Abstract
Flagellar motility has long been regarded as an important virulence factor. In Vibrio cholerae, the single polar flagellum is essential for motility as well as for proper attachment and colonization. In this study, we demonstrate that the novel flagellar protein FlgT is involved in anchoring the flagellum to the V. cholerae cell. A screen for novel colonization factors by use of TnphoA mutagenesis identified flgT. An in-frame deletion of flgT established that FlgT is required for attachment, colonization, and motility. Transmission electron microscopy revealed that while the flgT mutant is capable of assembling a phenotypically normal flagellum, the flgT population is mostly aflagellate compared to the wild-type population. Further analyses indicated that the flagellum of the flgT mutant is released into the culture supernatant from the cell upon completion of assembly. Additionally, hook basal body complexes appear to be released along with the filament. These results indicate that FlgT functions to stabilize the flagellar apparatus at the pole of the cell.
When humans ingest water or food that has been contaminated with Vibrio cholerae, an infection of the small intestine, known as the diarrheal disease cholera, can occur. V. cholerae is a Gram-negative, motile bacterium. Its single polar, sheathed flagellum functions as a means for the bacterium to move through its environment. Motility is essential for proper attachment and subsequent colonization of the intestinal epithelium (24, 28). It is well established that nonmotile mutants are less virulent than their motile counterparts. This is due to the fact that a fully functional flagellum is absolutely required for attachment and colonization (24, 28). We recently demonstrated that the reduced attachment phenotype displayed by nonmotile mutants is attributed to the inability of the bacterium to reach the epithelium (28), not from the previously supposed loss of adhesive properties (3, 12, 14, 36). More specifically, motility is necessary for the bacterium to swim through the mucus layer that protects the epithelial cells (3, 6, 14, 26). Passage through the mucus layer of the epithelium leads to flagellar breakage, which in turn signals to inform the V. cholerae cells that they have arrived at an appropriate site for colonization (26). Hence, motility has long been studied as a potential target for vaccine and drug therapies.
The flagellum is flexible and durable; it is made to withstand physical stress and force, and it is a powerful machine that is firmly anchored to the cell (8). The V. cholerae flagellum is protected by a membranous sheath, which appears contiguous with the outer membrane. At its most basic level, the flagellum consists of a base, a hook, and a filament. In actuality, the flagellum is a complex structure made up of dozens of different proteins (29). Flagellar biogenesis requires an enormous energy investment, and as such, it calls for a highly ordered and regulated process of assembly (1, 35). For example, there are six regions (I to VI) distributed throughout the large chromosome that contain the genes encoding motility and chemotaxis proteins (35). The genes are further categorized into four classes (I to IV). Transcription of these genes is regulated in a hierarchal manner, whereby transcription begins with the class I flagellar gene flrA, which encodes the master regulator, FlrA. In turn, FlrA activates the transcription of class II genes in a σ54-dependent manner (17, 35). Besides flagellar structural proteins, the class II gene products also include additional flagellar regulatory proteins, such as FlrB, FlrC, FlhF, FlhG, and FliA (σ28) (42). FlrC, together with σ54, activates the transcription of class III genes (35). Lastly, class IV genes are activated by σ28. The flagellar regulatory hierarchy also includes flagellar checkpoints that inhibit downstream gene expression.
In the present study, we investigate a novel motility gene, flgT, and the role of its product in stabilizing the flagellum of V. cholerae. Recently, flgT (VC2208) was discovered during a screen in the El Tor biotype strain C6706 to identify novel chemotactic motility genes (7). The class III flagellar gene flgT lies adjacent to the flgOP operon, within region I (7, 32, 42). The ΔflgT mutant displayed a severe defect in motility, and it was reported that very few cells were flagellated or motile (7). Since FlgT shares homology with the membrane-stabilizing protein TolB, Cameron et al. concluded that FlgT might play a role in flagellar sheath formation (7).
Here we report that a TnphoA mutagenesis study performed by our laboratory identified flgT as a novel colonization factor involved in epithelial cell attachment. The aim of this work was to investigate the role of FlgT in motility. Additionally, we establish that FlgT is involved in anchoring the flagellum of V. cholerae to the cell.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and DNA manipulations.
The strains and plasmids used in this study are listed in Table 1. All cultures were grown in LB medium supplemented with antibiotics at 37°C. Antibiotics were used at the following concentrations: streptomycin (Sm), 100 μg/ml; kanamycin (Km), 45 μg/ml; and ampicillin (Amp), 100 μg/ml. All DNA manipulations were performed using standard molecular and genetic techniques (4, 27). The ΔflgT and ΔflaA mutants were constructed by allelic exchange as previously described (37).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| S17-1λpir | Tpr SmrrecA thi pro hsdR M+ [RP4-2Tc::Mu::Kmr Tn7] (λpir) | 34; laboratory collection |
| Top-10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| V. cholerae strains | ||
| O395 | Classical Ogawa strain; Smr | Laboratory collection |
| KSK258 | O395 ΔlacZ3 | Karen Skorupski |
| BAJ43 | O395 ΔflgT | This study |
| BAJ61 | BAJ43(pBro26) | This study |
| RM414 | BAJ43(pBro37) | This study |
| RM421 | BAJ43(pBAD202) | This study |
| TJK189 | O395 ΔflaA | This study |
| Plasmids | ||
| pBAD202 | Kmr | Invitrogen |
| pBAD-TOPO | Ampr | Invitrogen |
| pBro26 | pBAD-TOPO FlgT-His6 | This study |
| pBro32 | pKAS32 ΔflgT | This study |
| pBro37 | pBAD202 FlgT-His6 | This study |
| pKAS32 | Allelic exchange vector; Ampr | 37 |
| pTK20 | pKAS32 ΔflaA | This study |
Transposon mutagenesis and mutant screen.
