2552 affected the association with mutant Der proteins. For this purpose, first, der(G414R), der(G424D), der(N469K), and der(T472A) mutant alleles were ligated into pIN vector (same as pINA except with a chloramphenicol-resistant cassette), yielding pINEcDerG414R, pINEcDerG424D, pINEcDerN469K, and pINEcDerT472A. Additionally, since Asn469 and Thr472 residues are located within the last 30 amino acid (aa) residues of E. coli Der but are not found in T. maritima Der (TmDer), pINEcDer460 encoding Der (aa 1 to 460; Der460) was cloned in order to examine the effect of the truncation on ribosome associations. These mutant plasmids were transformed into the der-deletion strain in which U2552 of 23S rRNA is methylated. This strain harbors a helper plasmid carrying the der gene, a temperature-sensitive replication origin, and an ampicillin-resistant gene (15). Transformants were plated on LB plates containing ampicillin, chloramphenicol, and kanamycin, and plates were incubated at 30°C. Colonies were picked and streaked on LB plates containing chloramphenicol and kanamycin, and then plates were incubated at 42°C to remove a helper plasmid. None of the transformants were able to form colonies at 42°C on LB plates containing ampicillin, confirming the loss of a helper plasmid (data not shown). As shown in Fig. 4A, pINEcDerG414R, pINEcDerN469K, and pINEcDerT472A complemented the der-deletion strain at 42°C; however, colony formation of the der-deletion strain was negatively affected by the pINEcDerG424D and pINEcDer460 plasmids. These results imply that methylation at the U2552 residue of 23S rRNA in intact 50S subunits induces a significant local rearrangement around methylation site.
FIG. 4.
Phenotypes of der-deletion strain expressing KH mutant Der. (A) The der-deletion strain was transformed with pINEcDer, pINEcDerV400A, pINEcDerG414R, pINEcDerG424D, pINEcDerN469K, pINEcDerT472A, or pINEcDer460 and plated on LB plates containing chloramphenicol (40 μg/ml) and kanamycin (35 μg/ml). Plates were first incubated at 30°C, and colonies formed at 30°C were streaked onto LB plates containing chloramphenicol and kanamycin; plates were then incubated at 42°C. (B) The nonspecific interaction of Der460 with ribosomes. Wild-type cells harboring pINAEcDer or pINAEcDer460 were cultured, and polysome samples were prepared in the presence of GDPNP, GDP, or Apo state. Polysome and Western blot analyses were carried out as described for Fig. 1. The wild-type cells harboring pINAEcDer or pINAEcDer460 were used as controls (shown in the first two lanes of the Western blot). An arrowhead indicates the endogenous Der in 50S subunit fractions, and the numbers indicate the ends of each fraction.
Peculiarly, Der460 was unable to support cell growth of a der-deletion strain; therefore, in order to further analyze the inability of Der460 to complement the der-deletion strain, pINAEcDer or pINAEcDer460 was transformed into wild-type E. coli cells and we performed polysome analysis followed by Western blot experiments. Surprisingly, in contrast to full-sized Der (Fig. 4B, panel a), Der460 interacted with polysomes as well as with ribosomal subunits in the presence of GDPNP (Fig. 4B, panel c). Note that the endogenous Der (arrowhead) coexisted with Der460 in 50S subunit fractions. Taking these results together, it is possible that both Der proteins are present at different 50S subunits or that the binding site of Der460 to 50S partially overlaps with that of full-length Der in the presence of GDPNP. Next, we tested whether this nonspecific interaction of Der460 with polysomes is nucleotide dependent. We carried out the same experiments as those represented by panels a and c of Fig. 4B, except that GDPNP was replaced with GDP or with no nucleotide (Apo state). Although to a lesser extent, Der460 still associated with polysomes. These data imply that the KH-like domain, including the last 30 aa residues, determines the specific association with 50S subunit.
As shown in Fig. 1, both the two G domains and the KH-like domain are essential for viability in ΔrrmJ as well as in the der-deletion strain (1, 15) and those G-domain mutant Der proteins exhibit an impaired ribosome association. All of the G-domain mutant Der proteins (Der GD-1, Der GD-1, and Der GD-12) exhibited substantially reduced binding to the 50S subunit (Fig. 1D), which supports the idea that cooperative binding of nucleotide to Der stimulates ribosome associations.