Transposon mutagenesis and selection of colonies for analysis were done as previously described (15, 43). The locations of the transposon insertion in mutants of interest were determined by chromosomal capture and DNA sequencing (38, 39). Transposon mutants were screened for defective epithelial cell attachment by use of the bacterial attachment assay described below. Approximately 400 candidate clones bearing random TnphoA insertions were each individually evaluated for the ability to attach to a monolayer of epithelial cells (15). Mutants defective for attachment were then screened for the ability to colonize the intestinal epithelium, using the infant mouse model of cholera as described below.
Tissue culture and bacterial attachment assay.
HT-29 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% Pen-Strep (Gibco) under 5% CO2, as previously described (5, 16). Overnight bacterial cultures were diluted 1:100 in phosphate-buffered saline (PBS) before incubation with confluent HT-29 monolayers. The bacterial cells were allowed to attach to the HT-29 cells for 1 h at 37°C. The HT-29 monolayers and attached bacteria were washed three times with PBS to remove unattached bacterial cells. The HT-29 cells were then lysed by incubating the coculture for 30 min in a 1% Triton X-100 solution. The resulting bacterial suspension was serially diluted and plated for enumeration of bacteria. LB agar plates supplemented with streptomycin and 5-bromo-4-chloro-3-indolyl-d-galactopyranoside (X-Gal) (40 μg/ml) were used for competitive attachment assays.
Infant mouse cholera model.
As previously described, overnight cultures were diluted 1:100, and each test strain was mixed with equal numbers (1:1) of an O395 ΔlacZ reference strain. Four- to 6-day-old CD-1 mice from mixed litters were orally inoculated with 50 μl of the bacterial mixture (16). The mice were then incubated at 30°C for 24 h. The bacteria were then recovered by homogenizing harvested intestines in 4 ml of LB broth containing 10% glycerol, using a Tissue Tearor homogenizer. The homogenate was serially diluted and plated on LB agar plates supplemented with streptomycin and 5-bromo-4-chloro-3-indolyl-d-galactopyramoside (X-Gal) (40 μg/ml) [test (output/input)/reference (output/input)]. Animal experiments were done in compliance with institutional animal care and use guidelines.
Motility assays and phase-contrast microscopy.
Motility plates (LB with 0.3% agar) were inoculated to test motility phenotypes. Colonies grown on solid medium were stabbed to the bottom of the soft agar plate by use of sterile round toothpicks. Plates were incubated at 37°C for 8 to 12 h. For visualizing motility, a few colonies from a fresh plate were suspended in 0.5 ml of LB, and 10 μl was placed on a glass slide for observation by phase-contrast microscopy. In triplicate, 1,000 cells were observed to calculate the percentage of motile ΔflgT cells.
Transmission electron microscopy (TEM) and immunoelectron microscopy.
Negatively stained samples were prepared as previously described, with modification (28). Briefly, overnight cultures were diluted 1:100 in fresh medium and grown to late log phase (optical density at 600 nm [OD600] of ∼2.0), unless otherwise stated. Formvar-coated grids were incubated with cultures for 2 min. The grids were then fixed in a 4% paraformaldehyde solution and washed three times for 1 min each in 0.1 M sodium cacodylate. Whole-cell preparations were negatively stained with 0.5% phosphotungstic acid (pH 6.5) for 2 min. Osmotically shocked cells were prepared by incubating cells in a 1% Triton X-100 solution (29) for 60 min.
Subcellular fractionation, SDS-PAGE, and immunoblot analysis.
Whole-cell extracts were prepared by suspending overnight cultures in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and were boiled for 10 min. A 2× SDS-PAGE buffer without 2-mercaptoethanol and bromophenol blue was used for protein estimation. Subcellular fractionation was performed as previously described (45). Protein concentrations of whole-cell extracts and subcellular fractions were determined with a bicinchoninic acid (BCA) protein assay kit (Pierce). Samples containing equal amounts of total protein were resolved by SDS-PAGE. Proteins were transferred to nitrocellulose membranes at 4°C. All primary antibodies were used at a dilution of 1:1,000. The horseradish peroxidase-conjugated secondary antibodies (anti-rabbit or anti-mouse) were used at a dilution of 1:10,000. ECL detection reagents were utilized to visualize reactive bands (Pierce). For the fractionation experiment, the samples were first probed with the anti-His antibody. After visualization, the membrane was stripped (Western Re-Probe; G-Bioscience) and reprobed with control antibodies to test the purity of the fractions (anti-FlgT, anti-VieA, anti-DsbA, anti-ToxR, and anti-OmpW).
For the flagellum (anti-flagellin) experiments, whole-cell extracts were prepared as follows. One milliliter of culture at an OD600 of 4.0 was centrifuged at 14,000 × g for 1 min. The supernatant was removed and placed in a fresh tube for further use. PBS was then added to the cell pellet, which was then suspended by 5 passes through a pipette. The tube was then gently inverted to ensure full suspension of the cells. The cells were then centrifuged, and the wash fraction was removed and stored in a fresh tube for further use. The cells were suspended in 500 μl of 2× SDS-PAGE buffer without 2-mercaptoethanol and bromophenol blue for protein estimation as described above. Whole-cell extracts were loaded according to equal amounts of total protein, whereas the supernatant and wash samples were first filter sterilized, using a 0.22-μm filter, to remove contaminating cells. Afterward, equal volumes of supernatant were loaded and further analyzed as mentioned above.
Protein purification and antiserum generation.
Escherichia coli Top-10 strains (Invitrogen) expressing pFlgT-His6 were grown overnight at 37°C, with shaking at 220 rpm, in LB medium supplemented with Amp at 100 μg/ml and with 0.02% arabinose. Cells were harvested by centrifugation at 7,000 × g for 15 min at 4°C and were suspended in an appropriate volume of 50 mM NaH2PO4 and 300 mM NaCl (pH 7.0). Cells were lysed via sonication, using an output setting of 23 W, for five rounds of five 10-s pulses, with each round separated by 1 min of recovery on ice. Lysates were centrifuged at 23,000 × g for 10 min at 4°C to remove cell debris. The tagged protein in the resulting supernatant was initially isolated using metal-affinity chromatography, eluting bound protein with 200 mM imidazole in 50 mM NaH2PO4 and 300 mM NaCl (pH 7.0) (Talon metal-affinity resin; BD Biosciences). This was followed by additional purification by gel filtration on a HiPrep Sephacryl S-100 column (Amersham Biosciences). Fractions were separated in a 12.5% SDS-containing polyacrylamide gel and stained with Coomassie blue for analysis. Fractions containing purified protein were concentrated by centrifugation in a Centriplus concentrator (Millipore) to 0.7 mg/ml in a buffer containing 50 mM NaH2PO4 and 300 mM NaCl (pH 7.0). Antibodies against FlgT were generated in rabbits, using FlgT-His6-tagged purified proteins (Pocono Rabbit Farm and Laboratory).
RESULTS
FlgT is required for attachment and colonization.
Using TnphoA mutagenesis, we identified flgT in an initial screen for putative colonization factors in the classical biotype strain O395 (17). We constructed an in-frame chromosomal deletion of the flgT gene to further examine the contribution of FlgT to surface attachment and colonization (Fig. 1).
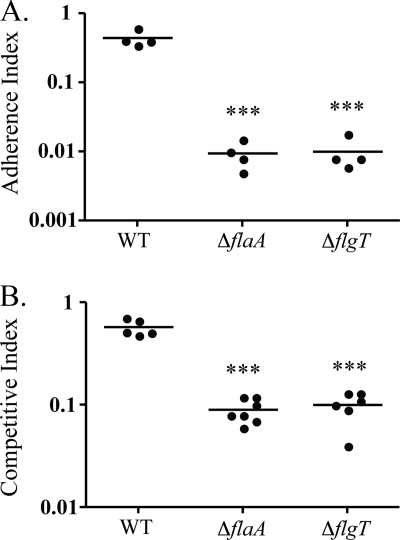
FIG. 1.
The ΔflgT mutant is defective in attachment and colonization. (A) V. cholerae strain O395 (WT) and ΔflaA and ΔflgT mutants were competed against an O395 ΔlacZ strain (KSK258) for in vitro attachment to HT-29 epithelial cells. Statistical analyses (unpaired Student's t test) were performed between each mutant and the WT, as indicated by asterisks. ***, P < 0.0001. (B) V. cholerae strain O395 (WT) and ΔflaA and ΔflgT mutants were competed against an O395 ΔlacZ strain (KSK258) for in vivo colonization of infant mice, using four to seven mice per group. Statistical analyses (unpaired Student's t test) were performed between each mutant and the WT, as indicated by asterisks. ***, P < 0.0001.
In an in vitro competitive attachment assay, the flgT mutant displayed a 40-fold decrease in attachment to epithelial cells compared to the wild type (WT) and was also similar to the aflagellate flaA mutant (17), which served as a negative control (Fig. 1A). Because initial attachment is important for colonization, we expected the flgT mutant to also be defective in colonizing the intestinal epithelium in the infant mouse cholera model. In this assay, we observed a 6-fold reduction in colonization of the flgT mutant compared to the WT (Fig. 1B).
FlgT is required for motility.
Because flgT belongs to flagellar gene cluster region I, divergently transcribed from the adjacent flgOP operon (7, 32, 35, 42), we further investigated the motility phenotype of the flgT mutant. In agreement with a recent report by Cameron et al., we found the ΔflgT strain to be nonmotile in 0.3% motility agar (7) (Fig. 2A). Expressing FlgT from a plasmid restored motility, confirming that FlgT is indeed required for motility. Additionally, Cameron et al. reported that motile cells were rarely seen via phase-contrast microscopy, and we too observed that the majority of the population was nonmotile; however, approximately 1 of every 1,000 ΔflgT cells (0.001%) was motile (7). Furthermore, the nonmotile phenotype of the flgT mutant helps to explain the attachment and colonization defects (Fig. 1).
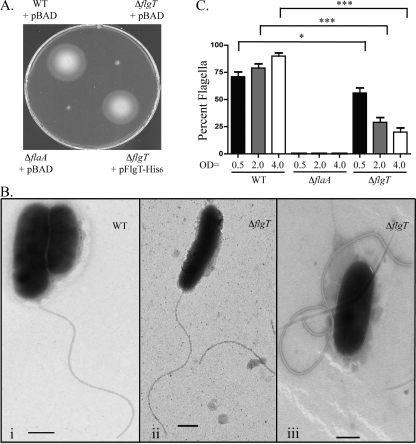
FIG. 2.