As mentioned earlier, upon nucleotide binding, Der appears to undergo conformational rearrangements for its ribosome association (22, 23). The KH mutant Der proteins isolated in this study had both G domains intact; notably, those KH mutations did not alter the GTPase activity of Der (data not shown). As shown by the comparisons of those KH mutations with TmDer and YphC structures, the mutations are located in the positively charged β-sheets (β14 and β15, respectively) of the KH-like domain (22, 23). Thus, Gly414 and Gly424 residues may be directly involved in Der associations with ribosome in ΔrrmJ cells. Alternatively, mutations at those residues may cause conformational changes in Der that inhibit its interaction with ribosomes. On the basis of TmDer X-ray structural analysis (23), GD-1 of E. coli Der is assumed to interact with the C-terminal 30 aa residues of Der that protrude toward GD-1 in the structure. Thus, it is tempting to speculate that, upon binding of GTP, the KH domain is released to specifically interact with 50S subunits.
Methylation at U2552 takes place at the A-loop of 23S rRNA that makes a direct interaction with aminoacyl (A)-site tRNA, and the function of RrmJ was thus found to be implicated in translation efficiency as well as accuracy (4, 28). Unlike other 23S rRNA mehtyltransferases (such as RrmA, RlmB, RumA, and RumB) that are dispensable (11, 19, 20), RrmJ seems to be crucial for ribosome function, probably because of the temporal and spatial importance of methylation at U2552 (3). Methylation at U2552 substantially modifies the A-loop fold of 23S rRNA (2); interestingly, KH mutant Der proteins are not defective in the der-deletion strain, suggesting that overexpressed Der may recognize the structural changes of the unmethylated A-loop region at U2552 in ΔrrmJ (Fig. 2 and 4).
50S subunits accumulated in ΔrrmJ were extremely unstable at lower Mg2+ concentrations, with underrepresented L5, L16, L18, L25, L27, L28, and L30 (12). Der-depleted cells also accumulated aberrant 50S subunits with a small amount of L2, L6, L9, and L18, suggesting that both Der and RrmJ are critical for ribosome assembly and integrity and share a mechanism to stabilize 50S subunits at a very late stage of 50S subunit maturation (15). These ribosomal instabilities were dependent on Mg2+ concentration in vitro (Fig. 3C and reference 15). Ribosomes, especially rRNAs, extensively interact with Mg2+ ions that neutralize the negative charges of rRNA backbone (17), and starvation of Mg2+ ions has a negative impact on the integrity and assembly of ribosomes (7, 8, 21). This may explain why starvation of Mg2+ in the absence of either RrmJ or Der in E. coli causes disintegration of 50S subunits.
Concluding remarks.
In this study, we demonstrated that GTP-induced conformational changes of Der are very important for its function with respect to the 50S subunit, and by using the ΔrrmJ strain we found that the KH-like domain is required for ribosome recognition. At present, it is unknown how Der and RrmJ proteins contribute to the structural integrity of 50S subunits in cells. Further studies of Der may provide us with an insight into the role of Der in ribosome biogenesis. Due to the unique primary sequences and topology of the KH-like domain in Der, it may be possible to design antibiotics that inhibit the binding of the KH-like domain to 50S subunits.
Acknowledgments
We thank Sangita Phadtare and Ursula Jacob for their scientific insights and kind gift of strain ΔrrmJ (HB23).
Footnotes
Published ahead of print on 19 February 2010.