FlgT is essential for motility and flagellar stability. (A) Zones of motility of V. cholerae WT, ΔflaA, ΔflgT, and ΔflgT/pFlgT-His6 strains in motility agar. (B) Transmission electron micrographs showing the polar flagella of the V. cholerae WT (i) and the ΔflgT strains. The two ΔflgT mutant cell types are shown in panels ii (flagellate) and iii (aflagellate). Grids were prepared at an OD600 of ∼2.0. Bar, 500 nm. (C) Graphical representation of percent flagellated cells of the V. cholerae WT, ΔflaA, and ΔflgT strains during mid-log (OD600 = 0.5), late-log (OD600 = 2.0), and stationary (OD600 = 4.0) phases. A total of 100 cells per strain were examined via transmission electron microscopy. Brackets indicate statistical comparisons (unpaired Student's t test) between the indicated data sets. ***, P < 0.0001; *, P < 0.05.
We next investigated the cause of the nonmotile phenotype of the flgT mutant. Using TEM, we assessed whether the ΔflgT strain produced a flagellum or whether it was aflagellate (OD600, ∼2.0). Upon initial examination, we observed two distinct cell types within the ΔflgT population (Fig. 2B). Although there were cells with polar flagella (Fig. 2B, panel ii), the majority of the cells were aflagellate (Fig. 2B, panel iii). Also, it is important that unattached flagella, which appeared normal in length and structure, were commonly observed elsewhere on the grids containing the mutant samples but not on the grids containing the WT samples. These observations imply that the deletion of flgT results in the loss of motility due to the fact that the flagellum does not remain associated with the cell.
In order to better understand the extent of the flagellar defect of the flgT mutant, we next quantified the proportions of the WT and ΔflgT populations that were flagellated (Fig. 2C). Over time, the number of flagellated cells within the WT population increased, with a maximum of 90% of the population being flagellated. In contrast, during mid-log-phase growth (OD600 = 0.5), the flgT mutant population appeared to be flagellated to some extent (56%); however, at late log phase (OD600 = 2.0), the proportion of flagellated cells decreased severely, to only 29%, making the flagellar defect more evident. Moreover, in stationary phase (OD600 = 4.0), the population of flagellated ΔflgT cells was reduced to 20%. As expected, the aflagellate flaA mutant population never produced flagella. These data indicate that the flgT mutant is defective in retaining its flagellum compared to the WT (90% versus 20% flagellation; 4.5-fold difference). Because unattached or released whole flagella were routinely observed during TEM, this strongly suggests that the reduced number of flagellated cells within the ΔflgT population was not due to a defect in flagellar assembly or production but rather to a defect in the ability to anchor the flagellum to the cell.
The flgT mutant releases its flagellum into the culture supernatant.
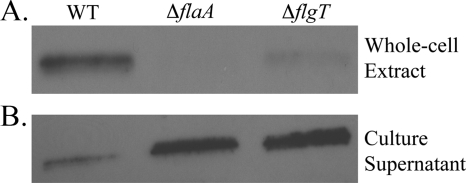
To test the hypothesis that FlgT is important for anchoring the flagellum to the cell, whole-cell extracts were examined for the loss of the flagellum from the flgT mutant compared to the WT. The flagellum of V. cholerae is comprised of 5 flagellins, FlaABCDE, of which only FlaA is required for filament assembly (17). Notably, flaA is a class III gene, while the flaBCDE genes belong to class IV (17, 35). An anti-flagellin antibody (30) was used as a means to identify flagella. This antibody recognizes all five flagellins, which share extensive homology with each other and run closely together in a protein gel, appearing as one large band in anti-flagellin immunoblots (Fig. 3) (17, 28, 30). As shown in Fig. 3A, the flgT mutant produced a barely detectable level of cell-associated flagellin compared to the WT. In the ΔflaA control, no flagellin was detected because this strain produces and secretes alternate flagellins into the culture supernatant that are not associated with the whole-cell extract (17, 46).
FIG. 3.
The flagellum produced by the ΔflgT mutant does not remain associated with the bacterial cell. (A) Anti-flagellin antiserum was used to probe whole-cell extracts prepared from the V. cholerae WT, ΔflaA, and ΔflgT strains at an OD600 of 4.0. (B) Anti-flagellin antiserum was used to probe the supernatants from the cultures that were used to prepare the whole-cell extracts shown in panel A. Results were similar at an OD600 of 2.0; however, the overall levels of flagellin were reduced (data not shown).
The supernatants of the above cultures were also monitored for the presence of flagellin. We hypothesized that since the flgT flagella were less strongly associated with the cells, there would be an increase of flagella present in the supernatant of the flgT mutant compared to the WT supernatant. As expected, the supernatant collected from the flgT mutant contained larger amounts of flagellin than did the WT supernatant (Fig. 3B). The increased expression and secretion of alternate flagellins by the flaA mutant were also observed in the culture supernatant, as previously reported (17, 46). The results presented thus far indicate that the flgT mutant can indeed assemble a flagellum; however, that flagellum does not remain associated with the cell and instead is released into the culture supernatant.
The flgT mutant releases the entire flagellum.
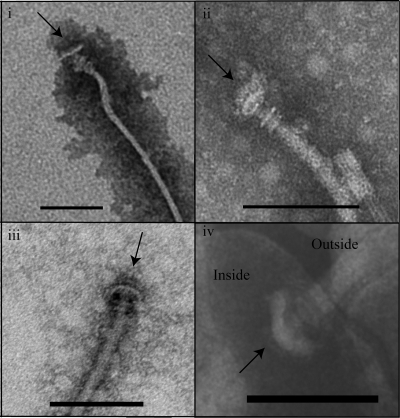
The evidence suggesting that FlgT plays a role in anchoring the flagellum to the cell prompted further ultrastructural investigation of the flgT mutant. TEM confirmed that full-length flagellar filaments were regularly released into the culture supernatant, as noted above (OD600, ∼2.0). The majority (84%) of the filaments remained associated with the hook, whereas only 16% of the filaments were also associated with the native hook basal bodies (HBBs), which are not normally visible because they are buried and well anchored within the cell (Fig. 4, panels i to iii). In our hands, the isolation of HBBs from WT V. cholerae proved to be extremely difficult and resulted in the purification of what ultimately appeared to be the rod substructure (data not shown). Only when WT cells were osmotically shocked could the HBB structure be visualized (Fig. 4, panel iv). These data definitively show that a large portion, if not all, of the flagellar apparatus is released into the supernatant by the flgT mutant. Importantly, the same structures were never observed in the WT supernatants. Collectively, these data further support the hypothesis that FlgT functions to anchor the flagellum to the cell.
FIG. 4.
Flagella with the HBB complex are released into the culture supernatant of the flgT mutant. (i to iii) Transmission electron micrographs of flagella released into the culture supernatant by the flgT mutant. (iv) Transmission electron micrograph of the polar flagellum of an osmotically shocked WT cell. Arrows point to the base of the flagellum. Grids were prepared at an OD600 of ∼2.0. Bar, 100 nm.
Membrane protrusions can be seen at the sites of flagellar release.
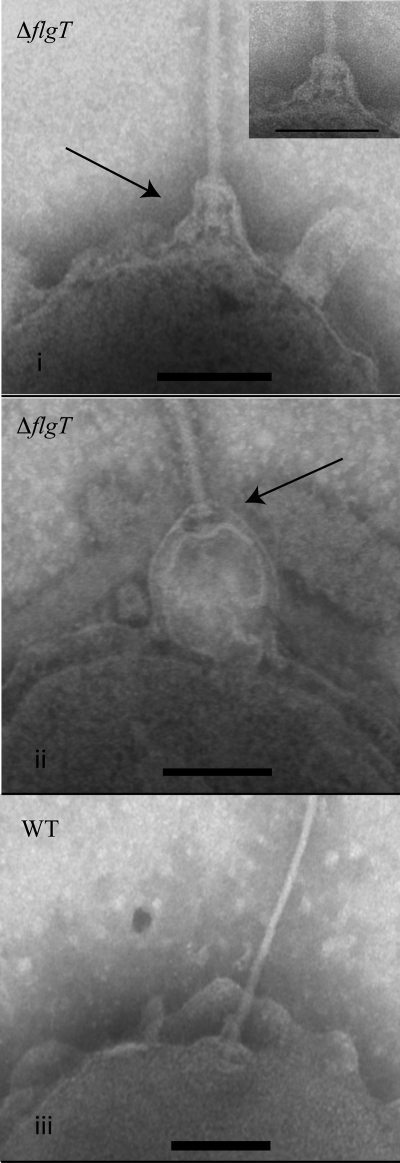
During our TEM studies, we were able to capture several images showing the flagellar apparatus being released from the cell (OD600, ∼2.0) (Fig. 5). In the flgT mutant, it appears as though the only entity holding the flagellum to the cell is the outer membrane (Fig. 5, panels i and ii). Outer membrane blebbing and the occurrence of membrane vesicles are common phenomena in Gram-negative organisms (21, 25). Interestingly, membrane blebbing also occurred with the WT strain, but the flagellum remained anchored to the cell (Fig. 5, panel iii). Membrane blebbing also occurred with nonflagellated cells (data not shown). These data suggest that either (i) membrane blebbing facilitates the release of the flagellum or (ii) as the flagellum is released from the cell due to the loss of proper anchoring, it remains associated with the membrane, thus resulting in a protrusion event.
FIG. 5.
FlgT is involved in anchoring the flagellum. (i and ii) Transmission electron micrographs of flagella being released from the flgT mutant. (iii) Transmission electron micrograph of the flagellar pole of the WT. Arrows indicate the flagellar apparatus. Grids were prepared at an OD600 of ∼2.0. Bar, 100 nm.
FlgT localizes to the periplasm.