REFERENCES
- 1.Bharat, A., M. Jiang, S. M. Sullivan, J. R. Maddock, and E. D. Brown. 2006. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 188:7992-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard, S. C., and J. D. Puglisi. 2001. Solution structure of the A loop of 23S ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A. 98:3720-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bügl, H., E. B. Fauman, B. L. Staker, F. H. Zheng, S. R. Kushner, M. A. Saper, J. C. A. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 4.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 5.Caldas, T., E. Binet, P. Bouloc, and G. Richarme. 2000. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271:714-718. [DOI] [PubMed] [Google Scholar]
- 6.Caldon, C. E., P. Yoong, and P. E. March. 2001. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 41:289-297. [DOI] [PubMed] [Google Scholar]
- 7.Chao, F. C., and H. K. Schachman. 1956. The isolation and characterization of a macro-molecular ribonucleoprotein from yeast. Arch. Biochem. Biophys. 61:220-230. [DOI] [PubMed] [Google Scholar]
- 8.Chao, F. C. 1957. Dissociation of macromolecular ribonucleoprotein of yeast. Arch. Biochem. Biophys. 70:426-431. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., D. L. Court, and X. H. Ji. 1999. Crystal structure of ERA: a GTPase-dependent cell cycle regulator containing an RNA binding motif. Proc. Natl. Acad. Sci. U. S. A. 96:8396-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghrayeb, J., H. Kimura, M. Takahara, H. Hsiung, Y. Masui, and M. Inouye. 1984. Secretion cloning vectors in Escherichia coli. EMBO J. 3:2437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafsson, C., and B. C. Persson. 1998. Identification of the rrmA gene encoding the 23S rRNA m(1)G745 methyltransferase in Escherichia coli and characterization of an m(1)G745-deficient mutant. J. Bacteriol. 180:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hager, J., B. L. Staker, H. Bügl, and U. Jakob. 2002. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 277:41978-41986. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y. P. J., G. V. T. Swapna, P. K. Rajan, H. P. Ke, B. Xia, K. Shukla, M. Inouye, and G. T. Montelione. 2003. Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J. Mol. Biol. 327:521-536. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, J., and M. Inouye. 2001. An essential GTPase, Der, containing double GTP-binding domains from Escherichia coli and Thermotoga maritima. J. Biol. Chem. 276:31415-31421. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, J., and M. Inouye. 2006. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 61:1660-1672. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, K., J. Alsina, J. Q. Chen, and M. Inouye. 2003. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 48:1005-1016. [DOI] [PubMed] [Google Scholar]
- 17.Klein, D. J., P. B. Moore, and T. A. Steitz. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10:1366-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner, C. G., T. Kobayashi, and M. Inouye. 1990. Isolation of subtilisin pro-sequence mutations that affect formation of active protease by localized random polymerase chain-reaction mutagenesis. J. Biol. Chem. 265:20085-20086. [PubMed] [Google Scholar]
- 19.Lövgren, J. M., and P. M. Wikstrom. 2001. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J. Bacteriol. 183:6957-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen, C. T., J. Mengel-Jorgensen, F. Kirpekar, and S. Douthwaite. 2003. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 31:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy, B. J. 1962. Effects of magnesium starvation on ribosome content of Escherichia coli. Biochim. Biophys. Acta 55:880-888. [Google Scholar]
- 22.Muench, S. P., L. Xu, S. E. Sedelnikova, and D. W. Rice. 2006. The essential GTPase YphC displays a major domain rearrangement associated with nucleotide binding. Proc. Natl. Acad. Sci. U. S. A. 103:12359-12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, V. L., J. Hwang, E. Fox, M. Inouye, and A. M. Stock. 2002. Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10:1649-1658. [DOI] [PubMed] [Google Scholar]
- 24.Sharma, M. R., C. Barat, D. N. Wilson, T. M. Booth, M. Kawazoe, C. Hori-Takemoto, M. Shirouzu, S. Yokoyama, P. Fucini, and R. K. Agrawal. 2005. Interaction of era with the 30S ribosomal subunit: implications for 30S subunit assembly. Mol. Cell 18:319-329. [DOI] [PubMed] [Google Scholar]
- 25.Spee, J. H., W. M. Devos, and O. P. Kuipers. 1993. Efficient random mutagenesis method with adjustable mutation frequency by use of PCR and dITP. Nucleic Acids Res. 21:777-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan, J., U. Jakob, and J. C. A. Bardwell. 2002. Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter, M., S. G. Clark, and A. D. Levinson. 1986. The oncogenic activation of human P21ras by a novel mechanism. Science 233:649-652. [DOI] [PubMed] [Google Scholar]
- 28.Widerak, M., R. Kern, A. Malki, and G. Richarme. 2005. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene 347:109-114. [DOI] [PubMed] [Google Scholar]