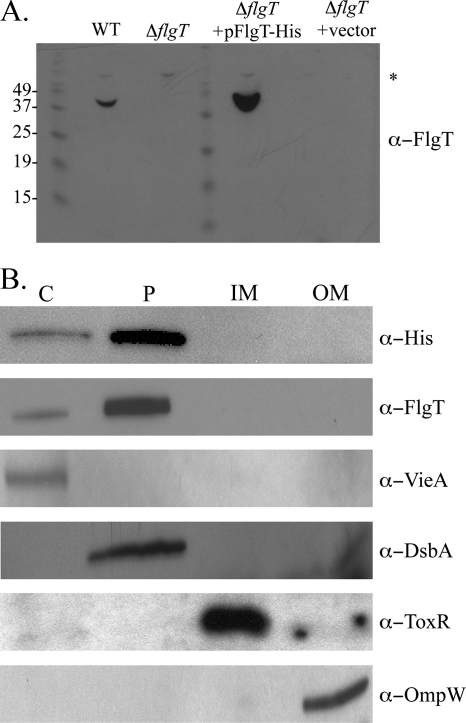
Because FlgT appears to anchor the flagellum to the cell, we were interested in determining the location of FlgT in the cell. Also, since FlgT is predicted to be a secreted protein and to share homology with the periplasmic proteins TolB (7) and MreC (40; HHpred), we utilized subcellular fractionation to purify the periplasm from the WT and ΔflgT strains and used an anti-FlgT antibody to determine whether FlgT was indeed a periplasmic protein as predicted. As shown in Fig. 6A, a band was detected in the periplasmic fraction of the WT strain, near the predicted molecular mass of FlgT, 42.6 kDa, but not in the periplasmic fraction of the ΔflgT strain. Likewise, we observed a prominent band in the periplasmic fraction of the ΔflgT mutant complemented with pFlgT.
FIG. 6.
Cellular localization of FlgT. (A) Anti-FlgT immunoblot of periplasmic fractions isolated from V. cholerae WT, ΔflgT, ΔflgT/pFlgT-His6, and ΔflgT/vector strains. The asterisk indicates a cross-reactive band. The apparent molecular weights of the protein ladder are listed to the left of the gel. (B) Subcellular fractionation was carried out on the ΔflgT/pFlgT-His6 strain. The cytoplasm (lane C), periplasm (lane P), inner membrane (lane IM), and outer membrane (lane OM) fractions were isolated and separated by SDS-PAGE, followed by anti-His and anti-FlgT immunoblotting. Anti-VieA, anti-DsbA, anti-ToxR, and anti-OmpW immunoblots were performed to confirm fraction purity.
To confirm the localization of FlgT in the periplasm, all subcellular fractions of the ΔflgT mutant complemented with pFlgT-His6 were analyzed for the presence of FlgT. This experiment confirmed that the majority of FlgT was present in the periplasmic fraction, as determined by anti-His and anti-FlgT immunoblot analyses (Fig. 6B). As controls, other known proteins were examined to ensure the purity of the isolated fractions. VieA was correctly located in the cytoplasm (23), and DsbA was appropriately located in the periplasm (33). Additionally, the inner membrane protein ToxR (31) and the outer membrane protein OmpW (9) were both associated with the proper membrane fractions. From these data, we concluded that FlgT is a periplasmic protein that functions to anchor the flagellum to the cell.
DISCUSSION
FlgT was identified using a screen for novel colonization factors. FlgT is essential for attachment, colonization, and motility. We recently reported that the phenotype of reduced attachment of nonmotile mutants to epithelial cells is attributed to the loss of flagellar motility (28). Similarly, Lee et al. demonstrated that colonization also requires functional motility (24). In light of the finding that the flgT mutant is nonmotile, this explains its phenotypes of reduced attachment and colonization. Flagellar motility aids in the ability of V. cholerae to come into contact with epithelial cells (3, 6, 14, 24, 26, 28). Therefore, most nonmotile mutants are defective in adherence and colonization because they do not make sufficient contact with the epithelium. Some nonmotile mutants such as rpoN, flrC, and flgP mutants do appear to have a role in colonization independent of their role in motility (18, 28, 32).
The flgT mutant is capable of assembling a flagellum; however, once assembly is complete, the flgT mutant releases its flagellum into the culture supernatant. This is indicated by the fact that whole-cell extracts are largely devoid of flagella, while the corresponding supernatants contain large amounts of flagella. Examination of the flgT mutant by TEM corroborated these findings. The flgT mutant appeared to be mostly aflagellate, which is in agreement with the findings of Cameron et al. (7). Full-length flagellar filaments were also present on the grids of the flgT mutant, in contrast to the case for the WT. Although Cameron et al. did not report unattached flagella in their samples, this may be due to a difference in the way their samples were prepared compared to ours (7). Cameron et al. gently pelleted and resuspended the bacterial cells prior to grid preparation. In contrast, we used wide pipette tips to gently pipette the cells directly from the culture onto the grids, minimally disrupting the cells. During the centrifugation step employed by Cameron et al., they may have removed the flagella that had been released into the supernatant, analogous to the method we used to prepare whole-cell extracts (Fig. 3).
Since flagellated cells within the flgT population were rarely observed, Cameron et al. performed a transcriptional analysis of the flgT mutant in order to understand the role of flgT in flagellar biosynthesis (7). From these experiments, it was determined that the flgT mutant is not able to initiate the transcription of class IV flagellar genes, which notably include the alternate flagellin genes flaBCDE as well as the motor genes motX, motA, and motB (none of which are required for the assembly of the flagellum [11, 17]).
The motor is comprised of two distinct components, the rotor and the stator. The stator components, MotA and MotB, form a channel through which ions (H+ or Na+, depending on the type of motor) can flow, resulting in the generation of force (20). V. cholerae, as well as other marine organisms, utilizes a sodium-driven motor, as opposed to a proton-driven motor, which is best studied using the organisms Salmonella and E. coli as model systems.
Besides the different ions (Na+ versus H+) that are required to generate force, another notable feature of the sodium-driven motor is an additional ring substructure known as the T ring (44). The MotX and MotY proteins make up the T ring, which is located between the MS and P rings of the basal body. The T ring promotes the localization and stabilization of MotA and MotB (10, 19, 20). In the absence of either MotY or MotX, the T ring is not formed and the MotA-MotB complex does not localize to the pole (44).
Compared to published images of HBBs isolated from various organisms, the HBBs of the flgT mutant appear most similar to those of Salmonella spp. (2, 41), not like those of Vibrio spp. (44). From these results, together with the transcriptional analysis provided by Cameron et al., we hypothesize that the T ring is lacking from the flagellum produced by the flgT mutant.
Since the flgT mutant is capable of expressing class III genes (motY) but not class IV genes (motX, motA, and motB), the T ring should not be formed (7). Interestingly, a ΔmotXYAB strain is flagellated like the WT, but it is nonmotile (11), indicating that the loss of the stator components does not affect the stability of the flagellum at the pole. These results further support our hypothesis that FlgT is required for anchoring the flagellum to the cell.
Membrane protrusions were observed at the sites of flagellar release of the flgT mutant. We were unable to determine whether the membrane protrusions are the cause or effect of flagellar release. It is likely that the flagellar substructures that are associated with the inner and outer membranes (the MS and L rings, respectively) continue to associate with the membrane and that as the flagellum is released, the membrane is pulled away from the cell, causing the appearance of a membrane protrusion. It also seems likely that FlgT could function to stabilize the membrane surrounding the flagellar pole. Evidence for this lies in the predicted homology between FlgT and the periplasmic protein TolB (7), which is important for membrane stability (13, 22). Collectively, the data presented here are consistent with the hypothesis that FlgT is an important anchoring component of the V. cholerae flagellum.
Because FlgT is required for flagellar anchoring at the pole, we attempted to determine whether or not FlgT is localized polarly. Unfortunately, we have been unable to detect FlgT by immunoelectron microscopy of thin-sectioned WT and ΔflgT samples (data not shown). If FlgT is localized polarly, then the cell would have to be sectioned directly through the pole in such a way that we could detect FlgT. The rare occurrence of such sections may contribute to our inability to detect FlgT by this method. In the future, immunofluorescence studies might provide better insight into the localization of FlgT.
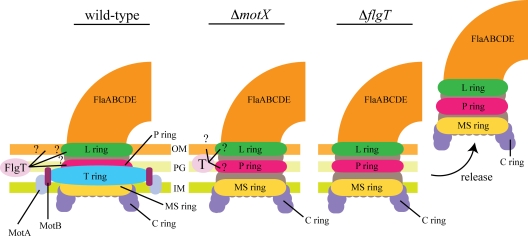
The information provided by the motor mutants (motX and motY) established the background from which we devised a model for the function of FlgT in flagellar anchoring (11, 44). Since the flgT mutant does not express motX, in theory it should be phenotypically similar to the ΔmotX strain with regard to the motor components (Fig. 7). As noted previously, the motX mutant does not produce a T ring and the MotA-MotB complex does not localize to the pole. Moreover, the motX mutant displays a normal flagellum that is anchored to the pole, similar to that of the WT (11, 28). Immunoblot analysis confirmed that the ΔmotX strain does express FlgT (data not shown). In the case of the flgT mutant, the flagellum is no longer anchored and is released from the cell. This suggests that FlgT is probably the major anchoring component of the flagellum of V. cholerae. However, the mechanism by which FlgT functions in the periplasm to provide anchoring support to the flagellar apparatus is not understood. Although orthologs of FlgT are present in other organisms that utilize a sodium-driven motor, such as other Vibrio spp. and Shewanella spp., none have been characterized. Because the T ring is not required for anchoring the flagellum, and because the MS, P, and L rings are not sufficient to anchor the flagellum in the flgT mutant, it is possible that FlgT interacts with different components of the HBB as well as with the outer membrane (Fig. 7). Pull-down experiments combined with cross-linking assays are under way. These experiments could identify FlgT-interacting proteins, which may offer information on how FlgT functions to stabilize the flagellum.
FIG. 7.
Role of FlgT in anchoring the flagellum to the V. cholerae cell body. For WT cells, the flagellum is securely anchored to the cell, which is also true for the motX mutant. Although the motX mutant lacks the stator components that make up the motor, flagellar stability is maintained, presumably via FlgT. In the case of the flgT mutant, the flagellum is similar to that of the motX mutant in that it does not assemble the T ring. However, unlike the case in the motX mutant, once flagellar assembly is complete, the flagellum is released from the cell of the flgT mutant, suggesting that FlgT plays a crucial role in anchoring the flagellum to the cell. It is hypothesized that FlgT could provide additional support to the preexisting anchoring components, such as the P and L rings. Another possibility is that FlgT stabilizes the membrane surrounding the flagellar apparatus. It also seems likely that FlgT functions as an indirect anchoring protein, such that FlgT forms a complex with another protein(s) in order to anchor the flagellar apparatus to the cell.
Acknowledgments
We thank Sarah M. Eck for a critical reading of the manuscript. We thank Louisa Howard for assistance with transmission electron microscopy. We give special thanks to Andrew Camilli, Debra Milton, and Johnny Peterson for the generous gifts of the anti-VieA, anti-flagellin, and anti-OmpW antibodies, respectively.
This work was supported by NIH grants AI025096 and AI039654 and by NSF grant OCN-0120677 to R.K.T. R.M.M. was also supported by NIH training grant T32AI07363.
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Apel, D., and M. G. Surette. 2008. Bringing order to a complex molecular machine: the assembly of the bacterial flagella. Biochim. Biophys. Acta 1778:1851-1858. [DOI] [PubMed] [Google Scholar]
- 2.Attmannspacher, U., B. E. Scharf, and R. M. Harshey. 2008. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol. Microbiol. 68:328-341. [DOI] [PubMed] [Google Scholar]
- 3.Attridge, S. R., and D. Rowley. 1983. The role of the flagellum in the adherence of Vibrio cholerae. J. Infect. Dis. 147:864-872. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 5.Benitez, J. A., R. G. Spelbrink, A. Silva, T. E. Phillips, C. M. Stanley, M. Boesman-Finkelstein, and R. A. Finkelstein. 1997. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells: an in vitro colonization model. Infect. Immun. 65:3474-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, S. M., and A. Camilli. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 101:5018-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, D. E., J. M. Urbach, and J. J. Mekalanos. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 105:8736-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnton, N. C., and H. C. Berg. 2008. Bacterial flagella are firmly anchored. J. Bacteriol. 190:8223-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, M., A. K. Chopra, J. M. Cantu, and J. W. Peterson. 1998. Antisera to selected outer membrane proteins of Vibrio cholerae protect against challenge with homologous and heterologous strains of V. cholerae. FEMS Immunol. Med. Microbiol. 22:303-308. [DOI] [PubMed] [Google Scholar]
- 10.Fukuoka, H., T. Wada, S. Kojima, A. Ishijima, and M. Homma. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825-835. [DOI] [PubMed] [Google Scholar]
- 11.Gosink, K. K., and C. C. Häse. 2000. Requirements for conversion of the Na(+)-driven flagellar motor of Vibrio cholerae to the H(+)-driven motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guentzel, M. N., and L. J. Berry. 1975. Motility as a virulence factor for Vibrio cholerae. Infect. Immun. 11:890-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry, T., S. Pommier, L. Journet, A. Bernadac, J. P. Gorvel, and R. Lloubes. 2004. Improved methods for producing outer membrane vesicles in Gram-negative bacteria. Res. Microbiol. 155:437-446. [DOI] [PubMed] [Google Scholar]
- 14.Jones, G. W., G. D. Abrams, and R. Freter. 1976. Adhesive properties of Vibrio cholerae: adhesion to isolated rabbit brush border membranes and hemagglutinating activity. Infect. Immun. 14:232-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863-866. [DOI] [PubMed] [Google Scholar]
- 16.Kirn, T. J., and R. K. Taylor. 2005. TcpF is a soluble colonization factor and protective antigen secreted by El Tor and classical O1 and O139 Vibrio cholerae serogroups. Infect. Immun. 73:4461-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 19.Koerdt, A., A. Paulick, M. Mock, K. Jost, and K. M. Thormann. 2009. MotX and MotY are required for flagellar rotation in Shewanella oneidensis MR-1. J. Bacteriol. 191:5085-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima, S., A. Shinohara, H. Terashima, T. Yakushi, M. Sakuma, M. Homma, K. Namba, and K. Imada. 2008. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. USA 105:7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo, K., A. Takade, and K. Amako. 1993. Release of the outer membrane vesicles from Vibrio cholerae and Vibrio parahaemolyticus. Microbiol. Immunol. 37:149-152. [DOI] [PubMed] [Google Scholar]
- 22.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 180:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Z., A. J. Clarke, and T. J. Beveridge. 1998. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 180:5478-5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Z., T. Miyashiro, A. Tsou, A. Hsiao, M. Goulian, and J. Zhu. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. USA 105:9769-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Martinez, R. M., M. N. Dharmasena, T. J. Kirn, and R. K. Taylor. 2009. Characterization of two outer membrane proteins, FlgO and FlgP, that influence Vibrio cholerae motility. J. Bacteriol. 191:5669-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarter, L. L. 2001. Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGee, K., P. Horstedt, and D. L. Milton. 1996. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J. Bacteriol. 178:5188-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Morris, D. C., F. Peng, J. R. Barker, and K. E. Klose. 2008. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J. Bacteriol. 190:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peek, J. A., and R. K. Taylor. 1992. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 89:6210-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Priefer, U. B., R. Simon, and A. Puhler. 1985. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J. Bacteriol. 163:324-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 38.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skorupski, K., and R. K. Taylor. 1997. Sequence and functional analysis of the gene encoding Vibrio cholerae cAMP receptor protein. Gene 198:297-303. [DOI] [PubMed] [Google Scholar]
- 40.Soding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, T., T. Iino, T. Horiguchi, and S. Yamaguchi. 1978. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J. Bacteriol. 133:904-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syed, K. A., S. Beyhan, N. Correa, J. Queen, J. Liu, F. Peng, K. J. Satchell, F. Yildiz, and K. E. Klose. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191:6555-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terashima, H., H. Fukuoka, T. Yakushi, S. Kojima, and M. Homma. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol. Microbiol. 62:1170-1180. [DOI] [PubMed] [Google Scholar]
- 45.Tripathi, S. A., and R. K. Taylor. 2007. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J. Bacteriol. 189:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xicohtencatl-Cortes, J., S. Lyons, A. P. Chaparro, D. R. Hernandez, Z. Saldana, M. A. Ledesma, M. A. Rendon, A. T. Gewirtz, K. E. Klose, and J. A. Giron. 2006. Identification of proinflammatory flagellin proteins in supernatants of Vibrio cholerae O1 by proteomics analysis. Mol. Cell. Proteomics 5:2374-2383. [DOI] [PubMed] [Google Scholar]